Abstract
NK cells are innate lymphoid cells important for immune surveillance, identifying and responding to stress, infection, and/or transformation. While conventional NK (cNK) cells circulate systemically, many NK cells reside in tissues where they appear to be poised to locally regulate tissue function. Here we tested the contribution of tissue-resident NK (trNK) cells to tissue homeostasis by studying ischemic injury in the mouse kidney. Parabiosis experiments demonstrate that the kidney contains a significant fraction of trNK cells under homeostatic conditions. Kidney trNK cells developed independent of NFIL3 and Tbet, and expressed a distinct cell surface phenotype as compared to cNK cells. Among these, trNK cells had reduced asialo-GM1 (AsGM1) expression relative to cNK cells, a phenotype observed in trNK cells across multiple organs and mouse strains. Strikingly, anti-AsGM1 antibody treatment, commonly used as NK cell-depleting regimen, resulted in a robust and selective depletion of cNKs, leaving trNKs largely intact. Using this differential depletion, we tested the relative contribution of cNK and trNK cells in ischemic kidney injury. Whereas anti-NK1.1 antibody effectively depleted both trNK and cNK cells and protected against ischemic-reperfusion injury, anti-AsGM1 antibody preferentially depleted cNK cells and failed to protect against injury. These data demonstrate unanticipated specificity of anti-AsGM1 antibody depletion on NK cell subsets and reveal a new approach to study the contributions of cNK and trNK cells in vivo. In total, these data demonstrate that trNK cells play a key role in modulating local responses to ischemic tissue injury in the kidney and potentially other organs.
Keywords: NK cells, asialo-GM1, tissue-resident NK, ischemia-reperfusion injury, kidney
INTRODUCTION
NK cells are rapidly responding lymphoid cells that have a central role in local and systemic immune surveillance. NK cell recognition and activation is mediated through engagement of multiple activating and inhibitory receptors, broadly referred to as NK receptors (NKRs), with the relative engagement of inhibitory and activating receptors influencing whether a cell is protected from, or becomes a target, for NK cell destruction (1–5). The specificity and sensitivity of NK cell recognition is thought to be initially acquired in the bone marrow and further refined in the periphery, a process known as education and licensing (6–10). Upon activation, NK cells can profoundly shape the immune response via cognate and non-cognate interactions, rapidly producing cytokines, chemokines, and/or directly inducing cell death (e.g. by perforin/granzyme-mediated apoptotic induction within target cells) (4, 11–14).
Genetic models have provided insights into NK cell development and their role as potent regulators of inflammation and immunity (15–21). However, given that many genetic models of NK cell deficiency are compounded by additional defects, studies often investigate the role of NK cells using antibody depletion methods. In vivo depletion studies of NK cells have been primarily done using: i) antibodies to NK1.1, a glycoprotein expressed on NK cells and subsets of γδ T cells, NKT, and CD8 T cells in certain strains of mice, or ii) antibodies to asialo-GM1 (AsGM1), a glycolipid highly expressed on NK cells, basophils and subsets of γδ T cells, NKT, and CD8 T cells (22). While anti-NK1.1 antibody depletion using the PK136 antibody clone works robustly in some strains of mice (e.g. C57BL/6J, SJL and NZB), this antibody does not work in other strains of mice due to allelic variation in the NKR-P1 gene that encodes NK1.1 (CD161) (23, 24). In contrast, anti-AsGM1 antibody robustly depletes NK cells across multiple strains of mice including C57BL/6J, BALB/c, DBA2 and 129 strains, and remains in use to this day (22, 25, 26).
Though NK cells were once considered to be a functionally homogeneous lymphocyte population (e.g. in contrast to the diversity of specificities and functionalities of T cells), there is now clear evidence that NK cells have the capacity to discriminate between diverse targets and to respond with diverse functions (27–36). In attempts to better understand this NK cell diversity, NK cells have been divided into subsets based on heterogeneous expression of various cell surface markers (e.g. TRAIL, Thy1, KLRG1, CCR2, CD11b, and CD27) or divergent functional properties (37–41). Recently, liver NK cells were shown to contain two distinct subsets, tissue-resident (trNK) and conventional (cNK), circulating NK cells, based on differences in trafficking and tissue-retention (42). Notably, further studies demonstrated that beyond these differences in trafficking, trNKs and cNKs have differences in cytokine production, cell surface proteins involved in cell adhesion and NK cell recognition, and distinct developmental requirements (43). While the contribution of trNK cells to shaping immune responses remains an active area of investigation, liver trNK cells are known to confer memory-like immune responses in a contact hypersensitivity model (42). The existence of both local and systemic NK cell reservoirs has significant implications in understanding the multi-faceted role of NK cells in regulating local and systemic immune responses.
Here we sought to investigate the characteristics and contribution of trNK cells to local tissue injury in the context of ischemia reperfusion injury. To study the role of NK cells in ischemia reperfusion injury (IRI), we used a mouse model of ischemic acute kidney injury (AKI), a common human pathology characterized by ischemia that can result from various etiologies including diabetes, major surgery, sepsis, and drug toxicity (44). While several previous studies have highlighted the importance of NK cells in ischemic AKI using patch-clamping models of AKI, models associated with intra-renal coagulation (45–47), the role of NK cells in ischemic AKI in the absence of intra-renal coagulation remains unknown. Notably, the dynamics and contributions of cNK and trNK cells in ischemic tissue injury remains undefined.
By studying trNK cells in the kidney, and multiple other organs, we found that trNK cells have variable and reduced asialo-GM1 (AsGM1) expression relative to cNK cells, such that trNK cells are relatively spared from depletion by anti-AsGM1 antibodies. Using this knowledge, we demonstrate that trNK cells are a potent mediator of IRI. These data identify a new method to study trNK cells in vivo and define a new physiological role for trNK cells, as a front-line player in the response to acute ischemic injury.
MATERIALS AND METHODS
Mice
B6 (C57BL/6J), NOD (NOD/ShiLtJ), BALB (BALB/cJ), 129 (129S1/SvImJ), and CD1dKO (B6.129S6-Del(3Cd1d2-Cd1d1)1Sbp/J, formerly named B6.129S6-Cd1d1/Cd1d2tm1Spb/J) mice were purchased from the Jackson Laboratory (Bar Harbor, ME) and briefly housed in a specific pathogen-free colony at the University of Colorado at AMC. CD1dKO (a gift from Dr. Laurent Gapin, University of Colorado, Anschutz Medical Campus) (78), B6, and BALB/c mice were also obtained from breeding colonies at the University of Colorado, Anschutz Medical Campus and maintained in a pathogen-free facility and were of 8–12 wk of age for experimental use. Mice were age- and sex-matched for experiments, with male mice uniformly used for general screening and IRI studies. Female mice were solely used for the analysis of trNK distribution in multiple organs. All experiments were approved by the Institutional Animal Care and Use Committee of the University of Colorado Denver, in accordance with the National Institutes of Health guidelines for use of live animals. The University of Colorado Denver, Anschutz Medical Campus is accredited by the American Association for Accreditation of Laboratory Animal Care. Tbet−/− (formal gene name Tbx21) mice were purchased from Jackson Laboratory and NFIL-3−/− mice (from Paul B. Rothman, Johns Hopkins University (43, 48)), were bred and housed in a pathogen-free facility, with procedures performed in accordance with the animal protocol approved by the Washington University School of Medicine (WUSM) Animal Studies Committee.
Induction of Ischemia reperfusion injury (IRI)
The hanging weight system was used as previously described (49). Briefly, mice were anesthetized with pentobarbital at 20mg/kg and body temperature was regulated using an anal probe during the entire surgery. For sham surgery, mice were subjected to a right kidney nephrectomy followed by recovery under a heat lamp. To induce IRI, nephrectomy of the right kidney was done, after which the left kidney was exposed and connective and adrenal tissue was gently separated. Next, the kidney was place in a Lucite cup and ischemia was induced for 30m by placing 6–0 nylon suture under the renal artery and applying hanging weights, to occlude blood flow. Following 30 minutes of ischemia, hanging weights were removed and the kidney was allowed to reperfuse. Mice were given saline, sutured closed and allowed to recover for the indicated time points of reperfusion (typically 4 and 24 hours of reperfusion).
Analysis of Renal Function
To assess kidney function, glomerular filtration rate (GFR) was measured as previously described (49). Briefly, mice were anesthetized with pentobarbital at 20mg/kg and mice were kept warm to maintain their body temp using an anal probe during entire surgery. Once mice were fully anesthesized, a catheter was inserted into the jugular vein with a second catheter inserted into the bladder to collect urine. Next, Inulin-FITC (Sigma) was infused at 800 microliters per minute. Blood and urine were collected every twenty minutes to measure GFR as described (49, 50).
Parabiosis
Parabiosis surgery was performed as previously described (42, 43). Briefly, a longitudinal skin incision was made on the flanks of both C57BL/6NCr (Ly5.2) and B6-Ly5.1/Cr age and weight matched female mice. Their elbows and knees were joined with dissolvable sutures and the incision closed with wound clips. Postoperative care included administration of buprenex for pain control, 5% dextrose and 0.9% sodium chloride for fluid replenishment. Nutritional gel packs were provided in each cage and Sulfatrim (antibiotic) in the drinking water for the duration of the experiment. Mice were sacrificed and analyzed fourteen days post surgery. The procedure was approved by the Animal Studies Committee at Washington University, St. Louis, MO.
