Abstract
DNA methylation has become increasingly recognized in the etiology of psychiatric disorders. Because brain tissue is not accessible in living humans, epigenetic studies are most often conducted in blood. Saliva is often collected for genotyping studies but is rarely used to examine DNA methylation because the proportion of epithelial cells and leukocytes varies extensively between individuals. The goal of this study was to evaluate whether saliva DNA is informative for studies of psychiatric disorders. DNA methylation (HumanMethylation450 BeadChip) was assessed in saliva and blood samples from 64 adult African Americans. Analyses were conducted using linear regression adjusted for appropriate covariates, including estimated cellular proportions. DNA methylation from brain tissues (cerebellum, frontal cortex, entorhinal cortex, and superior temporal gyrus) was obtained from a publically available dataset. Saliva and blood methylation was clearly distinguishable though there was positive correlation overall. There was little correlation in CpG sites within relevant candidate genes. Correlated CpG sites were more likely to occur in areas of low CpG density (i.e., CpG shores and open seas). There was more variability in CpG sites from saliva than blood, which may reflect its heterogeneity. Finally, DNA methylation in saliva appeared more similar to patterns from each of the brain regions examined overall than methylation in blood. Thus, this study provides a framework for using DNA methylation from saliva and suggests that DNA methylation of saliva may offer distinct opportunities for epidemiological and longitudinal studies of psychiatric traits.
Keywords: epigenetic, biomarker, oragene, EWAS, Human-Methylation450
INTRODUCTION
Epigenetic studies of psychiatric and behavioral traits have increased dramatically over the past decade [Pena et al., 2014; Smith et al., 2014b]. However, because brain tissue is not easily accessible in living humans, epigenetic studies are most often conducted in blood, which is readily available and potentially useful for biomarker identification. Indeed, DNA methylation of specific genes in blood has been suggested as potential biomarkers of major depressive disorder, bipolar disorder, and schizophrenia in adults [Fuchikami et al., 2011; Ghadirivasfi et al., 2011; Nohesara et al., 2011;Ebot Enaw JO SA, 2013; Ikegame et al., 2013c].
Genome-wide DNA methylation surveys in blood and brain have provided insight into genes whose regulation patterns vary in those with a history of child abuse, major depressive disorder, posttraumatic stress disorder, schizophrenia, and autism [Smith et al., 2011; Byrne et al., 2013; Mehta et al., 2013; Ladd-Acosta et al., 2013; Aberg et al., 2014; Wong et al., 2014]. While it is clear that DNA methylation and gene expression patterns are highly tissue-specific, numerous studies report correlation between blood and brain overall and at genes relevant to psychiatric disorders [Sullivan et al., 2006; Davies et al., 2012; Tylee et al., 2013]. However, epigenetic patterns in many tissues are static, making correlations alone of limited value. Regions of inter-individual variation, or those that respond to an environmental stimulus in a similar way, may be more informative [Relton and Davey, 2010; Rakyan et al., 2011; Heijmans and Mill, 2012; Mill and Heijmans, 2013]. For example, allele-specific methylation of FKBP5 responds to glucocorticoid stimulation in a similar way in blood and brain [Klengel et al., 2013].
Recent studies have explored the utility of other tissues for studies of complex traits. Lowe and colleagues recently compared the properties of blood and buccal cells [Lowe et al., 2013]. They found that DNA methylation from buccal cells, when compared to blood, is more likely to be hypomethylated and is enriched for (i) epithelial DNase I hypersensitivity sites; (ii) histone modifications indicative of activation; and (iii) disease-associated SNPs. Based on these observations, they concluded that buccal cells may be the best proxy tissue for the study of human diseases. Indeed buccal methylation from twins during childhood exhibits individual-specific environmental and stochastic influences, particularly among CpG poor regions such as shelves, shores, and open seas [van Dongen et al, 2014]. These CpG poor regions are enriched for tissue-specific differentially methylated regions that correlate with tissue-specific expression patterns [Slieker et al., 2013]. Further, examination of tissue-specific CpG sites suggests that DNA methylation from saliva, while most similar to blood, exhibits similarity to buccal cells as well. Thompson and colleagues also noted that DNA methylation from saliva and whole blood were more similar to each other when compared to lymphoblastoid cell lines, presumably due to the effects of transformation and culture conditions [Thompson et al., 2013].
Despite the interest in peripheral tissues other than blood, few studies have utilized saliva samples for genome-wide DNA methylation surveys [Ghadirivasfi et al, 2011; Essex et al., 2013; Melas et al., 2013; Yang et al, 2013]. Saliva is readily collectable, but the proportion of buccal epithelial cells and leukocytes varies between individuals and results in substantial cellular heterogeneity. Saliva also contains DNA from non-human sources (i.e., bacteria), which can result in varying concentrations of human DNA and potentially higher failure rates for genetic and epigenetic assays [Philibert et al, 2008; Abraham et al, 2012]. It is also unclear whether DNA methylation derived from saliva is informative for studies of psychiatric traits. Addressing these limitations will allow researchers who have collected saliva samples for genotyping to evaluate DNA methylation in their cohorts as well. The goal of this study is to develop a framework for examining saliva DNA methylation for epidemiologic studies of psychiatric disorders and to assess the relationship between methylation from saliva and other relevant tissues.
