SUMMARY
A decline in mitochondrial respiration represents the root cause of a large number of inborn errors of metabolism. It is also associated with common age-associated diseases and the aging process. To gain insight into the systemic, biochemical consequences of respiratory chain dysfunction, we performed a case-control, prospective metabolic profiling study in a genetically homogenous cohort of patients with Leigh syndrome French Canadian variant, a mitochondrial respiratory chain disease due to loss-of-function mutations in LRPPRC. We discovered 45 plasma and urinary analytes discriminating patients from controls, including classic markers of mitochondrial metabolic dysfunction (lactate and acylcarnitines), as well as unexpected markers of cardiometabolic risk (insulin and adiponectin), amino acid catabolism linked to NADH status (α-hydroxybutyrate), and NAD+ biosynthesis (kynurenine and 3-hydroxyanthranilic acid). Our study identifies systemic, metabolic pathway derangements that can lie downstream of primary mitochondrial lesions, with implications for understanding how the organelle contributes to rare and common diseases.
Graphical abstract
INTRODUCTION
Mitochondrial dysfunction is increasingly being recognized as a hallmark of rare and common age-associated diseases. Among inherited metabolic diseases, those affecting mitochondria are the most prevalent worldwide (1:5,000), frequently manifest in early childhood, and are associated with high morbidity and mortality (DiMauro, 2004; Torraco et al., 2009; Vafai and Mootha, 2013). These disorders may be caused by genetic lesions in either nuclear or mtDNA, which may disrupt numerous metabolic pathways housed in the mitochondria but most prominently the oxidative phosphorylation (OXPHOS) system (Debray et al., 2008; Munnich and Rustin, 2001). These disorders can impact virtually any organ system as a whole or in a tissue-specific manner. A subtle decline in OXPHOS is associated with skeletal muscle atrophy, type 2 diabetes, neurodegeneration, and the aging process itself (Vafai and Mootha, 2012), though the molecular basis and biochemical consequences are currently not known. There is growing interest in rare mitochondrial disorders, and studying them may shed insight into the role of the organelle to more common, age-associated disease.
Recently, tremendous progress has been achieved in the genetic characterization of rare mitochondrial disorders, yet their management remains challenging because of the difficulty to track their progression and the absence of proven therapies (DiMauro and Mancuso, 2007; Orsucci et al., 2009; Pfeffer et al., 2013; Schapira, 2012). This is due in part to our lack of understanding of the metabolic consequences of OXPHOS defects beyond ATP and the commonly reported high blood lactate. In this regard, the recent emergence of metabolomics technologies offers a means to systematically measure thousands of low-molecular-weight compounds in order to provide a global view of alterations in metabolic pathways induced by a given perturbation, whether resulting from a gene mutation or disease onset. These methods have been applied to numerous diseases such as diabetes and cardiovascular disease (for reviews, see Roberts and Gerszten, 2013 and Shah et al., 2012). However, only few studies have extended this method to mitochondrial disorders (Clarke et al., 2013; Leoni et al., 2012; Shaham et al., 2010), which pose significant challenges, owing to their genotypic and phenotypic heterogeneity combined with small patient populations.
To overcome these challenges, we performed metabolic profiling in a small but genetically defined cohort of patients with Leigh syndrome French Canadian variant (LSFC; MIM no. 220111). First described in 1993 (Merante et al., 1993; Morin et al., 1993), LSFC is a recessive disorder prevalent in the north-eastern region of Quebec (~1/2,000 births; carrier rate 1/23) (Merante et al., 1993; Morin et al., 1993). LSFC patients exhibit many hallmarks of mitochondrial disorders, including lactic acidosis and Leigh syndrome, a necrotizing encephalopathy (Debray et al., 2011; Finsterer, 2008; Morin et al., 1993). LSFC was the first human disorder whose underlying genetic locus was mapped via linkage disequilibrium (also known as genome-wide association study; Lee et al., 2001) and the underlying gene discovered via “integrative genomics” (Mootha et al., 2003). All known cases are due to missense (A354V) or deletion (C1277STOP) mutations in the nuclear leucine-rich pentatricopeptide-repeat-motif-containing (LRPPRC) gene, which encodes a mitochondrial localized RNA-binding protein. These mutations lead to a tissue-specific impact on mitochondrial OXPHOS complexes (Gohil et al., 2010; Mourier et al., 2014; Ruzzenente et al., 2012; Sasarman et al., 2010, 2015; Xu et al., 2004). More recently, LSFC fibroblasts were found to display multiple alterations in mitochondrial function and increased susceptibility to nutrient-induced cytotoxicity, particularly saturated fat (Burelle et al., 2015).
We recruited all known LSFC patients and, using a prospective, case-control study design, we performed metabolic profiling in plasma and urine using two different targeted mass spectrometric (MS)-based methods. We report a metabolic signature consisting of 45 compounds or ratios that distinguish LSFC patients from controls. The metabolites provide insight into the metabolic pathway derangements that ensue from a root mitochondrial lesion and provide a valuable resource for understanding the biochemical consequences and basis of mitochondrial dysfunction.
RESULTS
We performed metabolic profiling of all known living patients with LSFC. Eight of the nine patients are homozygotes for the A354V founder mutation, and one patient is compound heterozygous for this mutation and a premature stop (Mootha et al., 2003). Our study design is described in Experimental Procedures and depicted in Figure S1. Patients were prospectively matched with controls for gender, age, BMI, physical activity level, and nutritional status. Metabolic measurements encompassed standard clinical biochemical parameters and multiple metabolite classes in plasma and urine. These analyses generated two independent data sets, hereafter referred to as “platforms,” which were processed through a statistical workflow for quality control filtering, missing data imputation, principal component analysis (PCA), and permutation testing. Significance threshold was set at a false discovery rate of 10%, corresponding to a p value < 0.03 for Platform 1 and <0.023 for Platform 2. Characteristics of LSFC patients and controls are summarized in Table 1 and described in more detail in Table S1. As expected given the intended matching, there were no statistical differences between patients and controls for age, BMI, or physical activity (paired t test; α = 0.05).
Table 1.