Flow cytometric cell analysis
To quantify the number of NK and NKT cells, mice were perfused with PBS, kidney were harvested, and minced through a 70um filter. Digested tissue was washed with RPMI and then lymphocytes were purified using a 36% percoll gradient (GE Healthcare). Cell were stained in FACS buffer (PBS, 0.2% Fetal Bovine Serum, 0.01% sodium azide), anti-Fc receptor blockade CD16/32, and αCD3 PE-Cy7 (145-2C11), αNK1.1 PerCP-Cy5.5 (PK136), αNKp46 (29A1.4), and αCD45(30-F11). NK cells were defined as CD45+CD3−NK1.1+ events, with NKT cells defined as CD45+CD3+NK1.1+ events. For studies analyzing tissue-resident NK cells, mice were perfused with 15 mL of saline (excluding parabiotic studies and analysis of Tbet−/− and NFIL-3−/− mice) after which kidney, liver, lung, pancreas and uterus were harvested, minced and mechanically disrupted, followed by a 1 hr enzymatic digestion in collagenase D (1 mg/mL, Roche) at 37°C. Cells were then washed in complete RPMI 1640 (10% fetal calf serum, 1% penicillin/streptomycin). Cells were then stained with a cocktail of fluorescently-labeled antibodies from eBioscience: αCD3 e450 (145-2C11), αCD19 e450 (ebio1D3), αMHCII e450 (MS/114.15.2), αCD45 APC-eFluor780 (30-F11), αNK1.1 PerCP-Cy5.5 (PK136); or Biolegend: αNKp46 PE or PE-Cy7 (29A1.4), αCD49b PE-Cy7 (DX5), αCD49b (HMα2), αTRAIL PE, αCD44 FITC, αCD160 PE, αCD49a PE or APC, αAsGM1 AlexaFlour647 (poly21460); or BD Pharmingen αCD49a (Ha31/8). NK cells were identified as lymphocytes (by FSC and SSC) that were CD45+, Lineage negative (CD3−CD19−MHCII−), and either NK1.1+ or NKp46+. Tissue-resident NK (trNK) and cNK cells were identified as either CD49a+ or DX5+, respectively. Anti-AsGM1 antibody staining was done using 0.35 μg antibody per stain for 30 minutes. In a subset of studies, cNK cells were defined as NK cells that expressed CD49b+ CD49a− using the HMα2 clone, instead of the CD49b-reactive DX5 clone. Note that both antibodies recognize CD49b, though the DX5 clone may be less-sensitive than HMα2 for cells that express lower levels of CD49b (51). For NK, trNK versus cNK, and NKT cells composition after IgG or NK1.1 treatment, mice were treated with either IgG control antibody or NK1.1 on day −3 and −1. On day 0, mice were then subjected to hanging weight system for 30 minutes to induce IRI. After 1 day of reperfusion, kidney was harvested and lymphocytes were purified and analyzed using flow cytometry after stained as noted above with NKp46 being the NK cell marker. For parabiotic studies, cells were stained for CD45.1 (clone A20) and CD45.2 (clone 104), both from eBioscience. Gating strategies for flow cytometric analysis are indicated in figure legends. Samples were analyzed using a BD LSRII flow cytometer.
NK cell depletion
For NK1.1 depleting experiments, B6 and CD1d knock out mice were treated with either 200μg control α-mouse IgG2a (BioXCell C1.18.4, mouse IgG2a Isotype control) or α-NK1.1 (BioXCell PK136) on day −3 and −1. For Asialo-GM1 depleting experiments, B6 mice were treated with 800μg of either rabbit IgG (Southern Biotech) or AsGM1 (WAKO) on day −1. On day 0, mice were then subjected to hanging weight system for 30 minutes to induce IRI. After 1 day of reperfusion, kidney function was measured by GFR. NK cell depletion was verified by measuring NK cell percentages (CD45+CD3−NKp46+).
Detection of Cleaved Caspase-3
B6 mice were treated with IgG or NK1.1 or AsGM1 antibodies as above. Mice that received IgG or NK1.1 antibodies were exposed to sham (−I) or ischemic (+I) surgery and allowed to recover for 4 hours. AsGM1 treated mice only received ischemic surgery (+I). At 4 hours reperfusion, kidneys were harvested, fixed in 4% PFA and paraffin embedded. Five micron thick paraffin sections were deparaffinized, antigens unmasked and immunohistochemically stained for cleaved Caspase 3 (Cell Signaling, Danvers, MA; rabbit polyclonal; cat#: 9661, 1:1000 in TBST + 1% BSA w/v). Sections required antigen retrieval in BORG solution (Biocare Medical, Concord, CA; high pH; 10 minutes at 110°C) in the Decloaking chamber (Biocare). All incubations were performed on the autostainer (Benchmark XT; Ventana Medical Systems/Roche, Tucson, AZ) at an operating temperature of 37°C. Primary antibody was incubated for 32 minutes and detected with a modified I-VIEW DAB (Ventana) detection kit. The I-VIEW secondary antibody was replaced with a species specific polymer (Rabbit ImmPress; Full Strength; Vector Labs, Burlingame, CA; 8 minutes) and streptavidin-horseradish peroxidase replaced with diluted Rabbit ImmPress (1:2 dilution in PBS pH 7.6; 8 minutes). All sections were counterstained in Harris hematoxylin for 2 minutes, blued in 1% ammonium hydroxide (v/v), dehydrated in graded alcohols, cleared in xylene and coverglass mounted using synthetic resin. Quantitation of caspase-3 immuno-positive cells was determined by gathering the numerical total across the entire kidney section, at a 20X (0.26 mm2) magnification. Representative images were chosen based on the ratio of the number of caspase-3 positive cells in ischemic groups versus sham group. Differences between αNK1.1 and αAsGM1 groups were corroborated by a second, independent investigator who was blinded to the experimental groups.
Software and Statistical Analysis
Data analysis and plotting were done using Prism 5 (GraphPad Software). Flow cytometric data were analyzed using FlowJo version 10 (TreeStar), with data displayed as high-resolution pseudo-color dot plots showing outliers, plotted on a biexponential axis. Statistical analyses were performed using Prism 5, with unpaired student t-test or one-way ANOVA with Bonferroni’s post-test correction for multiple comparisons. Experimental values were considered to be statistically different if they were p<0.05.
RESULTS
Kidney conventional NK cells preferentially express Asialo-GM1 compared to tissue-resident NK cells
Recently, two subsets of NK cells were identified in the liver based on either tissue-retention (trNK) versus systemic circulation (cNK) (42). Further studies showed that these subsets could be identified as CD49a+DX5− (trNK) versus DX5+CD49a− (cNK) in the liver as well as multiple other organs (42, 43). While trNK cells were identified in multiple organs, the presence of trNK cells in the kidney has not been reported. To test this, we analyzed NK cells in the kidney and identified both CD49a+DX5− and CD49a−DX5+ NK subsets (Fig. 1A). In the unmanipulated kidney, CD49a+ NK cells were a minor, but readily detectable fraction (15–20%) of the NK cell pool (Fig. 1B). To demonstrate that CD49a+ NK cells in the kidney were tissue-resident we analyzed these populations in parabiotic pairs of mice. This analysis demonstrated that CD49a+ NK cells were present only in the host and not the other parabiont, indicating that CD49a+ NK cells did not systemically circulate between the two parabionts (Fig. 1C–D). In contrast, DX5+CD49a− NK cells were detectable in both parabionts, indicating that DX5+ NK cells were capable of systemic circulation, exchanging these cells between the two parabionts in the kidney (Fig. 1C–D). The percent chimerism in the parabiotic mice was determined by the expression of CD45.1 and CD45.2 in the spleen (Fig. 1E). These data demonstrate that the kidney contains both CD49a+ trNK cells and DX5+ cNK cells.
Figure 1. The kidney contains both conventional NK cells and tissue-resident NK cells characterized by differential AsGM1 expression.
B6 mice were perfused with PBS, and kidney tissue was analyzed for the presence of CD49a+ tissue-resident (trNK) versus DX5+ conventional (cNK) NK cells. (A) Representative flow cytometric analysis to identify NK cells (NK1.1+CD3−CD19−MHC classII−) that were either trNK cells (CD49a+DX5−) or cNK cells (DX5+CD49a−). (B) Percent distribution of trNK and cNK cell subsets in the kidney. (C–E) Negligible exchange of kidney CD49a+DX5− NK cells in parabiotic mice. WT B6 (CD45.2) mice were parabiosed to congenic B6 (CD45.1) mice. At day 14 post-surgery, the spleen and kidney were harvested and flow cytometry performed. (C) Representative dot plots of the kidney gated on live CD3−CD19−NK1.1+ cells followed by a CD45.1 gate (left panels) and CD45.2 (right panels) as indicated for each parabiont. The percentages of CD49a+DX5− and CD49a−DX5+ cells are indicated in the dot plots. (D) The percentages of CD49a+DX5− and DX5+CD49a− cells in the kidney from C are shown in the stacked bar graph which represents 8 parabiotic pairs done three independent times. (E) The chimerism in the parabiotic pairs is measured by the percentage of CD45.1 and CD45.2 expression in the spleen depicted in the stacked bar graph. (F) Representative expression levels of cell surface proteins comparing trNK (shaded gray) and cNK (open red) cells, defined as in A. (G) Representative histogram of Asialo-GM1 expression for trNK and cNK cell subsets, with quantification of median fluorescent intensity on right. Data depict mean +/− SEM, with each data set containing data from 3–5 mice per group from independent experiments. Statistical significance indicated by *, p<0.05, as determined by unpaired t-test.
trNK cells and cNK cells have differential expression of multiple proteins (42, 43). To investigate the cell surface phenotype of these two subsets in the kidney, we performed an analysis of proteins involved in NK cell activation and trafficking. These studies showed that in the kidney, trNK cells had uniform increased expression of CD44 (an activation and adhesion protein), CD160 (an activation-associated protein) and TRAIL (an apoptosis-inducing cell surface protein) relative to cNK cells, with a subset of trNK cells also expressing the chemokine receptor CCR2 (Fig. 1F). In contrast, trNK cells had uniformly low expression of CD62L (an adhesion protein) relative to cNK cells (Fig. 1F). trNK cells also had reduced expression of two inhibitory receptors relative to cNK cells, uniformly lacking expression of KLRG1 with a prominent fraction of trNK cells that also failed to express CD244 (Fig. 1F). Strikingly, kidney trNK cells had reduced expression of asialo-GM1 (Fig. 1G), a glycolipid moiety that has been a target for antibody-mediated depletion of NK cells in vivo (52). These data identify that trNK cells have a more activated phenotype with potentially reduced susceptibility to inhibitory signals compared to cNK cells. They further indicate that trNK cells express variable levels of AsGM1 and may be differentially susceptible to AsGM1-antibody mediated depletion in vivo.