MATERIALS AND METHODS
Participants
We evaluated adult African American subjects recruited as part of a larger study investigating the influence of genetic and environmental factors on response to stressful life events in a predominantly African American, urban population of low socioeconomic status (SES) [Gillespie et al, 2009]. Briefly, research participants were approached in the waiting rooms of the primary care clinic or obstetrical-gynecological clinic of a large urban, public hospital while either waiting for their medical appointments or while waiting with others who were scheduled for medical appointments. Subjects willing to participate provided written informed consent and participated in a verbal interview. All procedures in this study were approved by the Institutional Review Board of Emory University School of Medicine and the Grady Health Systems Research Oversight Committee.
DNA Methylation
Both saliva and blood samples were collected from each participant. Saliva was stored in Oragene DNA sample collection kits (DNA Genotek), and blood was collected in EDTA vacuum tubes. DNA was extracted using the Puregene Genomic DNA kit (Invitrogen, Carlsbad, CA). Samples were resolved on a 1% agarose gel to verify that the DNA was of high molecular weight (at least 2 kb) and quantified using PicoGreen (Invitrogen).
DNA methylation was interrogated for each sample using the HumanMethylation450 BeadChip (Illumina). Briefly, 1 µg of DNA was converted with sodium bisulfite, amplified, fragmented, and hybridized according to the manufacturer's instructions. The raw data is available in GEO under accession number GSE61653. One sample of the same female DNA was included on each BeadChip as a technical control throughout the experiment and assessed for reproducibility using the Pearson correlation coefficient. Beta values were generated with BeadStudio and were set to missing (no call) if detection P-values exceeded .001. CpGassoc [Barfield et al, 2012] was used to remove samples with probe detection call rates <95% and those with an average intensity value of either <50% of the experiment-wide sample mean or <2,000 arbitrary units (AU). In addition, CpG sites with missing data for >10% of samples were excluded from analysis. Finally, probes with known SNPs or that cross hybridize between autosomes and sex chromosomes were also removed [Chen et al, 2013]. No samples were removed from either dataset. However, 53,726 probes were removed from the saliva dataset, and 53,835 were removed from the blood dataset. Only probes that were common to both datasets were analyzed. Finally, Beta Mixture Quantile dilation (BMIQ) was used to normalize each dataset [Teschendorff et al., 2013]. Hierarchical clustering of each tissue type did not reveal any additional outliers. For each individual sample and CpG site, the signals from methylated (M) and unmethylated (U) bead types were used to calculate a beta value as β= M/(U + M).
Statistical Analysis
The method described by Houseman and colleagues was used to estimate the proportion of epithelial cells in saliva DNA and the proportion of lymphocytes and neutrophils in blood DNA [Houseman et al., 2012; Koestler et al., 2013]. We used data from buccal epithelial cells (GEO GSE46573) and FACS sorted leukocytes subtypes (GEO GSE35069) to identify tissue-specific CpG sites and then estimated the relative proportion of each cell type in our heterogeneous saliva and blood samples, respectively. This approach has been widely used and extensively validated [Adalsteinsson et al., 2012; Koestler et al., 2013; Liu et al., 2013; Sun et al., 2013].
To examine the similarities between DNA methylation in saliva and blood, we modeled saliva methylation as a linear function of blood methylation, adjusting for positional effects and the proportion of epithelial cells in each sample. We used a false discovery rate (FDR) procedure [Storey, 2002] to determine a set of associated CpG sites with FDR < .05. Next, we sought to compare DNA methylation in each tissue to that of four brain regions (cerebellum, frontal cortex, entorhinal cortex, and superior temporal gyrus) from a publically available dataset (GEO GSE43414) that has been previously described [Pidsley et al., 2013]. The raw data were quality controlled according to the strategy described above, resulting in the removal of one sample from the entorhinal cortex data and between 880–1,942 probes from each of four brain datasets. Only probes that passed QC in all datasets (N = 431,582) were included in the analysis. Euclidean distance was used to compare DNA methylation between tissues. To accomplish this, we calculated average methylation for each CpG site in each tissue. To compare two tissues, we calculated the difference between their methylation levels at each CpG site, and computed the square root of the squared sum of those differences to reflect the relative similarity between two tissues. Specifically, smaller Euclidean distances are indicative of greater similarity. In addition, we functionally classified based on average methylation levels corresponding to low (0 < β ≤ .2), intermediate (.2 < β < .8) or high (β ≥ .8) levels of methylation. Agreement was assessed with a chi square test consistent with the approach used by Davies and colleagues [Davies et al., 2012]. Finally, we identified methylation quantitative trait loci (meQTL) in this cohort by applying the approach described previously to the HumanMethylation450 data from the parent study [Smith etal., 2014a].