Clinical Characteristics of LSFC Patients and Matched Control Individuals
| Patients | Controls | |
|---|---|---|
| Gender (male:female) | 4:5 | 4:5 |
| Age, years | 22 (6–30) | 24 (5–32) |
| BMI in adults, kg/m2 | 23.5 (22.3–26.7) | 24.0 (21.6–29.2) |
| BMI for age in children, percentile | 72 (1–96) | 70 (2–92) |
| Serum glucose, mmol/l | 4.9 (4.2–5.5) | 4.7 (4.4–5.2) |
| Serum triglycerides, mmol/l | 1.32 (0.67–4.18); 3/9 | 1.12 (0.41–1.55); 1/9 |
| Serum total cholesterol, mmol/l | 4.74 (3.53–7.19); 3/9 | 4.39 (3.47–5.48); 1/9 |
| Serum LDL cholesterol, mmol/l | 2.76 (2.08–4.52); 3/8a | 2.45 (1.59–3.45); 2/9 |
| Serum HDL cholesterol, mmol/l | 1.01 (0.76–1.49); 2/9b | 1.46 (1.13–2.19) |
| Serum total cholesterol/HDL ratio | 4.23 (3.13–7.33)b | 3.11 (2.05–3.96) |
| Serum AST, U/l | 23 (14–30) | 24 (12–91); 1/9 |
| Serum ALT, U/l | 21 (14–51); 1/9 | 18 (14–58); 2/9 |
| Serum GGT, U/l | 11 (8–86); 1/9 | 13 (6–37) |
| Plasma insulin, mU/l | 10.3 (3.8–26.2)b | 4.79 (3.2–11.9) |
| Plasma adiponectin, ng/ml | 4,200 (1,304–10,410)b | 10,229 (3,580–37,804) |
Data presented as median (min-max) and, where applicable, proportion of abnormal results when measured as part of the routine biochemical assay. Subjects’ characteristics are described in more details in Table S1.
Data for LDL cholesterol were missing for one patient given that triglyceride level was too high.
p < 0.03 patients versus controls following permutation test (Platform 1).
Clinical Biochemical Assays in LSFC Patients versus Controls
Standard biochemical and hormonal assays, measured as part of Platform 1, showed that LSFC patients had significantly increased total cholesterol/HDL ratio and insulin levels, as well as reduced levels of HDL cholesterol and adiponectin. A trend for a significant increase in triglyceride levels was also observed (p = 0.039; Tables 1 and S2), whereas serum glucose was similar to controls (p = 0.320). Collectively, these changes would suggest an increased cardiometabolic risk (Leiter et al., 2011) in LSFC patients.
Global Metabolic Profiles in LSFC versus Controls
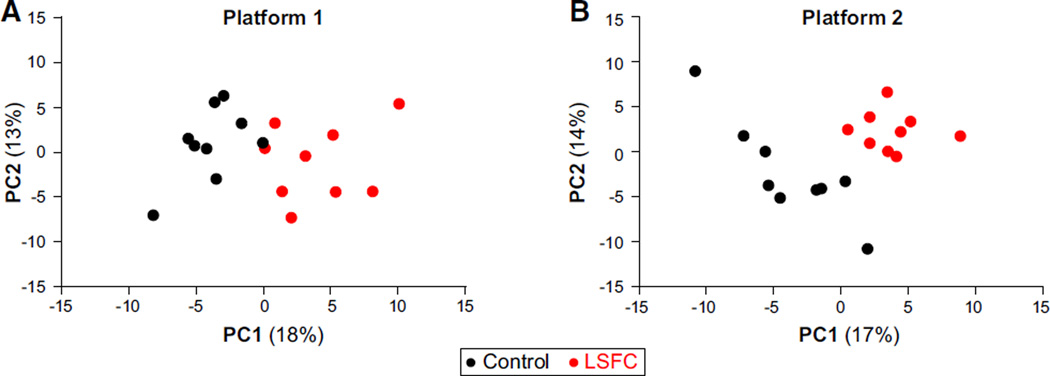
PCA was first applied to the post-quality control, imputed data set from each platform, namely 137 analytes including standard biochemistry and hormones for Platform 1 and 156 for Platform 2, with an overlap of 37 metabolites between the two platforms. As shown in Figure 1, the first PC from each platform fully discriminated patients from controls, indicating a distinct metabolic profile in LSFC patients. In each data set, the top two PCs accounted for 31% of the total variation (see Table S3 for loading scores). Metabolites contributing the most to the separation included lactate, β-hydroxybutyrate, acylcarnitines of various chain lengths, and α-hydroxybutyrate; to the best of our knowledge, the latter has not been previously reported in plasma from patients with an inherited mitochondrial disorder.
Figure 1. Principal-Component Analysis Discriminates LSFC Patients and Controls.
For each panel, the post-quality control, imputed data set was used to perform the analysis.
(A) Platform 1; PC1 and PC2 account for 18% and 13% of variation, respectively.
(B) Platform 2; PC1 and PC2 account for 17% and 14% of variation, respectively.
Loading scores are reported in Table S3; see also Figure S1 for an overview of analytes submitted to the PCA and Table S2 for raw data sets.