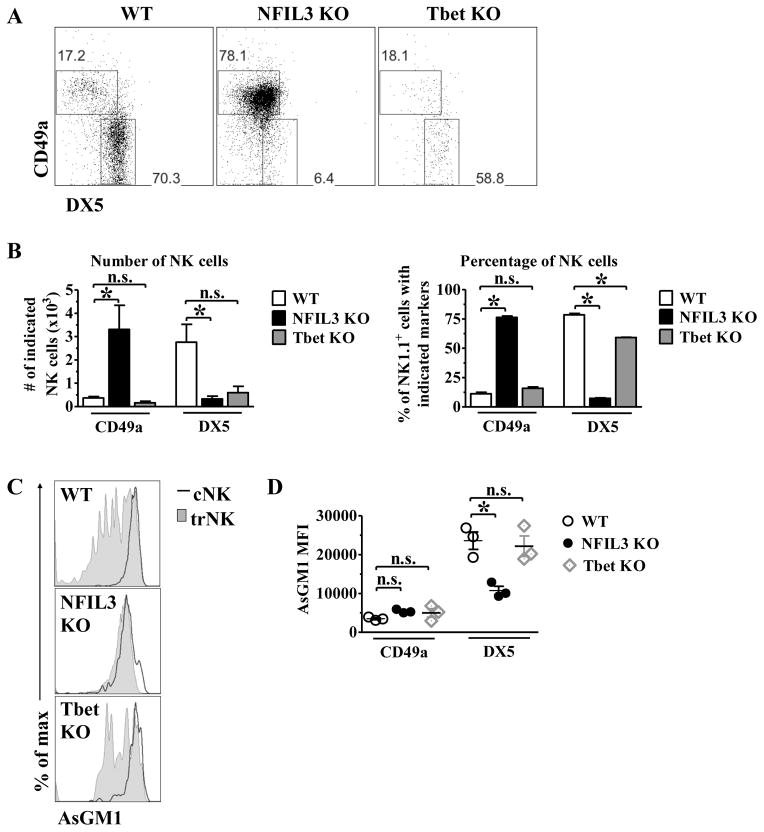
Kidney trNK cells develop independent of NFIL3 and Tbet
We next sought to determine the developmental requirements for kidney trNK cells. To do this, we investigated the relative frequency and abundance of trNK and cNK cell subsets in the kidney under baseline conditions, in mice deficient in either NFIL3, which is required for cNK cell development (53, 54), or Tbet (Tbx21), which contributes to NK cell maturation (38) and is required for the development of liver and skin but not uterine trNK cells (43). Kidney trNK cells were present at or above levels found in C57BL/6 mice both by total cell number and by frequency in NFIL3-deficient mice (Fig. 2A–B). Conversely, cNK cells were largely absent from the kidney in NFIL3-deficient mice, consistent with previous reports identifying NFIL3 as an essential regulator of cNK development (53, 54). trNK cells were also present at a normal frequency in Tbet-deficient mice, although Tbet-deficient mice had reduced number of NK cells, with ~50% the number of trNK cells and only ~20% the number of cNK cells relative to C57BL/6J mice (Fig. 2A–B). AsGM1 was expressed at lower levels in trNK cells relative to cNK cells in both NFIL3-deficient and Tbet-deficient mice, consistent with AsGM1 expression levels observed in trNK cells in wild-type mice (Fig. 2C–D). While differential AsGM1 expression was observed between trNK and cNK cells in all strains of mice, NFIL3-deficient mice have reduced AsGM1 expression in cNK cells and fewer AsGM1 low trNK cells than in wild-type mice (Fig. 2C–D). These studies show that kidney trNK cells develop independent of NFIL3 and Tbet.
Fig. 2. Kidney CD49a+DX5− NK cells develop independent of NFIL3 and Tbet.
The kidney was isolated from WT, NFIL3 KO, and Tbet KO mice, stained, and analyzed by flow cytometry. (A) Representative dot plots were gated on live CD45+CD3−CD19−NK1.1+ and show the percentage of CD49a and DX5 expression. (B) The bar graphs depict the number (left panel) and percentage (right panel) of live CD45+CD3−CD19−NK1.1+ that express CD49a and DX5 in the kidney of WT, NFIL3 KO and Tbet KO mice. (C) The histograms indicate the expression of AsGM1 on gated CD45+CD3−CD19−NK1.1+ CD49a+ (shaded gray histogram, trNK) and CD45+CD3−CD19−NK1.1+ DX5+ (red line histogram, cNK). (D) The graph represents the mean fluorescence intensity of AsGM1 on CD49a+ and DX5+ on NK cells of WT (open circles), NFIL3 KO (filled circles), and Tbet KO (grey diamonds) mice. Experiments were performed three independent times. Statistical significance indicated by *, p<0.05, as determined by unpaired t-test between trNK and cNK cells within the same genotype.
Asialo-GM1 is preferentially expressed on cNK cells compared to trNK cells across multiple strains and organs
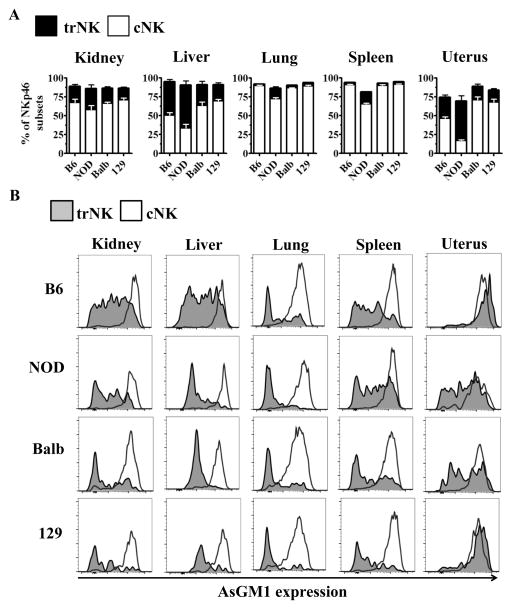
Given the broad use of AsGM1 NK depleting regimen on various strains of mice, we sought out to determine if preferential expression of AsGM1 on cNK cells was exclusive to the kidney and B6 mice. To determine whether trNK cells and cNK cells had differential expression of AsGM1 in additional tissues and genetic backgrounds, we analyzed these NK cell subsets in different strains of mice using NKp46, a common marker for NK cells (55). Notably, NKp46 is interchangeable with NK1.1 when identifying trNK and cNK cell subsets in the kidney of B6 mice (Supplemental Fig. 1).
When we analyzed the relative frequency of cNKs and trNKs across common inbred strains of mice (B6, NOD, BALB/c and 129), we found that trNK frequency was relatively comparable across different backgrounds, with NOD mice showing a slightly increased trNK frequency in some organs (Fig. 3A). While certain organs consistently had low frequencies of trNK cells (spleen, lung), other organs uniformly had elevated frequencies of trNKs (liver, kidney, and pancreas) regardless of background (Fig. 3A, Supplemental Fig. 1B–C). The frequency of uterine trNK cells varied dramatically between different strains of mice with BALB/c and 129 strains of mice having low frequencies of trNK cells compared to B6 and NOD mice (Fig. 3A).
Figure 3. Conventional and tissue-resident NK cells have differential expression of asialo-GM1 across multiple organs and genetic backgrounds.
B6, NOD, BALB/c, and 129 mice were analyzed for the frequency of trNK and cNK cell subsets and the AsGM1 expression in these subsets. (A) Frequency of trNK and cNK cells was determined in multiple organs in various strains based on NK markers described in Fig. 1A. (B) AsGM1 expression in trNK (shaded gray) and cNK (open red) cells, with organ analyzed indicated at the top of each column, and genetic background indicated at the left of each row. Each data set has 3–5 mice per group from independent experiments.
Next, we analyzed the expression of AsGM1 on cNK and trNK cells in various organs and genetic backgrounds. cNK cells uniformly expressed high levels of AsGM1 across all organs analyzed, regardless of strain (Fig. 3B, Supplemental Fig. 1D). In contrast, trNK cells had differential expression of AsGM1, with the majority of trNK cells expressing only low levels of AsGM1 in the kidney, spleen, liver, and lung (Fig. 3B). While a subset of trNK cells had comparable expression of AsGM1 relative to cNKs (e.g. in the liver of B6 mice), the only instance where trNK cells uniformly expressed high levels of AsGM1 comparable to cNK cells was in the uterus of some strains of mice (B6, 129) (Fig. 3B). Analysis of NK cell subsets using AsGM1 as a primary parameter relative to either CD49a or DX5 further identified CD49a+ trNK cells with low, intermediate and high levels of AsGM1 expression, varying by organ and genetic background (Supplemental Fig. 2). Taken together, these data show that cNK cells uniformly express high levels of AsGM1 and that trNK cells contain a phenotypically heterogeneous population characterized by variable, but reduced expression of AsGM1 across multiple organs and strains of mice.