RESULTS
There was no difference in the number of samples that failed or probes that were excluded from this analysis based on the source of DNA (saliva or blood). The estimated proportion of epithelial cells in saliva DNA ranged from 3–99% (median 26%). In blood, the estimated proportion of lymphocytes ranged from 25–70% (median 48%) and neutrophils ranged from 42–84% (median 58%). Note that because the proportions were independently estimated, they do not necessary total to 100%.
Correlation Between Saliva and Blood
Because saliva samples contain a heterogeneous mix of epithelial cells and leukocytes, DNA methylation in saliva may reflect that of blood, particularly in individuals with a higher proportion of leukocytes. To test this, we performed an unsupervised hierarchical clustering of all CpG sites in blood and saliva from the same subjects (Fig. 1). All blood samples clustered distinctly from saliva samples. Next, we tested whether each CpG site in saliva predicted that of blood using linear models adjusting for the proportion of epithelial cells in each sample. Overall, methylation of saliva and blood was positively correlated within individuals for 88.5% of CpG sites (Fig. 2). Correlated CpG sites were identified in almost every gene. They were more common in low-density CpG regions (CpG shores and open seas) and intergenic regions while uncorrelated sites were more common in CpG islands, promoters and gene bodies (P < 2.2 × 10−16 for all comparisons). Saliva significantly predicted blood methylation for 27.3% of CpG sites in a linear regression (FDR < .05; 2.1 × 10−68 < P < .021 Supplementary Table SI).
FIG. 1.
Tissue-specific DNA methylatlon patterns. Hierarchical clustering of each subject (x-axis) for all CpG sites (y-axis) segregates samples by tissue, with all saliva samples clustering to the left of the graph and all blood samples clustering to the right.
FIG. 2.
Correlation between saliva and blood. This histogram depicts the correiation coefficient comparing methylatian levels of blood and saliva for each of the CpG sites examined. Those that are shaded are positively correlated.
We then wanted to understand whether the similarities between blood and saliva samples are influenced by the estimated proportion of epithelial cells versus leukocytes. First, we characterized the association between saliva methylation and the estimated proportion of epithelial cells in each sample and found that methylation of 68.8% of all CpG sites differed relative to epithelial cell proportion (FDR < .05). Methylation of samples with the lowest proportion of epithelial cells based on a median split appears to resemble those of blood more than the methylation levels of samples with higher epithelial cell proportions (Supplementary Fig. 1).
Psychiatric Candidate Genes
We specifically considered 162 CpG sites from genes implicated in psychiatric disorders not only by genetic but also epigenetic studies (FKBP5, BDNF, NR3C1, and SLC6A4). Overall, 15 of 75 CpG sites (19.7%) in BDNF (2.23 × 10−14 < P < .022), 8 of 33 (24.2%) in FKBP5 (21.42 × 10−15 < P < .048), 5 of 40 (12,5%) in NR3C1 (.015 < P < .048), and 2 of 14 (14.3%) in SLC6A4 (2.49 × 10−11 < P < 5.19 × 10−8) associated between saliva and blood (Supplementary Table SI). Predictably, this relationship was more pronounced in subjects with higher proportions of leukocytes. Of the 17 CpG sites in these genes that have been associated with child abuse history [Weder et al., 2014], all but 2 (cg24650785 and cg17860381) exhibit tissue-specific methylation patterns.
Methylation Patterns in Saliva and Blood Compared With Brain Regions
Because buccal epithelial and brain cells are derived from the ectodermal layer during development, we predicted that DNA methylation in saliva may be more consistent with typical methylation patterns in the four brain regions tested. To test this, we categorized the average methylation levels of each CpG site based on low (0 < β ≤ .2), intermediate (.2 < β < .8), or high (β ≥ .8) levels of methylation. When comparing these functional classifications across tissue types, DNA methylation from saliva was ~3% more likely to agree with each of the brain regions than with methylation from blood (P < 2.2 × 10−16). Next, we calculated the average Euclidean distance for each CpG site in each tissue (Fig. 3a). Overall, comparisons of DNA methylation between saliva and the four brain regions appeared more similar than comparisons between blood and the four brain regions, a pattern that was even more pronounced in saliva samples containing higher proportions of epithelial cells. In contrast, overall methylation of saliva samples with the lowest proportion of epithelial cells (Euclidean distance of 41.6) more closely resembled methylation of blood compared to the samples with the highest proportion of epithelial cells (Euclidean distance of 54.4; note that smaller Euclidean distance indicates more similarly).
FIG. 3.
Genome-wide relationship between DNA methyl at ion in saliva, blood, and four brain regions. Euclidean distance was calculated for each tissue using (A) all CpG sites and [B) CpG sites that vary in saliva. For each plot, blood, and saliva were compared to each of the four brain regions. Smaller differences indicate more similarity between the compared tissues. CRBLM, cerebellum; FCTX, frontal cortex; EntCTX, entorhinal cortex; Gyrus, superior temporal gyrus.