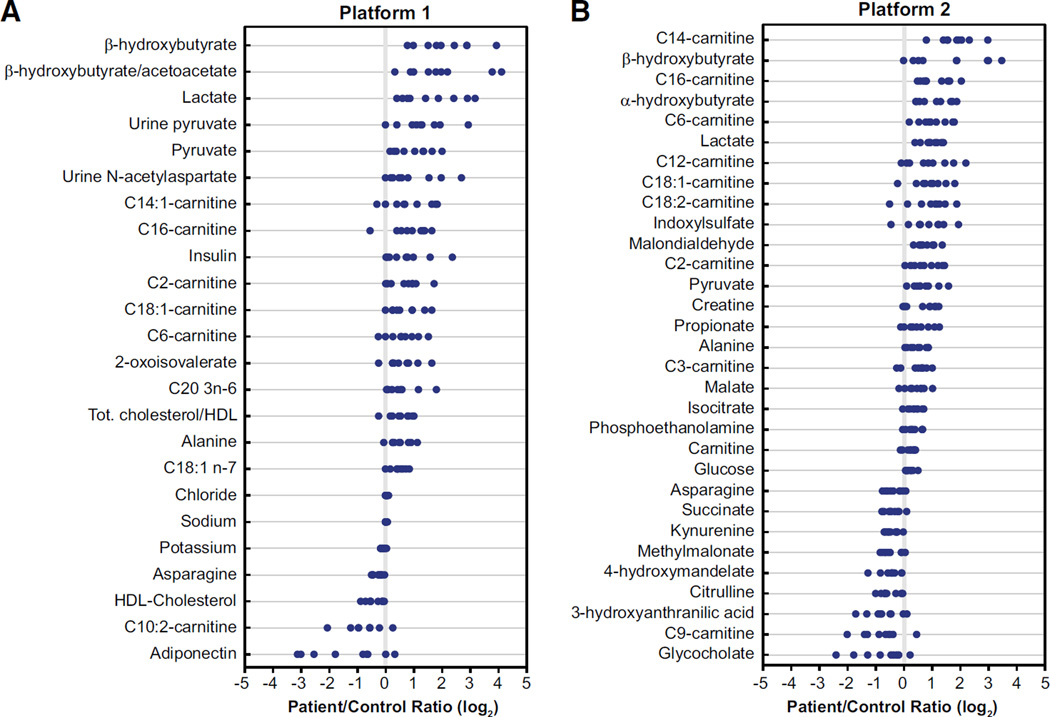
The two data sets were then independently subjected to permutation analyses, and statistical significance was reached between patients and controls for 24 analytes/ratios out of 137 in Platform 1 (p < 0.03) and for 31 analytes out of 156 in Platform 2 (p < 0.023), with an overlap of nine significant metabolites between both platforms (Table S4). All analytes are depicted as volcano plots in Figure S2 and listed in Table S2. Combining the results of both platforms, 46 single variables (representing 45 unique compounds/ratios) out of the 256 single variables submitted to the permutation test were statistically different between LSFC patients and controls, as depicted in Figure 2. Of note, the compound heterozygote was similar to the other LSFC patients (A354V homozygotes).
Figure 2. Individual Analytes with Statistical Significance in LSFC Patients versus Controls.
(A) Platform 1; (B) Platform 2. Each dot represents a log2-transformed patient/matched control ratio. Metabolites are ordered by mean log2-transformed patient/control ratio. See also Figure S2 for data presented as volcano plots, Figure S1 for a summary of measured analytes, and Table S2 for log2-transformed ratios and p values of all analytes.
Concurring with the multivariate PCA, the most striking differences were observed for (1) plasma β-hydroxybutyrate and lactate, as well as the β-hydroxybutyrate/acetoacetate ratio; (2) acylcarnitines, and more specifically myristoylcarnitine and palmitoylcarnitine (C14 and C16, respectively); and (3) α-hydroxybutyrate (Figure 2). Altogether, the changes in metabolite levels can be organized into a small handful of metabolic pathways. First, these include elevated NADH/NAD+ ratio (increased β-hydroxybutyrate, lactate, and β-hydroxybutyrate/acetoacetate ratio) and closely related alterations in citric acid cycle (CAC) reactions, including those catalyzing entry or removal of carbons from this cycle (increased isocitrate, malate, and propionate, albeit decreased succinate and methylmalonate). Then, elevated C2, C6, C12, C14, C14:1, C16, C18:1, and C18:2 acylcarnitine but decreased C9 and C10:2 species reflect disrupted fatty acid oxidation, whereas impaired biosynthesis is suggested by increased vaccenic acid (C18:1n-7), dihomo-γ-linolenic acid (C20:3n-6), and phosphoethaloamine. Of particular interest, there were also unexpected changes in plasma metabolites relevant to amino acid catabolism, further substantiating the complexity of metabolic perturbations linked to NAD+ metabolism. These include (1) an increase in α-hydroxybutyrate, a metabolite arising from the reduction of α-ketobutyrate formed from methionine and/or threonine, and (2) various intermediates of the tryptophan degradation pathways involved in NAD+ synthesis (kynurenine and 3-hydroxyanthranilic acid) or product of gut bacterial metabolism (indoxyl sulfate; Zhu et al., 2011). Finally, LSFC patients displayed increased plasma levels of creatine and decreased levels of the bile acid glycocholate, observations that were also made in patients affected by various OXPHOS defects (Pajares et al., 2013; Shaham et al., 2010) and with mutations in mitochondrial pantothenate kinase 2 (PANK2) (the rate-limiting enzyme in mitochondrial coenzyme A biosynthesis; Leoni et al., 2012), respectively.
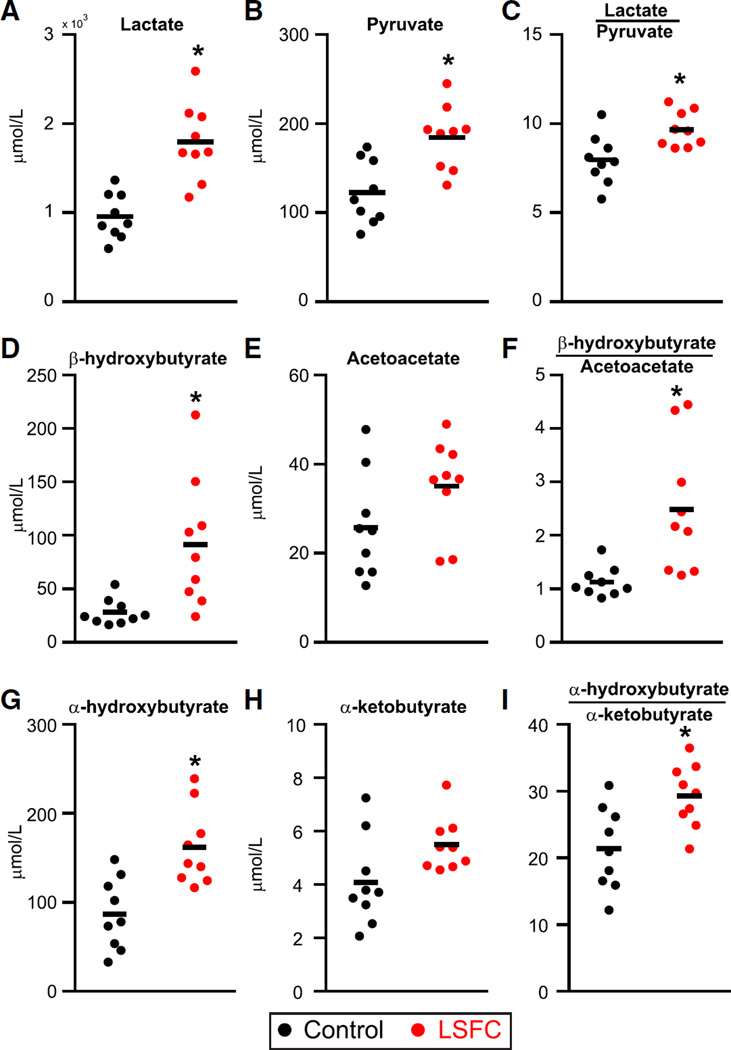
Quantitative Analyses of Metabolic Indices of NADH/NAD+
Because both PCA and permutation tests revealed that a perturbed redox status was a prominent feature of the distinctive metabolic signature in LSFC patients, an isotope dilution GC-MS method was developed in order to quantify with precision lactate and β-hydroxybutyrate as well as their corresponding oxidized counterparts, namely pyruvate and acetoacetate, respectively. In addition, we included α-hydroxybutyrate, a finding issued from the metabolic profiling on Platform 2 but that was not assessed on Platform 1, because we suspected that its elevation was also linked to the increased NADH/NAD+ ratio. Hence, it was measured along with its oxidized counterpart, namely α-ketobutyrate. The results of these analyses are consistent with the initial metabolic profiling data. In fact, the plasma concentration of lactate, pyruvate, β-hydroxybutyrate, and α-hydroxybutyrate, as well as the lactate/pyruvate and β-hydroxybutyrate/acetoacetate ratios, were all significantly higher in LSFC patients (Figure 3). Importantly, this analysis revealed that the α-hydroxybutyrate/α-ketobutyrate ratio was also significantly increased. Collectively, these quantitative measurements further support the findings of an elevated plasma level of α-hydroxybutyrate in LSFC patients, revealing an additional metabolic perturbation linked to the altered redox status in these patients.