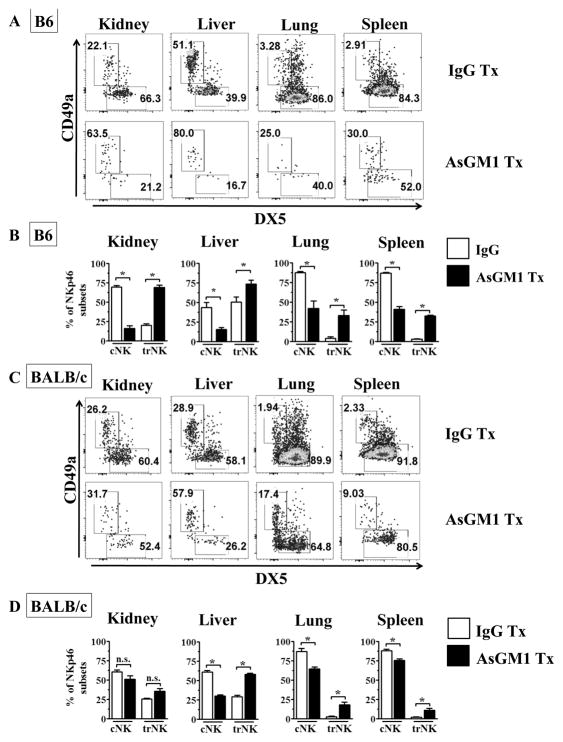
Asialo-GM1 antibody treatment preferentially depletes cNK cells in B6 and BALB/c mice and enhances the relative frequency of trNK cells
Anti-AsGM1 antibodies have been commonly used to deplete NK cells across diverse genetic backgrounds. Our data indicate that while AsGM1 is uniformly expressed at a high level in cNK cells, it is variably expressed in trNK cells, with a many trNK cells expressing low levels of AsGM1. We next sought to determine whether the presence of AsGM1low trNK cells may reduce the susceptibility of trNK cells to be depleted by AsGM1 antibody treatment in B6 mice. AsGM1 antibody treatment resulted in a pronounced decrease in the frequency and number of total NK cells across multiple organs, consistent with the overall higher frequency of cNK cells relative to trNK cells (Fig. 4A–B, Supplemental Fig. 3A). While AsGM1 treatment reduced the number of both cNK and trNK cells (Supplemental Fig. 3B), the magnitude of depletion between cNK cells and trNK cells profoundly differed. Indeed, when we measured the relative frequency of cNK cells and trNK cells, we found that AsGM1 antibody treatment resulted in a pronounced shift in the relative frequency of cNK cells and trNK cells within the kidney, liver, lung, and spleen (Fig. 4A–B). These data indicate that in B6 mice trNK cells have reduced susceptibility to AsGM1-based antibody depletion.
Figure 4. Asialo-GM1 antibody treatment preferentially depletes cNK cells in B6 and BALB/c mice to enhance the relative frequency of trNK cells.
B6 (A,B) and BALB/c (C,D) mice were treated rabbit IgG or Asialo-GM1 antibody day −1, and organs were harvested 1 day later, and analyzed for trNK and cNK cells (as in Fig. 1A). (A) Representative dot plots of NK cell subsets in B6 kidney, liver, lung, and spleen after IgG (top row) or AsGM1 (bottom row) treatment. (B) Frequency distribution of NK subsets in B6 mice across multiple organs based on CD49a and CD49b expression treated with IgG or AsGM1 antibody. (C) Representative dot plots of NK cell subsets in BALB/c kidney, liver, lung, and spleen after IgG (top row) or AsGM1 (bottom row) treatment. (D) Change in frequency distribution of NK subsets in BALB/c mice from multiple organs based on CD49a and CD49b expression treated with IgG or AsGM1. Each data set has 3–6 mice per group from independent experiments. Statistical significance indicated by *, p<0.05, as determined by unpaired t-test, comparing the frequency of either cNK or trNK in IgG versus AsGM1 antibody treated mice.
While AsGM1 antibody depletion is used in B6 mice, it is often used as a method to validate results obtained by NK1.1 antibody depletion. In contrast, certain strains of mice (e.g. BALB/c) do not express the PK136-reactive NK1.1 allele, such that AsGM1 antibody is the primary method to test the role for NK cells in vivo (23). Given that BALB/c mice also contain AsGM1low trNK cells, we next tested the selectivity of AsGM1 antibody depletion on cNK and trNK cells in this strain. AsGM1 antibody treatment again resulted in a large reduction in the frequency of NK cells and an overall decrease in cNK and trNK cell numbers (Supplemental Fig. 3A,C), with AsGM1 antibody treated mice showing a notable skewing in the relative frequency of trNK cells in the liver, lung and spleen (Fig. 4C–D). In total, these data indicate that AsGM1 antibody treatment preferentially depletes cNK cells, resulting in a relative sparing of trNK cells across multiple organs and two common inbred strains of mice.
Tissue-resident NK cells and conventional NK cells in IRI of the kidney
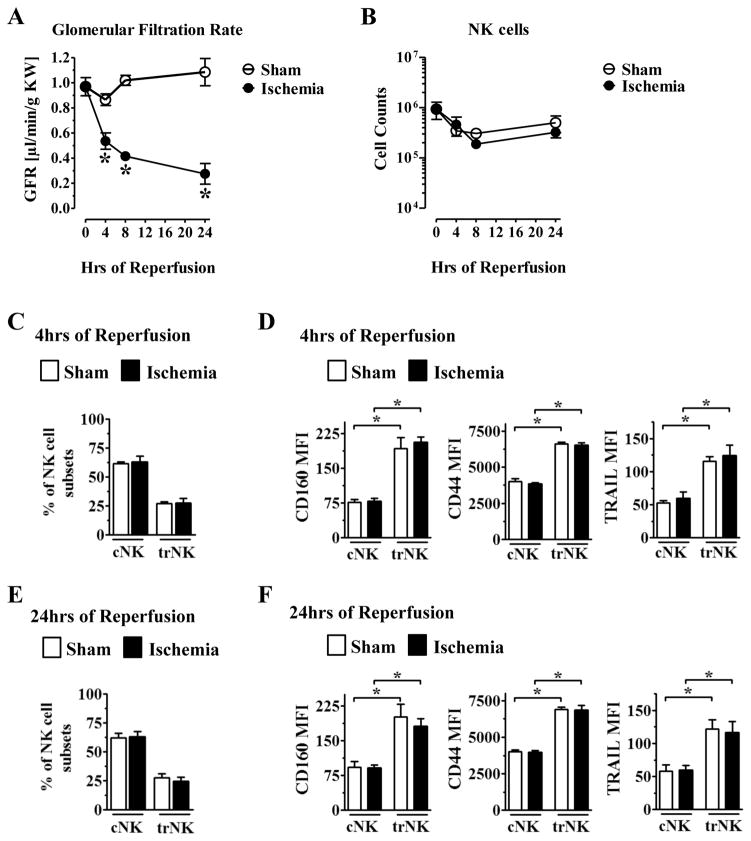
NK cells have been implicated in ischemia-reperfusion injury (IRI) in the kidney but the role of trNK and cNK cells is unclear (45–47). We sought to characterize the dynamics of trNK cells and cNK cells during ischemic injury in the kidney by employing the hanging weight system of IRI, a well-established model of acute kidney injury (49). In mice subjected to 30 minutes of ischemia, kidney function as measured by glomerular filtration rate (GFR) rapidly declined within 4 hours after reperfusion, with maximal loss of kidney function occurring by 24 hours (Fig. 5A). We next determined the total number of NK cells at 4, 8, and 24 hours post reperfusion, and found that there were no significant differences in the total number of NK cells present in the kidney following this acute ischemic injury (Fig. 5B). Given that NK cell numbers did not change dramatically, we next determined if there were changes in the relative distribution of trNK and cNK cells or their activation phenotype during this acute time course. At 4 and 24 hours post-reperfusion, there were no changes in the relative distribution of trNK cells and cNK cells in the kidney (Fig. 5C,E). Furthermore, when we analyzed the activation status of trNK cells and cNK cells during IRI, we found that trNK cells expressed higher levels of CD160, CD44, and TRAIL than cNK cells at both 4 and 24 hours, in both the naïve (sham) and ischemic kidney, with no discernable difference in either NK cell subset following ischemic injury (Fig. 5D,F). These distinct phenotypes mirrored differences observed in trNK and cNK cells in the steady state. Taken together, these data suggest that trNK cells and cNK cells have a relatively stable number and cell surface phenotype during acute ischemic kidney injury, with trNK cells showing evidence for a heightened activation state.
Figure 5. IRI in the kidney does not profoundly alter phenotype of trNK and cNK cells.
B6 mice were subjected to either sham surgery (white bars) or 30m of ischemia (black bars) and analyzed as described here. (A) After 4, 8, and 24hr of reperfusion, kidney function was measured by GFR. (B) B6 mice were subjected to IRI and after 4, 8, and 24hr of reperfusion, NK cells were quantified using percoll purification and analyzed by flow cytometric analysis by staining for CD45+CD3−NK1.1+ expression. (C) B6 mice were subjected to sham or 30m of ischemia and analyzed for the frequency of trNK and cNK cells at 4hrs after IRI. (D) Flow cytometric analysis of mean expression of activation markers on trNK versus cNK cells at 4hrs after IRI in the kidney. Each data point has 4–6 mice per group. (E) B6 mice were subjected to sham or 30m of IRI surgeries and analyzed for the frequency of trNK and cNK cells at 24hrs after IRI. (F) FACS analysis of mean expression of activation markers on trNK versus cNK at 24hrs after IRI in the kidney. Each data set has 4–6 mice per group from independent experiments. Statistical significance indicated by *, p<0.05, as determined by unpaired t-test, comparing either values from the same time point between sham and ischemia (A), or by one-way ANOVA with significance indicated between cNK and trNK (D, F).