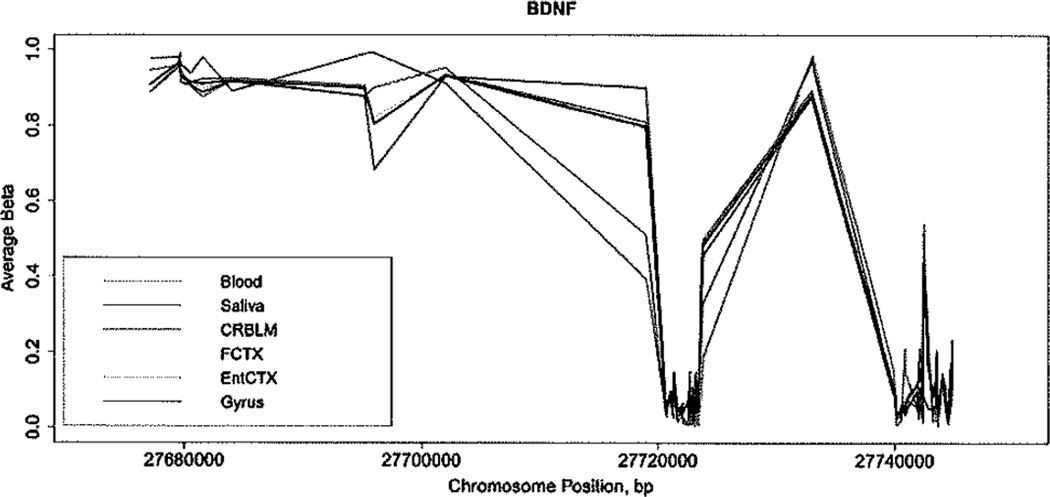
Recent studies suggest that DNA methylation of brain-derived neurotrophic factor (BDNF) may serve as a biomarker for multiple psychiatric traits [Fuchikami et al., 2011; Ebot Enaw JO SA, 2013; Ikegame et al., 2013a; Ikegame et al., 2013b; Kang et al., 2013]. Thus, we examined BDNF gene methylation as an example target gene to compare across tissues. We observed that the average DNA methylation levels of BDNF in saliva are also closer to those of each brain region than the average levels in blood for the majority of CpG sites across this gene (Fig. 4).
FIG. 4.
Relationship between DNA methylation in saliva, blood, and brain regions for BDNF. Average DNA methylation at each CpG site Is plotted by tissue.
Inter-Individual Variation
Regions of inter-individual variation have gained increased attention in studies of complex traits [Relton and Davey, 2010; Rakyan et al., 2011; Heijmans and Mill, 2012; Mill and Heijmans, 2013; Pena et al., 2014]. In this study, we denned a set of variable CpG sites (vCpGs) as those with variances ≥0.01. Saliva had almost three times as many vCpGs when compared to blood (10.7% vs. 3.5%, respectively). Of the 46,015 vCpGs in saliva, 51.3% have methylation levels that correlate with blood (P < .05). Similarly, CpG sites that vary in saliva are more likely to vary in each of the four brain tissues than would be expected by chance (14.9–19.1%; P < 1 × 10−16). Next, using only saliva vCpGs, we recalculated Euclidean distances (Fig 3b) and again observed more similarity between saliva and the four brain regions than between blood and the brain regions, particularly among saliva samples with more epithelial cells.
Regions of inter-individual variation are more likely to be influenced by genetic variation [van Dongen et al., 2012, 2014]. While we removed all probes containing known SNPs, methylation quantitative trait loci (meQTL) may still have substantial influence. Of the 46,015 vCpGs in saliva, methylation of 13,106 (28.5%) associated with one or more SNPs (P < .001) within a larger region of 50kb, and that estimate increased to 33,897 (73.7%) when we considered nominal associations (P < .05) between CpG sites and nearby SNPs.
DISCUSSION
The results of this study provide a framework for using saliva DNA for epigenetic studies of psychiatric traits. The foremost barrier to using DNA derived from saliva is that the source of that DNA is heterogeneous. Our results suggest that variation in the proportion of epithelial cells between individuals substantially influences DNA methylation measurements. While this is not surprising, it is imperative that future studies estimate the proportion of epithelial cells in saliva samples and control for that variation. In addition to epithelial cells and leukocytes, saliva also contains bacteria, which may increase error rates for assays that require a precise amount of human DNA. In this study, DNA from saliva was similar in quality to that of blood, and there was no difference in sample or probe performance. It should be noted that we used 1 µg of saliva DNA to account for variation potentially attributable to bacterial DNA contamination. It may be possible to add less for future studies as this assay appears to be relatively robust to variation in amount and quality of DNA used [Beyan et al., 2012; Ghantous et al., 2014; Moran et al., 2014].