Figure 3. Quantitative Profiling of Metabolites Reflective of NADH/NAD+ Redox Status.
Metabolites are shown in scatter dot plots with line indicating the mean. *p < 0.05 patients versus controls. See also Table S2 for raw data.
DISCUSSION
In this prospective study, we applied a comprehensive targeted metabolomics approach to a genetically homogenous cohort of patients with an inherited mitochondrial respiratory chain disorder. Despite a small sample size, the current case-control study design enabled the identification of a distinct metabolic signature, consisting of 45 compounds spanning indices of cardiometabolic risk, disrupted NADH/NAD+, as well as lipid and amino acid metabolism.
Markers of Increased Global Cardiometabolic Risk
Much to our surprise, a disturbed cholesterol profile along with alterations in insulin and adiponectin levels were found, suggesting an increased global cardiometabolic risk in LSFC patients despite their relatively young age (Leiter et al., 2011). Although diabetes has been strongly associated with mutations in some mtDNA-encoded genes (Maassen et al., 2004), dyslipidemia has only been previously reported in few other cohorts of patients with respiratory chain disorders (Clarke et al., 2013; Finsterer et al., 2001; Kaufmann et al., 2009). In LSFC patients, increased cardiometabolic risk is further supported by the higher plasma levels of the fatty acids vaccenic and dihomo-γ-linolenic acids (C18:1n-7 and C20:3n-6, respectively), which have both been recognized as markers of insulin resistance (Lovejoy et al., 2001; Zulyniak et al., 2012). Interestingly, phosphoethanolamine, of which plasma levels were also increased, may also be associated with these alterations. This metabolite is an intermediate in the biosynthesis of phosphatidylethanolamine through the CDP-ethanolamine pathway (“Kennedy pathway”), and transgenic mice heterozygous for Pcyt2—a gene involved in this pathway that leads to increased hepatic phosphoethanolamine synthesis—display progressive hypertriglyceridemia, insulin resistance, as well as liver steatosis (Fullerton et al., 2009), which is also a characteristic found in LSFC patients (Morin et al., 1993).
Disturbances in Pathways Related to Energy Metabolism and the NADH/NAD+ Ratio
As expected, lactate, pyruvate, and alanine were elevated in LSFC patients as compared with controls, as it has often been reported in patients with mitochondrial diseases, although these metabolites might not be specific to mitochondrial dysfunction (Debray et al., 2008; Haas et al., 2008; Smeitink et al., 2006). Our quantitative analyses further substantiated elevated plasma lactate/pyruvate and β-hydroxybutyrate/acetoacetate ratios, hence reflecting an accumulation of reducing equivalents in the cytosol and mitochondria, respectively. Elevations in blood CAC intermediates and saturated even chain acylcarnitines have previously been reported in patients with mitochondrial disorders (Haas et al., 2008; Smeitink et al., 2006). The elevation in acylcarnitines of all chain lengths in LSFC patients reflects a global disturbance of mitochondrial fatty acid β-oxidation, suggesting an impaired lipid-handling capacity (Adams et al., 2009; Houten and Wanders, 2010). This concurs with our recent finding of an increased susceptibility to palmitate-induced cytotoxicity in LSFC fibroblasts and is consistent with a tissue-specific defect in complex IV alone (for e.g., fibroblasts; Burelle et al., 2015) or combined with complex I (muscle and liver; Sasarman et al., 2010, 2015). The situation is likely to differ for complex I defects alone, which still enable entry of reducing equivalents arising from fatty acid oxidation at complex II and electron flux through complexes II–V and for which a positive effect of high-fat diets has been observed (Schiff et al., 2011).
Our systematic metabolomics approach also revealed an unexpected decrease in decadienoylcarnitine (C10:2) and nonanoylcarnitine (C9). The decrease in C10:2-carnitine, a partial oxidation product of the essential fatty acid linoleate (C18:2n-6), would suggest an increase in 2,4-dienoyl-CoA reductase activity that may be driven by an elevated NADPH/NADP ratio resulting from mitochondrial redox perturbation (Miinalainen et al., 2009). As for C9-carnitine, the metabolic origin of this compound is unclear, although it was suggested to be formed from the combined peroxisomal and mitochondrial catabolism of phytanic and pristanic acids, two branched chain fatty acids (Verhoeven et al., 1998). A decrease in C9-carnitine has also been reported in fibroblasts from patients with mitochondrial trifunctional protein deficiency (Roe et al., 2006), a fatty acid oxidation disorder. The latter findings emphasize the diversity of perturbations in fatty acid oxidation, which can result from mitochondrial dysfunction.