NK1.1 depleting antibody protects mice from kidney dysfunction during IRI
The use of anti-NK1.1 antibody as a depleting strategy for NK and NKT cells is well established in the field. While anti-NK1.1 depleting studies have been shown to be protective in some studies of ischemic kidney injury (47), the role of NK cells in the hanging-weight model of kidney injury remains to be elucidated. To test this, B6 mice were treated with either isotype or αNK1.1 depleting antibody (administered on day −3 and −1), subjected to IRI on day 0 and allowed to recover for 1 day. Depletion of NK cells (CD45+CD3−NKp46+) was confirmed in the kidney by flow cytometry (Fig. 6A), with NK1.1 antibody treatment resulting in an equivalent depletion of both cNK cells and trNK cells (Fig. 6B,C). Whereas isotype treated animals subjected to IRI had a pronounced impairment in kidney function as measured by GFR, αNK1.1 treated mice showed a clear improvement in kidney function with GFR values nearly restored to values measured from sham, non-ischemic mice (Fig. 6D).
Figure 6. NK1.1 antibody treatment effectively depletes trNK and cNK cells in the kidney and attenuates kidney damage.
B6 mice were treated with IgG or NK1.1 antibody on day −3, −1, subjected to IRI on day 0 and analyzed. (A) The frequency of NK cells in the kidneys of B6 mice that were treated with IgG or NK1.1 antibody and subjected to sham (−I) or IRI (+I) surgeries at 1 day post-reperfusion. (B–C) B6 ischemic kidneys treated with either IgG or NK1.1 antibody were analyzed by flow cytometric analysis for distribution of trNK versus cNK cells, with frequencies plotted in panel C. (D) Quantification of GFR in B6 and CD1d KO mice that received either IgG or NK1.1 antibody treatment in sham (−I) or ischemic (+I) conditions. Each data set has 3–6 mice per group from independent experiments. Statistical significance indicated by *, p<0.05, as determined by unpaired t-test (C), comparing IgG and NK1.1 treated mice or by one-way ANOVA with Bonferroni’s post-test correction for multiple comparisons (A, D). n.s., no significant difference.
The NK1.1 antigen is expressed by both NK and NKT cells, both which have been implicated in acute kidney injury (AKI) (45, 47, 56, 57). To test the role of NKT cells in this model, we quantified the number of NKT cells (CD45+CD3+NK1.1+), at 4, 8 and 24 hours post-IRI and found no significant changes in total numbers (Supplemental Fig. 4A–B). Importantly, when comparing the number of NK and NKT cells in the ischemic kidney, NK cells were much more abundant than NKT cells (Supplemental Fig. 4C).
Given that the NK1.1 antibody can deplete both NK and NKT cells, we sought to test the impact of NK1.1 antibody treatment on mice genetically deficient in NKT cells (e.g. Jα18- or CD1d-deficient mice). While mice deficient in the T cell receptor gene segment Jα18 have a deficit in NKT cells, recent studies have shown that these mice also have major alterations in T cell receptor repertoire (58). Based on this, we tested CD1d1 x CD1d2-deficient mice, which are defective in NKT cell development, without other known defects in lymphocyte development (59, 60). As expected, αNK1.1 antibody treatment profoundly depleted NK cells in CD1d-deficient mice (Supplemental Fig. 4D). Whereas isotype-treated CD1d-deficient mice subjected to ischemia showed decreased GFR, similar to wild-type mice, αNK1.1 antibody treatment significantly improved GFR in both CD1d-deficient, and wild-type, ischemic mice (Fig. 6D). Based on these data, we conclude that the primary protective effect of αNK1.1 antibody treatment results from depletion of NK cells, not NKT cells.
Asialo-GM1 preferentially ablates cNK cells, showing trNK cells promote AKI
While the above data identify NK cells as mediators of ischemic injury, they do not discriminate between the relative contributions of trNK cells and cNK cells, since NK1.1 antibody treatment equivalently depletes both NK cell subsets. Given the differential expression of AsGM1 on trNK cells and cNK cells in the kidney, and their differential susceptibility to AsGM1 antibody depletion, we hypothesized that AsGM1 antibody treatment may allow us to dissect the relative contributions of trNK cells and cNK cells to ischemic kidney injury. To test this, we treated B6 mice with αAsGM1 antibody depletion and subjected these mice to either sham (non-ischemia) or ischemic injury. Anti-AsGM1 antibody treatment resulted in a profound reduction of kidney NK cells in these mice, comparable to the magnitude of depletion observed following αNK1.1 antibody treatment (compare Fig. 6B, Fig. 7A). Consistent with our previous data, αAsGM1 antibody resulted in a prominent increase in the relative frequency of trNK cells in the ischemic kidney (Fig. 7A,B).
Figure 7. Asialo-GM1 antibody preferentially depletes cNK cells, showing trNK cells promote AKI.
B6 mice were treated with IgG or AsGM1 antibody on day −1 or treated with NK1.1 antibody day −3 and −1, then subjected to sham (−I) or IRI (+I) surgeries on day 0 and analyzed at day +1. (A) Representative dot plots of kidneys 24 hours after IRI from B6 mice that were treated with IgG or AsGM1 antibody, and analyzed for the frequency of NKp46+ NK cells (left) and the frequency of trNK and cNK cells in either IgG (top) or AsGM1 (bottom) treated mice. (B) Quantification of the frequency of trNK and cNK cells after IgG or AsGM1 antibody treatment in ischemic kidneys. (C) B6 mice treated with IgG, AsGM1, or NK1.1 antibody were given sham (−I) or IRI (+I) surgeries and glomerular filtration rate was measured 1 day post surgery. Each data set has 5–8 mice per group from independent experiments. Statistical significance indicated by *, p<0.05, as determined by unpaired t-test (B), comparing IgG and AsGM1 treated mice or by one-way ANOVA with Bonferroni’s post-test correction for multiple comparisons (C).
Next, we tested the consequence of antibody treatment of kidney injury and function. First, we examined the consequence of αNK1.1 and αAsGM1 antibody treatment on the early induction of apoptosis after ischemic injury, measured by active caspase-3 staining at 4 hours post-reperfusion. These studies showed that αNK1.1 antibody-injected mice had a significantly decreased number of apoptotic cells relative to αAsGM1 antibody-injected mice (Supplemental Fig. 4E–I). To interrogate the consequence of these antibodies on kidney function at 24 hours reperfusion, we measured kidney function by GFR, a gold standard for kidney function, comparing isotype, αAsGM1, and αNK1.1 antibody-injected mice. Strikingly, while αNK1.1 antibody treatment conferred protection, αAsGM1 antibody treatment failed to protect mice from IRI despite similar reduction in overall NK cells percentages (Fig. 7C). Given that αAsGM1 poorly depletes trNK cells, in contrast to αNK1.1 antibody treatment, these data identify that trNK cells are sufficient to promote IRI in the kidney.
DISCUSSION
NK cells are sentinels in the immune response that rapidly detect changes in the cellular environment to produce cytokines and to induce target-cell apoptosis. While NK cells have been implicated in diverse responses (e.g. virus infection, DNA damage, cellular transformation and ischemia-reperfusion injury), a common trigger of NK cells is the detection of cellular stress which either increases NK cell activating ligands or decreases NK cell inhibitory ligands (61). Recent data indicate that NK cells not only circulate, but that a subset of tissue-resident NK (trNK) cells are retained in situ (42, 43). Here, we hypothesized that trNK cells function as important mediators in ischemia-reperfusion injury by being poised for immediate response to tissue injury. To test this, we studied this process in the kidney and found that: i) trNK cells are present in the kidney, ii) trNK in the kidney, and other tissues, have reduced expression of AsGM1, a common target in NK cell depletion studies, and iii) anti-AsGM1 antibody depletion preferentially spares kidney trNK cells. Based on this knowledge, we directly compared how two different methods of NK cell depletion that differed in their ability to deplete trNK cells impacted IRI. Our studies show that although trNK cells are numerically minor, these cells are potent mediators of ischemic tissue injury.
NK cells are regulated by the relative engagement of activating and inhibitory NK cell receptors by their respective ligands on neighboring cells (61). A common theme of these responses is that cellular stress in target cells can either increase activating ligands (e.g. MICA in humans and Rae-1 in mice) or decrease inhibitory receptor ligands (e.g. MHC class I). In this context, ischemia-reperfusion injury, the process of transient oxygen insufficiency (i.e. hypoxia) to a tissue, is associated with the induction of cellular stress markers such as MICA (in humans) and Rae-1 (in mice), which are potent activating ligands for activating NK cells (45, 62–64), placing NK cells as central mediators of ischemic tissue injury.
Here, we studied the role of NK cells in IRI, by using a mouse model of ischemic acute kidney injury (AKI), a common human pathology characterized by ischemia that can result from various etiologies including diabetes, major surgery, sepsis, and drug toxicity (44). While previous studies highlighted the importance of NK cells in ischemic AKI using patch-clamping models of AKI, which are associated with intra-renal coagulation (45–47), the role of cNK and trNK cells in ischemic injury has not yet been studied.