The relationship between DNA from saliva and other tissues relevant for psychiatric disorders is also not clear. Though there appeared to be correlation overall, the majority of correlated CpG sites had limited variance in either saliva or blood. The lack of dynamically regulated CpG sites on the HumanMethylation450 BeadChip has been noted previously [Ziller et al., 2013] as well as an underrepresentation of intergenic regions and regions in putative regulatory regions. Despite this, CpG sites whose methylation levels correlated between blood and saliva were more likely to occur in CpG shores and open seas, areas that are reportedly more likely to contain individual-level environmental and stochastic influences [van Dongen et al., 2014], However, when only variable CpG sites were considered, there were substantial differences between DNA methylation levels measured in saliva versus blood despite the fact that leukocytes were present in both tissue types. Consistent with reports of tissue specific differences in DNA methylation [Rakyan et al., 2008; Byun et al., 2009; Ghosh et al., 2010], our results suggest that blood and saliva have relatively little epigenetic similarity overall or in candidate genes relevant for psychiatric disorders. Methylation differences in BDNF, FKBP5, NR3C1, and SLC6A4 have been reported in the brain, blood cells, and other tissues [Weaver et al., 2004; McGowan et al., 2009; Sugawara et al., 2011; Ikegame et al., 2013a; Ewald et al., 2014; Guidotti et al., 2014]. The results of this study present a distinct opportunity for peripheral studies of psychiatric traits by expanding the tissues available for epidemiological and biomarker studies. It may also be informative for investigators evaluating specific mechanistic hypotheses. For example, immune dysregulation has been increasingly recognized in those with specific psychiatric disorders, and DNA methylation differences have been linked to immune function in the blood of those with schizophrenia, depression, and posttraumatic stress disorder [Uddin et al., 2010, 2011; Smith et al., 2011; Frydecka etal., 2014; Liu etal., 2014].
The results of this study also suggest that DNA methylation from saliva may be more similar to that of brain tissues on average than DNA methylation from blood. This observation may reflect common developmental origins, or it may simply reflect the increased variation of methylation levels in saliva DNA. The latter is consistent with a report suggesting buccal cells may be preferable to blood as proxy tissue for human diseases that are not related to blood [Lowe et al., 2013]; similar to the findings in buccal cells, we observed a higher proportion of variable CpG sites in saliva than in blood, potentially because of increased cellular heterogeneity in saliva samples. It is also interesting to note that many of the vCpGs were influenced by genetic variation outside of the probe, and such meQTLs appear to be highly correlated across tissues (i.e., blood and brain) as well as age and race groups [Smith et al., 2014a]. However, these results should be interpreted with caution. Because the blood and saliva samples were not obtained from the same individuals as the brain samples, we are not able to go beyond a descriptive analysis of different brain regions in this study. The relationship between epigenetic characteristics of different tissues should be evaluated in the same individuals to control for inter-individual variation in genetic and environmental factors, recognizing that the most meaningful areas will be those that vary in response to specific exposures.
There are a number of additional limitations to consider. In this study, we were not able to evaluate oral hygiene practices that may influence cellular proportions or DNA methylation patterns in saliva samples. Studies that use the approach we outline should thoroughly evaluate whether biological factors, such as oral hygiene practices, independently influence DNA methylation in their cohort. Specifically, DNA from bacterial or other non-human sources may range substantially between individuals and influence assay quality [Philibert et al., 2008]. Another important limitation is that we estimated the proportion of epithelial cells in each subject using data from buccal swabs, which may sample cells from layers beneath the epithelium including the stratum granulosum and spinosum. The blood and saliva samples used in this study were obtained from a biobank, and buccal swabs were not available. Ideally, the study would utilize samples collected for this study that could be sorted for a single relevant cell type, which is always preferable to using heterogeneous cell mixtures, such as whole blood and saliva. However, that is not always possible for large epidemiological surveys of biobanked samples. While this underlies the need for approaches to use saliva in DNA methylation studies, epithelial cells from saliva should not be considered equivalent to buccal swabs without empirical evidence. Finally, because the DNA was bisulfite converted, we were unable to distinguish between methylcytosine and hydroxymethylcytosine, which may obscure regulatory relationships in this dataset. However, the proportion of 5-hydroxymethylcytosine (hmC) is low in peripheral tissues (<0.1%) so that the effects of potentially confounding the two types of modification may have less impact on the comparisons between blood and saliva [Kriaucionis and Heintz, 2009; Globisch et al., 2010; Shahal et al., 2014].
Our results provide a framework for methylation studies in DNA extracted from saliva and highlight the importance of controlling for the proportion of epithelial and leukocyte cells. This presents an attractive opportunity for investigators that have already collected salivary DNA for genetic studies of psychiatric traits. Expanding both the number and types of samples available for large epidemiological investigations will likely accelerate discovery of how genetic and environmental risk factors contribute to the development of psychiatric disorders.