Perturbations in Amino Acid Metabolic Pathways
Beyond markers of mitochondrial energy metabolic pathways, other unexpected changes reflecting perturbations in the metabolism of several amino acids were observed in LSFC patients. These include higher plasma levels of α-hydroxybutyrate. However, its oxidized metabolite counterpart, namely α-ketobutyrate, which arises from the catabolism of the essential amino acids threonine and/or methionine in the cytosol (Landaas, 1975), did not differ; consequently, the α-hydroxybutyrate/α-ketobutyrate ratio was also significantly elevated. Whereas these findings likely represent another consequence of an elevated NADH/NAD+ ratio, the higher level of α-hydroxybutyrate also highlights major critical perturbations in its metabolism. The latter is depicted in Figure 4 along with changes in closely related metabolites. Similarly to pyruvate, α-ketobutyrate is at a metabolic crossroad between the cytosol and the mitochondria. It may be transported to the mitochondria and subjected to oxidative decarboxylation to form propionyl-CoA; these steps are likely catalyzed by the pyruvate transporter and the branched-chain α-keto acid dehydrogenase complex (Jakobs et al., 1977; Paxton et al., 1986; Smith and Strang, 1958; Steele et al., 1984). Alternatively, α-ketobutyrate may be reduced to α-hydroxybutyrate by lactate dehydrogenase or β-hydroxybutyrate dehydrogenase (Rosalki and Wilkinson, 1960).
Figure 4. Pathway Relationship of Major Metabolic Alterations in LSFC Patients.
This scheme depicts pathways related to reported perturbations in cytosolic and mitochondrial NADH accumulation, disrupted citric acid cycle (CAC), fatty acid β-oxidation, and amino acid metabolism, with a specific emphasis on those linked to α-hydroxybutyrate formation. Metabolites whose levels increased are indicated in red and with an upward arrow; those whose levels decreased are indicated in green and with a downward arrow. Metabolites whose levels did not change significantly are indicated in black with a sideways arrow. Those metabolites that were not measured are indicated in black (with no arrow). See also Figure S1 for an overview of analytes measured in the study and Table S2 for raw data and p values.
A few studies have reported increased α-hydroxybutyrate in patients with inherited metabolic diseases, such as lactic acidosis (Pettersen et al., 1973) or maple syrup urine disease (Jakobs et al., 1977; Smith and Strang, 1958). More recently, this metabolite has been reported to represent a strong and early marker of insulin resistance and glucose intolerance (Adams, 2011; Ferrannini et al., 2013; Fiehn et al., 2010; Gall et al., 2010; Xu et al., 2013) and was suggested as a potential biomarker for the diagnosis of metabolic syndrome (Demine et al., 2014; Lin et al., 2014). Factors proposed to play a role in α-hydroxybutyrate accumulation in plasma include perturbations in branched-chain amino acid catabolism, hepatic glutathione synthesis, or redox status (Gall et al., 2010; Landaas, 1975). Whereas the exact mechanisms underlying its elevation in LSFC patients remain to be elucidated, the present findings do not support a central role for branched-chain amino acids as proposed for insulin-resistant subjects (Adams, 2011; Newgard, 2012), because their levels were similar between LSFC patients and controls. Furthermore, whereas impaired glutathione metabolism has been reported in children with classical Leigh syndrome (Pastore et al., 2013), levels of reduced and oxidized glutathione in our LSFC cohort were similar to controls, suggesting that oxidative stress was not increased. This observation is consistent with our recent finding in LSFC fibroblasts (Burelle et al., 2015) but contrasts with those reported for other OXPHOS defects, namely complex I and III (Diaz et al., 2012; Distelmaier et al., 2009; Koene et al., 2011). It is noteworthy that the plasma level of malondialdehyde was significantly increased in LSFC patients, albeit this is not a specific oxidative stress marker, because it is also generated by cyclo-oxygenases in thromboxane metabolism (Kadiiska et al., 2005). An accelerated methionine metabolism through the S-adenosylmethionine cycle in LSFC patients is, however, suggested by the increased plasma levels of creatine, which may reflect a low intracellular energy status (Shaham et al., 2010). This may also suggest a perturbation in methylation reactions, which could impact on signaling pathways regulating cellular survival or mitochondrial biogenesis such as Sirt1 and/or PGC-1α (Guéant Rodriguez et al., 2013).
Our results also demonstrate alterations in the metabolism of tryptophan (reduced kynurenine and 3-hydroxyanthranilic acid) and aspartate (increased N-acetylaspartate), which have been linked to neurodegeneration (Sas et al., 2007; Tan et al., 2012), an important clinical feature of LSFC. Among intermediates of the tryptophan catabolic pathways that lead to NAD+ synthesis, kynurenine and 3-hydroxyanthranilic acid were significantly modified (Figure 4), although tryptophan itself or other metabolites of this pathway were unchanged. As for N-acetylaspartate, it is found exclusively in neurons and arises from the condensation of aspartate and acetyl-CoA in the mitochondria. Its increased level in urine has been reported in patients with aspartoacylase deficiency (Canavan disease), a leukodystrophy characterized by neurodegeneration and early death (Al-Dirbashi et al., 2007). In the brain, its elevation has been reported as a marker of compromised neuronal integrity, but reduction suggests neurodegenerative disorders (Knapman et al., 2012; Moreno et al., 2001).
Conclusions
This study highlights the power of metabolomics when applied in a prospective case-control design to a genetically homogenous form of Leigh syndrome presenting with many of the hallmark features of mitochondrial respiratory chain disease. Results uncovered a LSFC metabolic signature including both classic and unexpected metabolites, which collectively highlight the complexity of perturbations that may result from mitochondrial dysfunction but also have implications for understanding potential contributions of mitochondrial dysfunction to common disease. Altogether, this signature concurs with the reported tissue-specific impact of mutated LRPPRC on mitochondrial OXPHOS complexes (Mourier et al., 2014; Sasarman et al., 2010, 2015) and function in LSFC fibroblasts (Burelle et al., 2015), while representing also possible targets for intervention. Future studies will be needed in order to further delineate the underlying metabolic mechanisms and further characterize whether the identified metabolites may be relevant as biological markers for the assessment of severity, prediction of lactic acidosis crises, monitoring of disease progression, or response to interventions in patients with LSFC and, possibly, other inherited mitochondrial disorders.