Our identification of trNK cells as an important cell type in mediating IRI damage emphasizes how local NK cell responses can trigger tissue injury in the absence of NK cell recruitment. It is also notable that trNK cells express low levels of two inhibitory receptors KLRG1 and CD244, suggesting that trNK cells may have an increased propensity towards cytotoxicity resulting from decreased inhibitory signaling. Importantly, KLRG1- NK cells have been reported in other studies that were not conventional NK cells (CD3−NK1.1+DX5−), suggesting that in some contexts, absence of KLRG1 may be another marker for trNK cells (65–67). Additionally, CCR2 was exclusively on a subset of CD49a+ trNK cells in unmanipulated and ischemic kidney (data not shown), consistent with reports of CCR2+ NK cells during lung inflammation (39); these data suggest that trNK cells may play a role in the CCL2/CCR2 chemokine axis important for monocyte recruitment. Note that while our studies highlight a role for trNK cells in the early stages of acute ischemic injury, at this time, we have not focused on later stages of ischemic injury when recruitment of conventional NK cells may further enhance tissue damage (29).
Asialo-GM1 was identified as a NK cell marker over 30 years ago (68, 69), and depleting regimens using anti-asialo-GM1 have been critical in showing the requirement of NK cells in diverse experimental settings (70–75). Indeed, while AsGM1 antibody depletion is not uniquely restricted to NK cells (e.g. (22)), this method remains in use today, both to deplete NK cells in mouse strains that do not express the PK136-reactive NK1.1 allele (e.g. BALB/c), and as a complementary approach to anti-NK1.1 antibody studies in NK1.1-expressing mice (e.g. C57BL/6).
Our data indicating differential expression of AsGM1 on subsets of trNK cells provide a new level of consideration in the use of AsGM1 antibody depletion. First, by comparing and contrasting the impact of AsGM1- and NK1.1-based depletion methods on trNK and cNK cells and IRI, our studies illustrate a new approach to distinguish trNK and cNK cell functions in vivo. Second, our data identify an unanticipated heterogeneity of AsGM1 expression among trNK cells. While AsGM1 antibody significantly depletes cNK cells, this antibody regimen is unable to deplete at least a subset of trNK cells. As such, AsGM1 antibody results in incomplete depletion of trNK cells, an especially important consideration when studying tissues enriched in trNK cells (e.g. kidney, liver, pancreas, and uterus). Notably, a simple approach to understanding the utility and limitations of AsGM1 antibody depletion studies can be obtained by analyzing AsGM1 expression on conventional and trNK cells in the organ of interest. Our studies also emphasize the importance of analyzing NK cell depletion studies not only in the blood (which lacks trNK cells) but also in situ. Notably, analysis of NK cell depletion in the spleen and/or peripheral blood (which is devoid of trNK cells) or use of DX5 as an NK marker (which is not expressed on trNK cells) may significantly over-estimate the efficiency of antibody-based depletion in various tissues by failing to consider trNK cell reservoirs. Given these caveats, we consider it likely that the functional contribution of trNK cells remains under-appreciated, especially in studies based solely on AsGM1-depleting antibodies.
Depending on the organ and strain of mouse, trNK cells represent a sizable fraction of the NK cell pool with the long-term capacity to monitor and influence tissue function. While the function of trNK cells remains an active area of investigation, trNK cells appear to have unique capabilities, demonstrated both by their ability to directly facilitate delayed type hypersensitivity reactions (42) and by their unique cytokine and cytolytic profile (43). Given that trNK cells are retained in tissue and poised for activation, our data identify trNK cells as an early player in the IRI response in the kidney. While future studies are required to determine the functional mechanisms by which trNK cells promote IRI, it is clear that trNK cells are sufficient to induce tissue damage even after AsGM1 antibody depletion reduced the overall number of trNK cells.
Based on these data, we hypothesize that trNK cells may be major mediators of ischemic injury in multiple organs. Curiously, trNK cells have tissue-specific differences in terms of their development, with Tbet-dependent liver and skin trNK cells, in contrast to Tbet- and NFIL3-independent kidney and uterine trNK cells (43). While developmental and functional commonalities between kidney and uterine trNK cells remain to be further explored, it is notable that uterine trNK cells appear to regulate spiral artery arborization in the uterus (76). It is tempting to speculate that kidney trNK cells may also be involved in vascular changes, whether during ischemic injury or during development.
An important future goal in understanding trNK cells will be to determine the functional heterogeneity of trNK cells, both within an individual tissue and between different tissue types. Based on our studies, we propose that AsGM1 expression can serve as one additional parameter that may provide insights into trNK cell subsets. Examples of this include the heterogeneity of AsGM1 expression within trNK cells in the kidney, with some cells expressing little to no AsGM1 in contrast to other AsGM1high trNK cells. One notable exception to the heterogeneous, reduced expression of AsGM1 on trNK cells is in uterine trNK cells in B6 mice, consistent with an increasing literature suggesting unique developmental and functional properties of uterine NK cells (43, 76, 77). While the biologic consequence of differential expression of AsGM1 remains unknown, in the future it will be important to determine whether AsGM1 serves as an indicator of trNK maturation, localization and/or directly shapes the functionality of trNK cells.
trNK cells are a recently described subset of NK cells, whose function(s) remain an active area of investigation. By studying trNK cells in the kidney and other organs, our data identify an unanticipated differential expression of AsGM1 on trNK cells in multiple organs and strains of mice. These data emphasize that AsGM1 expression may provide a powerful new approach by which to gain fundamental new insights into NK cell development and function. Furthermore, our studies provide the first evidence that trNK cells play a central role in ischemic tissue injury.
Supplementary Material
Acknowledgments
This research was funded by National Institutes of Health Grants R01-DK097075, R01-HL092188, R01-HL098294, POI-HL114457, and R01-HL119837 (to H.K.E.); R01-AI093637 (to D.H.); American Heart Association National Scientist Development Grant #13SDG14510023 (to E.T.C.); and NIH grant R01-AI106561 and the Howard Hughes Medical Institute (to W.M.Y.). The University of Colorado Denver Histology Shared Resource is supported in part by the Cancer Center Support Grant (P30CA046934).
The authors thank Dr. Laurent Gapin, Jingjing Zhang, and Kathryn D. Tuttle for the kind gift of CD1dKO mice, Dr. Ron G. Gill and Marilyne Coulombe for the kind gift of AsGM1 antibody, Dr. Almut Grenz and Uladzimir Shabeka for training in the use of the hanging weight model, and Tom Nguyen, Melissa Ledezma and Kristann Magee for expert technical assistance with mice. We thank Liping Yang for assistance with the parabiotic studies. The authors appreciate the contribution to this research made by E. Erin Smith, HTL(ASCP)CMQIHC, Allison Quador, HTL(ASCP)CM and Jessica Arnold HTL(ASCP)CM of the University of Colorado Denver Histology Shared Resource. Contents are the authors’ sole responsibility.
References
- 1.Bryceson YT, March ME, Ljunggren HG, Long EO. Synergy among receptors on resting NK cells for the activation of natural cytotoxicity and cytokine secretion. Blood. 2006;107:159–166. doi: 10.1182/blood-2005-04-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiesa S, Tomasello E, Vivier E, Vely F. Coordination of activating and inhibitory signals in natural killer cells. Molecular immunology. 2005;42:477–484. doi: 10.1016/j.molimm.2004.07.030. [DOI] [PubMed] [Google Scholar]
- 3.Vivier E, Nunes JA, Vely F. Natural killer cell signaling pathways. Science. 2004;306:1517–1519. doi: 10.1126/science.1103478. [DOI] [PubMed] [Google Scholar]
- 4.Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nature immunology. 2008;9:503–510. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- 5.Yokoyama WM. Inhibitory receptors signal activation. Immunity. 2008;29:515–517. doi: 10.1016/j.immuni.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 6.Narni-Mancinelli E, Ugolini S, Vivier E. Tuning the threshold of natural killer cell responses. Current opinion in immunology. 2013;25:53–58. doi: 10.1016/j.coi.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 7.Thielens A, Vivier E, Romagne F. NK cell MHC class I specific receptors (KIR): from biology to clinical intervention. Current opinion in immunology. 2012;24:239–245. doi: 10.1016/j.coi.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 8.Rahim MM, Tu MM, Mahmoud AB, Wight A, Abou-Samra E, Lima PD, Makrigiannis AP. Ly49 receptors: innate and adaptive immune paradigms. Frontiers in immunology. 2014;5:145. doi: 10.3389/fimmu.2014.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper MA, Colonna M, Yokoyama WM. Hidden talents of natural killers: NK cells in innate and adaptive immunity. EMBO reports. 2009;10:1103–1110. doi: 10.1038/embor.2009.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raulet DH, Vance RE. Self-tolerance of natural killer cells. Nature reviews Immunology. 2006;6:520–531. doi: 10.1038/nri1863. [DOI] [PubMed] [Google Scholar]