ACKNOWLEDGMENTS
The authors gratefully acknowledge the participants of the study and the Grady Trauma Project staff for their assistance with participant recruitment and data collection. The authors thank Anne Löschner and Maik Ködel for excellent technical support and Jessica Lam for editorial support. This research was supported by the National Institutes of Health Grants MH071537 and MH096764 (K.J.R.), MH085806 (A.K.S.), and HD071982 (B.D.B.). This work was also supported by the Brain and Behavior Foundation (A.K.S.), the Howard Hughes Medical Institute (K.J.R.), the Max Planck S o ciety (E.B.B), the Behrens–Weise foundation (E.B.B), and the European Union under European Research Council GA no. 281338 (E.B.B).
Grant sponsor: National Institutes of Health; Grant numbers: MH071537, MH096764, MH085806, HD071982; Grant sponsor: Brain and Behavior Foundation; Grant sponsor: Howard Hughes Medical Institute; Grant sponsor: Max Planck Society; Grant sponsor: Behrens–Weise Foundation; Grant sponsor: European Research Council; Grant number: 281338.
REFERENCES
- Aberg KA, McClay JL, Nerella S, Clark S, Kumar G, Chen W, Khachane AN, Xie L, Hudson A, Gao G, Harada A, Hultman CM, Sullivan PF, Magnusson PK, van den Oord EJ. Methylome-wide association study of schizophrenia: Identifying blood biomarker signatures of environmental insults. JAMA Psychiatry. 2014;71(3):255–264. doi: 10.1001/jamapsychiatry.2013.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham JE, Maranian MJ, Spiteri I, Russell R, Ingle S, Luccarini C, Earl HM, Pharoah PP, Dunning AM, Caldas C. Saliva samples are a viable alternative to blood samples as a source of DNA for high through-put genotyping. BMC Med Genomics. 2012;5:19. doi: 10.1186/1755-8794-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adalsteinsson BT, Gudnason H, Aspelund T, Harris TB, Launer LJ, Eiriksdottir G, Smith AV, Gudnason V. Heterogeneity in white blood cells has potential to confound DNA methylation measurements. PLoS ONE. 2012;7(10):e46705. doi: 10.1371/journal.pone.0046705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barfield RT, Kilaru V, Smith AK, Conneely KJM. CpGassoc: An R function for analysis of DNA methylation microarray data. Bioinformatics. 2012;28(9):1280–1281. doi: 10.1093/bioinformatics/bts124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyan H, Down TA, Ramagopalan SV, Uvebrant K, Nilsson A, Holland ML, Gemma C, Giovannoni G, Boehm BO, Ebers GC, Lernmark A, Cilio CM, Leslie RD, Rakyan VK. Guthrie card methylomics identifies temporally stable epialleles that ate present at birth in humans. Genome Res. 2012;22(11):2138–2145. doi: 10.1101/gr.134304.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne EM, Carrillo-Roa T, Henders AK, Bowdler L, McRae AP, Heath AC, Martin NG, Montgomery GW, Krause L, Wray NR. Monozygotic twins affected with major depressive disorder have greater variance in methylation than their unaffected co-twin. Transl Psychiatry. 2013;3:e269. doi: 10.1038/tp.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun HM, Siegmund KD, Pan F, Weisenberger DJ, Kanel G, Laird PW, Yang AS. Epigenetic profiling of somatic tissues from human autopsy specimens identifies tissue- and individual-specific DNA methylation patterns. Hum Mol Genet. 2009;18(24):4808–4817. doi: 10.1093/hmg/ddp445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YA, Lemirc M, Choufani S, Butcher DT, Grafodatskaya D, Zanke BW, Gallinger S, Hudson TJ, Weksberg R. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8(2):203–209. doi: 10.4161/epi.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies MN, Volta M, Pidsley R, Lunnon K, Dixit A, Lovestone S, Coarfa C, Harris RA, Milosavljevic A, Troakes C, Al-Sarraj S, Dobson R, Schalkwyk LC, Mill J. Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biol. 2012;13(6):R43. doi: 10.1186/gb-2012-13-6-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebot Enaw JO, Smith AK. Biomarker development for brain-based disorders: Recent progress in psychiatry. J Neurol Psychol. 2013;1(2):7. doi: 10.13188/2332-3469.1000006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essex MJ, Boyce WT, Hertzman C, Lam LL, Armstrong JM, Neumann SM, Kobor MS. Epigenetic vestiges of early developmental adversity: Childhood stress exposure and DNA methylation in adolescence. Child Dev. 2013;84(1):58–75. doi: 10.1111/j.1467-8624.2011.01641.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald ER, Wand GS, Seifuddin F, Yang X, Tamashiro KL, Potash JB, Zandi P, Lee RS. Alterations in DNA methylation of Fkbp5 as a determinant of biood-brain correlation of glucocorticoid exposure. Psychoneuroendocrinology. 2014;44:112–122. doi: 10.1016/j.psyneuen.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydecka D, Karpinski P, Misiak B. Unravelling immune alterations in schizophrenia: Can DNA methylation provide clues. Epigenomics. 2014;6(3):245–247. doi: 10.2217/epi.14.26. [DOI] [PubMed] [Google Scholar]