EXPERIMENTAL PROCEDURES
Subjects, Sample Collection, and Genotyping
LSFC patients were included based on medical history and genetic testing for the A354V and C1277STOP mutations in LRPPRC. Matching criteria for controls included gender, age (±2 years for children <18 years; ±5 years for adults), BMI (±10 percentiles of BMI for age for children; ±3 kg/m2 for adults), and physical activity level (see the Supplemental Experimental Procedures for details). Carrying A354V or C1277STOP mutation was an exclusion criterion for controls. The protocol was approved by the Human Ethics and Research Committee of the Centre de Santé et de Services Sociaux de Chicoutimi. Written informed consent was obtained for all study participants or their legal guardians, and assent was obtained when applicable. Metabolic profiling was performed on venous blood and urine samples collected after an overnight fast of minimum 12 hr, during which water was allowed. Genotyping was performed on saliva samples. Samples were processed as described in the Supplemental Experimental Procedures.
Metabolite Profiling and Quantitative Analysis
The overall workflow for metabolic profiling is depicted in Figure S1 and described in detail in the Supplemental Experimental Procedures. This was performed using a combination of standard biochemical and hormonal assays as well as established targeted GC-MS and LC-MS methods, altogether encompassing 407 analytes, which were treated as two distinct platforms. Platform 1 included clinical laboratory assays, hormonal assays, and MS-based profiling of amino acids, fatty acids, organic acids, and acylcarnitines. For Platform 2, two LC-MS methods were used to profile polar metabolites. After obtaining data from Platforms 1 and 2, a quantitative isotope dilution GC-MS method was developed to quantify plasma metabolites specifically reflecting cellular redox state, based on a previously published method (Lauzier et al., 2013).
Data Mining and Statistical Analysis
Results from Platform 1 and Platform 2 were treated as two distinct data sets to which the statistical workflow was applied using R 3.0.2 (R Foundation for Statistical Computing) for quality control filtering, log2-transformation, and missing data imputation (see the Supplemental Experimental Procedures). Raw data for all measured metabolites are provided in Table S2. The post-quality control, imputed data sets were submitted to PCA (SIMCA-P+ 13.0; Umetrics) and permutation test between the two groups. All possible distinct permutations were conducted within patients/control pairs, for a total of 256 (28). The significance threshold was determined according to the estimation of true and false positives and was established in order to correspond to an estimated false discovery rate of 10%. This led to a p threshold of <0.03 for Platform 1 and 0.023 for Platform 2. For the quantitative profiling of selected metabolites, p values were generated by a permutation test, and a threshold of 0.05, corresponding to a false discovery rate of 5.7%, was used to control for type 1 error.
Supplementary Material
Highlights.
A metabolic signature is revealed in patients with a genetic mitochondrial disorder
Profiling of 407 plasma/urine analytes identified 45 distinctive markers
Markers reflect changes in cardiovascular risk as well as NAD+ lipid and amine metabolism
Markers also include metabolites linked to neurodegeneration
ACKNOWLEDGMENTS
This work was supported by the Canadian Institutes of Health Research (CIHR) (grant no. 102168 awarded to C.D.R., C.L., J.D.R., and E.A.S.), NIH (R01DK081457 to V.K.M.), Association de l’acidose lactique (AAL), Fonds de la Recherche en Santé du Québec, Fondation du Grand Défi Pierre Lavoie, and Corporation de recherche et d’action sur les maladies héréditaires (COR-AMH). V.K.M. is an Investigator of the Howard Hughes Medical Institute. J.D.R. holds the Canada Research Chair in Genetics and Genomic Medicine. R.S. is supported by an NIH T32 grant through the Harvard Medical School Genetics Training Program. We thank B. Maranda for helpful discussions, J. Landry for subject recruitment and sample collection, I. Robillard Frayne for GC-MS expertise, C. Beauchamp for genotyping, S. Cherkaoui for bioinformatics, K.A. Pierce and K. Bullock for metabolic profiling at the Broad Institute, and T. Gagnon at Centre hospitalier universitaire de Sherbrooke. We gratefully acknowledge LSFC patients, their families, and the AAL (http://www.aal.qc.ca). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
CONSORTIA
At the time of recruitment, the members of the LSFC Consortium were, in alphabetical order, Azadeh Aliskashani, Bruce G. Allen, Chantale Aubut, Claudine Beauchamp, Chantal Bemeur, Yan Burelle, Guy Charron, Lise Coderre, Christine Des Rosiers, Sonia Desch^enes, François Labarthe, Jeannine Landry, Catherine Laprise, Geneviève Lavallée, Pierre Lavoie, Bruno Maranda, Charles Morin, Yvette Mukaneza, Tamiko Nishimura, John D. Rioux, Marie-Ève Rivard, Florin Sasarman, Eric A. Shoubridge, Jessica Tardif, Julie Thompson Legault, Nancy Tremblay, Vanessa Tremblay-Vaillancourt, Luc Vachon, and Josée Villeneuve. Affiliations are listed in the Supplemental Information along with individual author contributions.
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, two figures, and four tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2015.09.054.