- 11.Crouse J, Xu HC, Lang PA, Oxenius A. NK cells regulating T cell responses: mechanisms and outcome. Trends in Immunology. 2015;36(1):49–58. doi: 10.1016/j.it.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 12.Soderquest K, Walzer T, Zafirova B, Klavinskis LS, Polic B, Vivier E, Lord GM, Martin-Fontecha A. Cutting edge: CD8+ T cell priming in the absence of NK cells leads to enhanced memory responses. Journal of immunology. 2011;186:3304–3308. doi: 10.4049/jimmunol.1004122. [DOI] [PubMed] [Google Scholar]
- 13.Champsaur M, Lanier LL. Effect of NKG2D ligand expression on host immune responses. Immunological reviews. 2010;235:267–285. doi: 10.1111/j.0105-2896.2010.00893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Topham NJ, Hewitt EW. Natural killer cell cytotoxicity: how do they pull the trigger? Immunology. 2009;128:7–15. doi: 10.1111/j.1365-2567.2009.03123.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seillet C, Huntington ND, Gangatirkar P, Axelsson E, Minnich M, Brady HJ, Busslinger M, Smyth MJ, Belz GT, Carotta S. Differential requirement for Nfil3 during NK cell development. Journal of immunology. 2014;192:2667–2676. doi: 10.4049/jimmunol.1302605. [DOI] [PubMed] [Google Scholar]
- 16.Luevano M, Madrigal A, Saudemont A. Transcription factors involved in the regulation of natural killer cell development and function: an update. Frontiers in immunology. 2012;3:319. doi: 10.3389/fimmu.2012.00319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun JC, Ma A, Lanier LL. Cutting edge: IL-15-independent NK cell response to mouse cytomegalovirus infection. Journal of immunology. 2009;183:2911–2914. doi: 10.4049/jimmunol.0901872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vosshenrich CA, Ranson T, Samson SI, Corcuff E, Colucci F, Rosmaraki EE, Di Santo JP. Roles for common cytokine receptor gamma-chain-dependent cytokines in the generation, differentiation, and maturation of NK cell precursors and peripheral NK cells in vivo. Journal of immunology. 2005;174:1213–1221. doi: 10.4049/jimmunol.174.3.1213. [DOI] [PubMed] [Google Scholar]
- 19.Townsend MJ, Weinmann AS, Matsuda JL, Salomon R, Farnham PJ, Biron CA, Gapin L, Glimcher LH. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity. 2004;20:477–494. doi: 10.1016/s1074-7613(04)00076-7. [DOI] [PubMed] [Google Scholar]
- 20.Kennedy MK, Glaccum M, Brown SN, Butz EA, Viney JL, Embers M, Matsuki N, Charrier K, Sedger L, Willis CR, Brasel K, Morrissey PJ, Stocking K, Schuh JC, Joyce S, Peschon JJ. Reversible defects in natural killer and memory CD8 T cell lineages in interleukin 15-deficient mice. The Journal of experimental medicine. 2000;191:771–780. doi: 10.1084/jem.191.5.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DiSanto JP, Muller W, Guy-Grand D, Fischer A, Rajewsky K. Lymphoid development in mice with a targeted deletion of the interleukin 2 receptor gamma chain. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:377–381. doi: 10.1073/pnas.92.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishikado H, Mukai K, Kawano Y, Minegishi Y, Karasuyama H. NK cell-depleting anti-asialo GM1 antibody exhibits a lethal off-target effect on basophils in vivo. Journal of immunology. 2011;186:5766–5771. doi: 10.4049/jimmunol.1100370. [DOI] [PubMed] [Google Scholar]
- 23.Carlyle JR, Mesci A, Ljutic B, Belanger S, Tai LH, Rousselle E, Troke AD, Proteau MF, Makrigiannis AP. Molecular and genetic basis for strain-dependent NK1.1 alloreactivity of mouse NK cells. Journal of immunology. 2006;176:7511–7524. doi: 10.4049/jimmunol.176.12.7511. [DOI] [PubMed] [Google Scholar]
- 24.Mesci A, Ljutic B, Makrigiannis AP, Carlyle JR. NKR-P1 biology: from prototype to missing self. Immunologic research. 2006;35:13–26. doi: 10.1385/IR:35:1:13. [DOI] [PubMed] [Google Scholar]
- 25.Monnier J, Zabel BA. Anti-asialo GM1 NK cell depleting antibody does not alter the development of bleomycin induced pulmonary fibrosis. PloS one. 2014;9:e99350. doi: 10.1371/journal.pone.0099350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okazaki A, Hiraga N, Imamura M, Hayes CN, Tsuge M, Takahashi S, Aikata H, Abe H, Miki D, Ochi H, Tateno C, Yoshizato K, Ohdan H, Chayama K. Severe necroinflammatory reaction caused by natural killer cell-mediated Fas/Fas ligand interaction and dendritic cells in human hepatocyte chimeric mouse. Hepatology. 2012;56:555–566. doi: 10.1002/hep.25651. [DOI] [PubMed] [Google Scholar]
- 27.Min-Oo G, Lanier LL. Cytomegalovirus generates long-lived antigen-specific NK cells with diminished bystander activation to heterologous infection. The Journal of experimental medicine. 2014;211:2669–2680. doi: 10.1084/jem.20141172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu X, I, Weiss D, Zhang HH, Singh SP, Wynn TA, Wilson MS, Farber JM. Conventional NK cells can produce IL-22 and promote host defense in Klebsiella pneumoniae pneumonia. Journal of immunology. 2014;192:1778–1786. doi: 10.4049/jimmunol.1300039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gan Y, Liu Q, Wu W, Yin JX, Bai XF, Shen R, Wang Y, Chen J, La Cava A, Poursine-Laurent J, Yokoyama W, Shi FD. Ischemic neurons recruit natural killer cells that accelerate brain infarction. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:2704–2709. doi: 10.1073/pnas.1315943111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parham P, Moffett A. Variable NK cell receptors and their MHC class I ligands in immunity, reproduction and human evolution. Nature reviews Immunology. 2013;13:133–144. doi: 10.1038/nri3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar P, Thakar MS, Ouyang W, Malarkannan S. IL-22 from conventional NK cells is epithelial regenerative and inflammation protective during influenza infection. Mucosal immunology. 2013;6:69–82. doi: 10.1038/mi.2012.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peppa D, Gill US, Reynolds G, Easom NJ, Pallett LJ, Schurich A, Micco L, Nebbia G, Singh HD, Adams DH, Kennedy PT, Maini MK. Up-regulation of a death receptor renders antiviral T cells susceptible to NK cell-mediated deletion. The Journal of experimental medicine. 2013;210:99–114. doi: 10.1084/jem.20121172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakayama M, Takeda K, Kawano M, Takai T, Ishii N, Ogasawara K. Natural killer (NK)-dendritic cell interactions generate MHC class II-dressed NK cells that regulate CD4+ T cells. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:18360–18365. doi: 10.1073/pnas.1110584108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun JC, Beilke JN, Bezman NA, Lanier LL. Homeostatic proliferation generates long-lived natural killer cells that respond against viral infection. The Journal of experimental medicine. 2011;208:357–368. doi: 10.1084/jem.20100479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cooper MA, Yokoyama WM. Memory-like responses of natural killer cells. Immunological reviews. 2010;235:297–305. doi: 10.1111/j.0105-2896.2010.00891.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chauveau A, Aucher A, Eissmann P, Vivier E, Davis DM. Membrane nanotubes facilitate long-distance interactions between natural killer cells and target cells. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:5545–5550. doi: 10.1073/pnas.0910074107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kupz A, Scott TA, Belz GT, Andrews DM, Greyer M, Lew AM, Brooks AG, Smyth MJ, Curtiss R, 3rd, Bedoui S, Strugnell RA. Contribution of Thy1+ NK cells to protective IFN-gamma production during Salmonella typhimurium infections. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:2252–2257. doi: 10.1073/pnas.1222047110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gordon SM, Chaix J, Rupp LJ, Wu J, Madera S, Sun JC, Lindsten T, Reiner SL. The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity. 2012;36:55–67. doi: 10.1016/j.immuni.2011.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Helden MJ, Zaiss DM, Sijts AJ. CCR2 defines a distinct population of NK cells and mediates their migration during influenza virus infection in mice. PloS one. 2012;7:e52027. doi: 10.1371/journal.pone.0052027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hayakawa Y, Smyth MJ. CD27 dissects mature NK cells into two subsets with distinct responsiveness and migratory capacity. Journal of immunology. 2006;176:1517–1524. doi: 10.4049/jimmunol.176.3.1517. [DOI] [PubMed] [Google Scholar]
- 41.Takeda K, Hayakawa Y, Smyth MJ, Kayagaki N, Yamaguchi N, Kakuta S, Iwakura Y, Yagita H, Okumura K. Involvement of tumor necrosis factor-related apoptosis-inducing ligand in surveillance of tumor metastasis by liver natural killer cells. Nature medicine. 2001;7:94–100. doi: 10.1038/83416. [DOI] [PubMed] [Google Scholar]
- 42.Peng H, Jiang X, Chen Y, Sojka DK, Wei H, Gao X, Sun R, Yokoyama WM, Tian Z. Liver-resident NK cells confer adaptive immunity in skin-contact inflammation. The Journal of clinical investigation. 2013;123:1444–1456. doi: 10.1172/JCI66381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sojka DK, Plougastel-Douglas B, Yang L, Pak-Wittel MA, Artyomov MN, Ivanova Y, Zhong C, Chase JM, Rothman PB, Yu J, Riley JK, Zhu J, Tian Z, Yokoyama WM. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. eLife. 2014;3:e01659. doi: 10.7554/eLife.01659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bonventre JV, Yang L. Cellular pathophysiology of ischemic acute kidney injury. The Journal of clinical investigation. 2011;121:4210–4221. doi: 10.1172/JCI45161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang ZX, Wang S, Huang X, Min WP, Sun H, Liu W, Garcia B, Jevnikar AM. NK cells induce apoptosis in tubular epithelial cells and contribute to renal ischemia-reperfusion injury. Journal of immunology. 2008;181:7489–7498. doi: 10.4049/jimmunol.181.11.7489. [DOI] [PubMed] [Google Scholar]