- Fuchikami M, Morinobu S, Segawa M, Okamoto Y, Yamawaki S, Ozaki N, Inoue T, Kusumi I, Koyama T, Tsuchiyama K, Terao T. DNA methylation profiles of the brain-derived neurotrophic factor (BDNF) gene as a potent diagnostic biomarker in major depression. PLoS ONE. 2011;6(8):e23881. doi: 10.1371/journal.pone.0023881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghadirivasfi M, Nohesara S, Ahmadkhaniha HR, Eskandari MR, Mostafavi S, Thiagalingam S, Abdolmateky HM. Hypomethylation of the serotonin receptor type-2A Gene (HTR2A) at T102C polymorphic site in DNA derived from the saliva of patients with schizophrenia and bipolar disorder. Am J Med Genet B Neuropsychiatr Genet. 2011;156B(5):536–545. doi: 10.1002/ajmg.b.31192. [DOI] [PubMed] [Google Scholar]
- Ghantous A, Saffery R, Cros MP, Ponsonby AL, Hirschfeld S, Kasten C, Dwyer T, Herceg Z, Hernandez-Vargas H. Optimized DNA extraction from neonatal dried blood spots: Application in methylome profiling. BMC Biotechnol. 2014;14:60. doi: 10.1186/1472-6750-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Yates AJ, Fruhwald MC, Miecznikowski JC, Plass C, Smiraglia D. Tissue specific DNA methylation of CpG islands in normal human adult somatic tissues distinguishes neural from non-neural tissues. Epigenetics. 2010;5(6):527–538. doi: 10.4161/epi.5.6.12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie CF, Bradley B, Mercer K, Smith AK, Conneely K, Gapen M, Weiss T, Schwartz AC, Cubells JF, Ressler KJ. Trauma exposure and stress-related disorders in inner city primary care patients. Gen Hosp Psychiatry. 2009;31(6):505–514. doi: 10.1016/j.genhosppsych.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Globisch D, Munzel M, Muller M, Michalakis S, Wagner M, Koch S, Bruckl T, Biel M, Carell T. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS ONE. 2010;5(12):e15367. doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidotti A, Auta J, Davis JM, Dong E, Gavin DP, Grayson DR, Sharma RP, Smith RC, Tueting P, Zhubi A. Toward the identification of peripheral epigenetic biomarkers of schizophrenia. Neurogenet. 2014;28(1–2):41–52. doi: 10.3109/01677063.2014.892485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijmans BT, Mill J. Commentary: The seven plagues of epigenetic epidemiology. Int J Epidemiol. 2012;41(1):74–78. doi: 10.1093/ije/dyr225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, Wiencke JK, Kelsey KT. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegame T, Bundo M, Murata Y, Kasai K, Kato T, Iwamoto K. DNA methylation of the BDNF gene and its relevance to psychiatric disorders. J Hum Genet. 2013a;58(7):434–438. doi: 10.1038/jhg.2013.65. [DOI] [PubMed] [Google Scholar]
- Ikegame T, Bundo M, Sunaga F, Asai T, Nishimura F, Yoshikawa A, Kawamura Y, Hibino H, Tochigi M, Kakiuchi C, Sasaki T, Kato T, Kasai K, Iwamoto K. DNA methylation analysis of BDNF gene promoters in peripheral blood cells of schizophrenia patients. Neurosci Res. 2013b;77(4):208–214. doi: 10.1016/j.neures.2013.08.004. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Kim JM, Lee JY, Kim SY, Bae KY, Kim SW, Shin IS, Kim HR, Shin MG, Yoon JS. BDNF promoter methylation and suicidal behavior in depressive patients. J Affect Disord. 2013;151(2):679–685. doi: 10.1016/j.jad.2013.08.001. [DOI] [PubMed] [Google Scholar]