REFERENCES
- Adams SH. Emerging perspectives on essential amino acid metabolism in obesity and the insulin-resistant state. Adv. Nutr. 2011;2:445–456. doi: 10.3945/an.111.000737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams SH, Hoppel CL, Lok KH, Zhao L, Wong SW, Minkler PE, Hwang DH, Newman JW, Garvey WT. Plasma acylcarnitine profiles suggest incomplete long-chain fatty acid beta-oxidation and altered tricarboxylic acid cycle activity in type 2 diabetic African-American women. J. Nutr. 2009;139:1073–1081. doi: 10.3945/jn.108.103754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Dirbashi OY, Rashed MS, Al-Mokhadab MA, Al-Qahtani K, Al-Sayed MA, Kurdi W. Stable isotope dilution analysis of N-acetylaspartic acid in urine by liquid chromatography electrospray ionization tandem mass spectrometry. Biomed. Chromatogr. 2007;21:898–902. doi: 10.1002/bmc.815. [DOI] [PubMed] [Google Scholar]
- Burelle Y, Bemeur C, Rivard ME, Thompson Legault J, Boucher G, Morin C, Coderre L, Des Rosiers C LSFC Consortium. Mitochondrial vulnerability and increased susceptibility to nutrient-induced cytotoxicity in fibroblasts from leigh syndrome French canadian patients. PLoS ONE. 2015;10:e0120767. doi: 10.1371/journal.pone.0120767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke C, Xiao R, Place E, Zhang Z, Sondheimer N, Bennett M, Yudkoff M, Falk MJ. Mitochondrial respiratory chain disease discrimination by retrospective cohort analysis of blood metabolites. Mol. Genet. Metab. 2013;110:145–152. doi: 10.1016/j.ymgme.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debray FG, Lambert M, Mitchell GA. Disorders of mitochondrial function. Curr. Opin. Pediatr. 2008;20:471–482. doi: 10.1097/MOP.0b013e328306ebb6. [DOI] [PubMed] [Google Scholar]
- Debray FG, Morin C, Janvier A, Villeneuve J, Maranda B, Laframboise R, Lacroix J, Decarie JC, Robitaille Y, Lambert M, et al. LRPPRC mutations cause a phenotypically distinct form of Leigh syndrome with cytochrome c oxidase deficiency. J. Med. Genet. 2011;48:183–189. doi: 10.1136/jmg.2010.081976. [DOI] [PubMed] [Google Scholar]
- Demine S, Reddy N, Renard P, Raes M, Arnould T. Unraveling biochemical pathways affected by mitochondrial dysfunctions using metabolomic approaches. Metabolites. 2014;4:831–878. doi: 10.3390/metabo4030831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz F, Garcia S, Padgett KR, Moraes CT. A defect in the mitochondrial complex III, but not complex IV, triggers early ROS-dependent damage in defined brain regions. Hum. Mol. Genet. 2012;21:5066–5077. doi: 10.1093/hmg/dds350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMauro S. Mitochondrial diseases. Biochim. Biophys. Acta. 2004;1658:80–88. doi: 10.1016/j.bbabio.2004.03.014. [DOI] [PubMed] [Google Scholar]
- DiMauro S, Mancuso M. Mitochondrial diseases: therapeutic approaches. Biosci. Rep. 2007;27:125–137. doi: 10.1007/s10540-007-9041-4. [DOI] [PubMed] [Google Scholar]
- Distelmaier F, Visch HJ, Smeitink JA, Mayatepek E, Koopman WJ, Willems PH. The antioxidant Trolox restores mitochondrial membrane potential and Ca2+ -stimulated ATP production in human complex I deficiency. J. Mol. Med. 2009;87:515–522. doi: 10.1007/s00109-009-0452-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrannini E, Natali A, Camastra S, Nannipieri M, Mari A, Adam KP, Milburn MV, Kastenmüller G, Adamski J, Tuomi T, et al. Early metabolic markers of the development of dysglycemia and type 2 diabetes and their physiological significance. Diabetes. 2013;62:1730–1737. doi: 10.2337/db12-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiehn O, Garvey WT, Newman JW, Lok KH, Hoppel CL, Adams SH. Plasma metabolomic profiles reflective of glucose homeostasis in non-diabetic and type 2 diabetic obese African-American women. PLoS ONE. 2010;5:e15234. doi: 10.1371/journal.pone.0015234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finsterer J. Leigh and Leigh-like syndrome in children and adults. Pediatr. Neurol. 2008;39:223–235. doi: 10.1016/j.pediatrneurol.2008.07.013. [DOI] [PubMed] [Google Scholar]
- Finsterer J, Jarius C, Eichberger H. Phenotype variability in 130 adult patients with respiratory chain disorders. J. Inherit. Metab. Dis. 2001;24:560–576. doi: 10.1023/a:1012415810881. [DOI] [PubMed] [Google Scholar]
- Fullerton MD, Hakimuddin F, Bonen A, Bakovic M. The development of a metabolic disease phenotype in CTP:phosphoethanolamine cytidylyltransferase-deficient mice. J. Biol. Chem. 2009;284:25704–25713. doi: 10.1074/jbc.M109.023846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall WE, Beebe K, Lawton KA, Adam KP, Mitchell MW, Nakhle PJ, Ryals JA, Milburn MV, Nannipieri M, Camastra S, et al. RISC Study Group. alpha-hydroxybutyrate is an early biomarker of insulin resistance and glucose intolerance in a nondiabetic population. PLoS ONE. 2010;5:e10883. doi: 10.1371/journal.pone.0010883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohil VM, Nilsson R, Belcher-Timme CA, Luo B, Root DE, Mootha VK. Mitochondrial and nuclear genomic responses to loss of LRPPRC expression. J. Biol. Chem. 2010;285:13742–13747. doi: 10.1074/jbc.M109.098400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guéant Rodriguez RM, Spada R, Pooya S, Jeannesson E, Moreno Garcia MA, Anello G, Bosco P, Elia M, Romano A, Alberto JM, et al. Homocysteine predicts increased NT-pro-BNP through impaired fatty acid oxidation. Int. J. Cardiol. 2013;167:768–775. doi: 10.1016/j.ijcard.2012.03.047. [DOI] [PubMed] [Google Scholar]