- 46.Zhang ZX, Shek K, Wang S, Huang X, Lau A, Yin Z, Sun H, Liu W, Garcia B, Rittling S, Jevnikar AM. Osteopontin expressed in tubular epithelial cells regulates NK cell-mediated kidney ischemia reperfusion injury. Journal of immunology. 2010;185:967–973. doi: 10.4049/jimmunol.0903245. [DOI] [PubMed] [Google Scholar]
- 47.Kim HJ, Lee JS, Kim JD, Cha HJ, Kim A, Lee SK, Lee SC, Kwon BS, Mittler RS, Cho HR, Kwon B. Reverse signaling through the costimulatory ligand CD137L in epithelial cells is essential for natural killer cell-mediated acute tissue inflammation. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E13–22. doi: 10.1073/pnas.1112256109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kashiwada M, Levy DM, McKeag L, Murray K, Schroder AJ, Canfield SM, Traver G, Rothman PB. IL-4-induced transcription factor NFIL3/E4BP4 controls IgE class switching. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:821–826. doi: 10.1073/pnas.0909235107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grenz A, Hong JH, Badulak A, Ridyard D, Luebbert T, Kim JH, Eltzschig HK. Use of a hanging-weight system for isolated renal artery occlusion. Journal of visualized experiments: JoVE. 2011 doi: 10.3791/2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lorenz JN, Gruenstein E. A simple, nonradioactive method for evaluating single-nephron filtration rate using FITC-inulin. The American journal of physiology. 1999;276:F172–177. doi: 10.1152/ajprenal.1999.276.1.F172. [DOI] [PubMed] [Google Scholar]
- 51.Arase H, Saito T, Phillips JH, Lanier LL. Cutting edge: the mouse NK cell-associated antigen recognized by DX5 monoclonal antibody is CD49b (alpha 2 integrin, very late antigen-2) Journal of immunology. 2001;167:1141–1144. doi: 10.4049/jimmunol.167.3.1141. [DOI] [PubMed] [Google Scholar]
- 52.Kasai M, Yoneda T, Habu S, Maruyama Y, Okumura K, Tokunaga T. In vivo effect of anti-asialo GM1 antibody on natural killer activity. Nature. 1981;291:334–335. doi: 10.1038/291334a0. [DOI] [PubMed] [Google Scholar]
- 53.Kamizono S, Duncan GS, Seidel MG, Morimoto A, Hamada K, Grosveld G, Akashi K, Lind EF, Haight JP, Ohashi PS, Look AT, Mak TW. Nfil3/E4bp4 is required for the development and maturation of NK cells in vivo. The Journal of experimental medicine. 2009;206:2977–2986. doi: 10.1084/jem.20092176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gascoyne DM, Long E, Veiga-Fernandes H, de Boer J, Williams O, Seddon B, Coles M, Kioussis D, Brady HJ. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nature immunology. 2009;10:1118–1124. doi: 10.1038/ni.1787. [DOI] [PubMed] [Google Scholar]
- 55.Walzer T, Blery M, Chaix J, Fuseri N, Chasson L, Robbins SH, Jaeger S, Andre P, Gauthier L, Daniel L, Chemin K, Morel Y, Dalod M, Imbert J, Pierres M, Moretta A, Romagne F, Vivier E. Identification, activation, and selective in vivo ablation of mouse NK cells via NKp46. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:3384–3389. doi: 10.1073/pnas.0609692104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li L, Huang L, Sung SS, Lobo PI, Brown MG, Gregg RK, Engelhard VH, Okusa MD. NKT cell activation mediates neutrophil IFN-gamma production and renal ischemia-reperfusion injury. Journal of immunology. 2007;178:5899–5911. doi: 10.4049/jimmunol.178.9.5899. [DOI] [PubMed] [Google Scholar]
- 57.Kim HJ, Lee JS, Kim A, Koo S, Cha HJ, Han JA, Do Y, Kim KM, Kwon BS, Mittler RS, Cho HR, Kwon B. TLR2 signaling in tubular epithelial cells regulates NK cell recruitment in kidney ischemia-reperfusion injury. Journal of immunology. 2013;191:2657–2664. doi: 10.4049/jimmunol.1300358. [DOI] [PubMed] [Google Scholar]
- 58.Bedel R, Matsuda JL, Brigl M, White J, Kappler J, Marrack P, Gapin L. Lower TCR repertoire diversity in Traj18-deficient mice. Nature immunology. 2012;13:705–706. doi: 10.1038/ni.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sonoda KH, Exley M, Snapper S, Balk SP, Stein-Streilein J. CD1-reactive natural killer T cells are required for development of systemic tolerance through an immune-privileged site. The Journal of experimental medicine. 1999;190:1215–1226. doi: 10.1084/jem.190.9.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Exley MA, Bigley NJ, Cheng O, Shaulov A, Tahir SM, Carter QL, Garcia J, Wang C, Patten K, Stills HF, Alt FW, Snapper SB, Balk SP. Innate immune response to encephalomyocarditis virus infection mediated by CD1d. Immunology. 2003;110:519–526. doi: 10.1111/j.1365-2567.2003.01779.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL, Yokoyama WM, Ugolini S. Innate or adaptive immunity? The example of natural killer cells. Science. 2011;331:44–49. doi: 10.1126/science.1198687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen GE, Wu H, Ma J, Chadban SJ, Sharland A. Toll-like receptor 4 engagement contributes to expression of NKG2D ligands by renal tubular epithelial cells. Nephrology, dialysis, transplantation: official publication of the European Dialysis and Transplant Association - European Renal Association. 2011;26:3873–3881. doi: 10.1093/ndt/gfr234. [DOI] [PubMed] [Google Scholar]
- 63.Wei L, Lu J, Feng L, Long D, Shan J, Li S, Li Y. HIF-1alpha accumulation upregulates MICA and MICB expression on human cardiomyocytes and enhances NK cell cytotoxicity during hypoxia-reoxygenation. Life sciences. 2010;87:111–119. doi: 10.1016/j.lfs.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 64.Maurus CF, Schmidt D, Schneider MK, Turina MI, Seebach JD, Zund G. Hypoxia and reoxygenation do not upregulate adhesion molecules and natural killer cell adhesion on human endothelial cells in vitro. European journal of cardio-thoracic surgery: official journal of the European Association for Cardio-thoracic Surgery. 2003;23:976–983. doi: 10.1016/s1010-7940(03)00146-5. discussion 983. [DOI] [PubMed] [Google Scholar]
- 65.Cortez VS, Fuchs A, Cella M, Gilfillan S, Colonna M. Cutting edge: Salivary gland NK cells develop independently of Nfil3 in steady-state. Journal of immunology. 2014;192:4487–4491. doi: 10.4049/jimmunol.1303469. [DOI] [PubMed] [Google Scholar]
- 66.Tessmer MS, Reilly EC, Brossay L. Salivary gland NK cells are phenotypically and functionally unique. PLoS pathogens. 2011;7:e1001254. doi: 10.1371/journal.ppat.1001254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Beilke JN, Meagher CT, Hosiawa K, Champsaur M, Bluestone JA, Lanier LL. NK cells are not required for spontaneous autoimmune diabetes in NOD mice. PloS one. 2012;7:e36011. doi: 10.1371/journal.pone.0036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kasai M, Iwamori M, Nagai Y, Okumura K, Tada T. A glycolipid on the surface of mouse natural killer cells. European journal of immunology. 1980;10:175–180. doi: 10.1002/eji.1830100304. [DOI] [PubMed] [Google Scholar]
- 69.Young WW, Jr, Hakomori SI, Durdik JM, Henney CS. Identification of ganglio-N-tetraosylceramide as a new cell surface marker for murine natural killer (NK) cells. Journal of immunology. 1980;124:199–201. [PubMed] [Google Scholar]
- 70.Verma S, Loewendorf A, Wang Q, McDonald B, Redwood A, Benedict CA. Inhibition of the TRAIL death receptor by CMV reveals its importance in NK cell-mediated antiviral defense. PLoS pathogens. 2014;10:e1004268. doi: 10.1371/journal.ppat.1004268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Selathurai A, Deswaerte V, Kanellakis P, Tipping P, Toh BH, Bobik A, Kyaw T. Natural killer (NK) cells augment atherosclerosis by cytotoxic-dependent mechanisms. Cardiovascular research. 2014;102:128–137. doi: 10.1093/cvr/cvu016. [DOI] [PubMed] [Google Scholar]
- 72.Hall LJ, Murphy CT, Hurley G, Quinlan A, Shanahan F, Nally K, Melgar S. Natural killer cells protect against mucosal and systemic infection with the enteric pathogen Citrobacter rodentium. Infection and immunity. 2013;81:460–469. doi: 10.1128/IAI.00953-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sung JM, Lee CK, Wu-Hsieh BA. Intrahepatic infiltrating NK and CD8 T cells cause liver cell death in different phases of dengue virus infection. PloS one. 2012;7:e46292. doi: 10.1371/journal.pone.0046292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Etogo AO, Nunez J, Lin CY, Toliver-Kinsky TE, Sherwood ER. NK but not CD1-restricted NKT cells facilitate systemic inflammation during polymicrobial intra-abdominal sepsis. Journal of immunology. 2008;180:6334–6345. doi: 10.4049/jimmunol.180.9.6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alba A, Planas R, Clemente X, Carrillo J, Ampudia R, Puertas MC, Pastor X, Tolosa E, Pujol-Borrell R, Verdaguer J, Vives-Pi M. Natural killer cells are required for accelerated type 1 diabetes driven by interferon-beta. Clinical and experimental immunology. 2008;151:467–475. doi: 10.1111/j.1365-2249.2007.03580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Moffett A, Colucci F. Uterine NK cells: active regulators at the maternal-fetal interface. The Journal of clinical investigation. 2014;124:1872–1879. doi: 10.1172/JCI68107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tessier DR, Yockell-Lelievre J, Gruslin A. Uterine Spiral Artery Remodeling: The Role of Uterine Natural Killer Cells and Extravillous Trophoblasts in Normal and High-Risk Human Pregnancies. American journal of reproductive immunology. 2014 doi: 10.1111/aji.12345. [DOI] [PubMed] [Google Scholar]
- 78.Chen YH, Chiu NM, Mandal M, Wang N, Wang CR. Impaired NK11 T cell development and early IL-4 production in CD1-deficient mice. Immunity. 1997;6:459–467. doi: 10.1016/s1074-7613(00)80289-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.