- Klengel T, Mehta D, Anacker C, Rex-Haffner M, Pruessner JC, Pariante CM, Pace TW, Mercer KB, Mayberg HS, Bradley B, Nemeroff CB, Holsboer F, Heim CM, Ressler KJ, Rein T, Binder EB. Allele-specific FKBP5 DNA demethylation mediates gene-childhood trauma interactions. Nat Neurosci. 2013;16(1):33–41. doi: 10.1038/nn.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koestler DC, Christensen B, Karagas MR, Marsit CJ, Langevin SM, Kelsey KT, Wiencke JK, Houseman EA. Blood-based profiles of DNA methylation predict the underlying distribution of ceil types: A validation analysis. Epigenetics. 2013;8(8):816–826. doi: 10.4161/epi.25430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethyl-cytosine is present in Purkinje neurons and the brain. Science. 2009;324(5929):929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladd-Acosta C, Hansen KD, Briem E, Fallin MD, Kaufmann WE, Feinberg AP. Common DNA methylation alterations in multiple brain regions in autism. Mol Psychiatry. 2013;19(8):862–871. doi: 10.1038/mp.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu I, Chen J, Ehrlich S, Walton E, White T, Perrone-Bizzozero N, Bustillo J, Turner JA, Calhoun1 VD. Methylation patterns in whole blood correlate with symptoms in schizophrenia patients. Schizophr Bull. 2014;40(4):769–776. doi: 10.1093/schbul/sbt080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Aryee MJ, Padyukov L, Fallin MD, Hesselberg E, Runarsson A, Reinius L, Acevedo N, Taub M, Ronninger M, Shchetynsky K, Scheynius A, Kere J, Alfredsson L, Klareskog L, Ekstrom TJ, Feinberg AP. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat Biotechnol. 2013;31(2):142–147. doi: 10.1038/nbt.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe R, Gemma C, Beyan H, Hawa MI, Bazeos A, Leslie RD, Montpetit A, Rakyan VK, Ramagopalan SV. Buccals are likely to be a more informative surrogate tissue than blood for epigenome-wide association studies. Epigenetics. 2013;8(4):445–454. doi: 10.4161/epi.24362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan PO, Sasaki A, D’Alessio AC, Dymov S, Labonte B, Szyf M, Turecki G, Meaney MJ. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12(3):342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta D, Klengel T, Conneely KN, Smith AK, Altmann A, Pace TW, Rex-Haffner M, Loeschner A, Gonik M, Mercer KB, Bradley B, Muller-Myhsok B, Ressler KJ, Binder EB. Childhood maltreatment is associated with distinct genomic and epigenetic profiles in posttraumatic stress disorder. Proc Natl Acad Sci U S A. 2013;110(20):8302–8307. doi: 10.1073/pnas.1217750110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melas PA, Wei Y, Wong CC, Sjoholm LK, Aberg E, Mill J, Schalling M, Forsell Y, Lavebratt C. Genetic and epigenetic associations of MAOA and NR3C1 with depression and childhood adversities. Int J Neuropsychopharmacol. 2013;16(7):1513–1528. doi: 10.1017/S1461145713000102. [DOI] [PubMed] [Google Scholar]
- Mill J, Heijmans BT. From promises to practical strategies in epigenetic epidemiology. Nat Rev Genet. 2013;14(8):585–594. doi: 10.1038/nrg3405. [DOI] [PubMed] [Google Scholar]
- Moran S, Vizoso M, Martinez-Cardus A, Gomez A, Matias-Guiu X, Chiavenna SM, Fernandez AG, Esteller M. Validation of DNA methylation profiling in formalin-fixed paraffin-embedded samples using the Infinium HumanMethylation450 Microarray. Epigenetics. 2014;9(6):829–833. doi: 10.4161/epi.28790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohesara S, Ghadirivasfi M, Mostafavi S, Eskandari MR, Ahmadkhaniha H, Thiagalingam S, Abdolmaleky HM. DNA hypomethylation of MB-COMT promoter in the DNA derived from saliva in schizophrenia and bipolar disorder. J Psychiatr Res. 2011;45(11):1432–1438. doi: 10.1016/j.jpsychires.2011.06.013. [DOI] [PubMed] [Google Scholar]
- Pena CJ, Bagot RC, Labonte B, Nestler EJ. Epigenetic Signaling in Psychiatric Disorders. J Mol Biol. 2014;426(20):3389–3412. doi: 10.1016/j.jmb.2014.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philibert RA, Zadorozhnyaya O, Beach SR, Brody GH. Comparison of the genotyping results using DNA obtained from blood and saliva. Psychiatr Genet. 2008;18(6):275–281. doi: 10.1097/YPG.0b013e3283060f81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidsley R, CC YW, Lunnon, Mill J, Schalkwyk LC. A data-driven approach to preprocessing Illumina 450K methyiation array data. BMC Genomics. 2013;14:293. doi: 10.1186/1471-2164-14-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakyan VK, Down TA, Balding DJ, Beck S. Epigenome-wide association studies for common human diseases. Nat Rev Genet. 2011;12(8):529–541. doi: 10.1038/nrg3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakyan VK, Down TA, Thorne NP, Flicek P, Kulesha E, Graf S, Tomazou EM, Backdahl L, Johnson N, Herberth M, Howe KL, Jackson DK, Miretti MM, Fiegler H, Marioni JC, Birney E, Hubbard TJ, Carter NP, Tavare S, Beck S. An integrated resource for genome-wide identification and analysis of human tissue-specific differentially methylated regions (tDMRs) Genome Res. 2008;18(9):1518–1529. doi: 10.1101/gr.077479.108. [DOI] [PMC free article] [PubMed] [Google Scholar]