- Haas RH, Parikh S, Falk MJ, Saneto RP, Wolf NI, Darin N, Wong LJ, Cohen BH, Naviaux RK Mitochondrial Medicine Society’s Committee on Diagnosis. The in-depth evaluation of suspected mitochondrial disease. Mol. Genet. Metab. 2008;94:16–37. doi: 10.1016/j.ymgme.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houten SM, Wanders RJ. A general introduction to the biochemistry of mitochondrial fatty acid β-oxidation. J. Inherit. Metab. Dis. 2010;33:469–477. doi: 10.1007/s10545-010-9061-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobs C, Solem E, Ek J, Halvorsen K, Jellum E. Investigation of the metabolic pattern in maple syrup urine disease by means of glass capillary gas chromatography and mass spectrometry. J. Chromatogr. B Biomed. Sci. Appl. 1977;143:31–38. [Google Scholar]
- Kadiiska MB, Gladen BC, Baird DD, Graham LB, Parker CE, Ames BN, Basu S, Fitzgerald GA, Lawson JA, Marnett LJ, et al. Biomarkers of oxidative stress study III. Effects of the nonsteroidal anti-inflammatory agents indomethacin and meclofenamic acid on measurements of oxidative products of lipids in CCl4 poisoning. Free Radic. Biol. Med. 2005;38:711–718. doi: 10.1016/j.freeradbiomed.2004.10.024. [DOI] [PubMed] [Google Scholar]
- Kaufmann P, Engelstad K, Wei Y, Kulikova R, Oskoui M, Battista V, Koenigsberger DY, Pascual JM, Sano M, Hirano M, et al. Protean phenotypic features of the A3243G mitochondrial DNA mutation. Arch. Neurol. 2009;66:85–91. doi: 10.1001/archneurol.2008.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapman A, Kaltwasser SF, Martins-de-Souza D, Holsboer F, Landgraf R, Turck CW, Czisch M, Touma C. Increased stress reactivity is associated with reduced hippocampal activity and neuronal integrity along with changes in energy metabolism. Eur. J. Neurosci. 2012;35:412–422. doi: 10.1111/j.1460-9568.2011.07968.x. [DOI] [PubMed] [Google Scholar]
- Koene S, Willems PH, Roestenberg P, Koopman WJ, Smeitink JA. Mouse models for nuclear DNA-encoded mitochondrial complex I deficiency. J. Inherit. Metab. Dis. 2011;34:293–307. doi: 10.1007/s10545-009-9005-x. [DOI] [PubMed] [Google Scholar]
- Landaas S. The formation of 2-hydroxybutyric acid in experimental animals. Clin. Chim. Acta. 1975;58:23–32. doi: 10.1016/0009-8981(75)90481-7. [DOI] [PubMed] [Google Scholar]
- Lauzier B, Vaillant F, Merlen C, Gélinas R, Bouchard B, Rivard ME, Labarthe F, Dolinsky VW, Dyck JR, Allen BG, et al. Metabolic effects of glutamine on the heart: anaplerosis versus the hexosamine biosynthetic pathway. J. Mol. Cell. Cardiol. 2013;55:92–100. doi: 10.1016/j.yjmcc.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N, Daly MJ, Delmonte T, Lander ES, Xu F, Hudson TJ, Mitchell GA, Morin CC, Robinson BH, Rioux JD. A genomewide linkage-disequilibrium scan localizes the Saguenay-Lac-Saint-Jean cytochrome oxidase deficiency to 2p16. Am. J. Hum. Genet. 2001;68:397–409. doi: 10.1086/318197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiter LA, Fitchett DH, Gilbert RE, Gupta M, Mancini GB, McFarlane PA, Ross R, Teoh H, Verma S, Anand S, et al. Cardiometabolic Risk Working Group: Executive Committee. Cardiometabolic risk in Canada: a detailed analysis and position paper by the cardiometabolic risk working group. Can. J. Cardiol. 2011;27:e1–e33. doi: 10.1016/j.cjca.2010.12.054. [DOI] [PubMed] [Google Scholar]
- Leoni V, Strittmatter L, Zorzi G, Zibordi F, Dusi S, Garavaglia B, Venco P, Caccia C, Souza AL, Deik A, et al. Metabolic consequences of mitochondrial coenzyme A deficiency in patients with PANK2 mutations. Mol. Genet. Metab. 2012;105:463–471. doi: 10.1016/j.ymgme.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Vicente Gonçalves CM, Dai L, Lu HM, Huang JH, Ji H, Wang DS, Yi LZ, Liang YZ. Exploring metabolic syndrome serum profiling based on gas chromatography mass spectrometry and random forest models. Anal. Chim. Acta. 2014;827:22–27. doi: 10.1016/j.aca.2014.04.008. [DOI] [PubMed] [Google Scholar]
- Lovejoy JC, Champagne CM, Smith SR, DeLany JP, Bray GA, Lefevre M, Denkins YM, Rood JC. Relationship of dietary fat and serum cholesterol ester and phospholipid fatty acids to markers of insulin resistance in men and women with a range of glucose tolerance. Metabolism. 2001;50:86–92. doi: 10.1053/meta.2001.19440. [DOI] [PubMed] [Google Scholar]
- Maassen JA, ’T Hart LM, Van Essen E, Heine RJ, Nijpels G, Jahangir Tafrechi RS, Raap AK, Janssen GM, Lemkes HH. Mitochondrial diabetes: molecular mechanisms and clinical presentation. Diabetes. 2004;53(Suppl 1):S103–S109. doi: 10.2337/diabetes.53.2007.s103. [DOI] [PubMed] [Google Scholar]
- Merante F, Petrova-Benedict R, MacKay N, Mitchell G, Lambert M, Morin C, De Braekeleer M, Laframboise R, Gagné R, Robinson BH. A biochemically distinct form of cytochrome oxidase (COX) deficiency in the Saguenay-Lac-Saint-Jean region of Quebec. Am. J. Hum. Genet. 1993;53:481–487. [PMC free article] [PubMed] [Google Scholar]
- Miinalainen IJ, Schmitz W, Huotari A, Autio KJ, Soininen R, Ver Loren van Themaat E, Baes M, Herzig KH, Conzelmann E, Hiltunen JK. Mitochondrial 2,4-dienoyl-CoA reductase deficiency in mice results in severe hypoglycemia with stress intolerance and unimpaired ketogenesis. PLoS Genet. 2009;5:e1000543. doi: 10.1371/journal.pgen.1000543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lepage P, Miller K, Bunkenborg J, Reich M, Hjerrild M, Delmonte T, Villeneuve A, Sladek R, Xu F, et al. Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc. Natl. Acad. Sci. USA. 2003;100:605–610. doi: 10.1073/pnas.242716699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno A, Ross BD, Blüml S. Direct determination of the N-acetyl-L-aspartate synthesis rate in the human brain by (13)C MRS and [1-(13)C]glucose infusion. J. Neurochem. 2001;77:347–350. doi: 10.1046/j.1471-4159.2001.t01-1-00282.x. [DOI] [PubMed] [Google Scholar]
- Morin C, Mitchell G, Larochelle J, Lambert M, Ogier H, Robinson BH, De Braekeleer M. Clinical, metabolic, and genetic aspects of cytochrome C oxidase deficiency in Saguenay-Lac-Saint-Jean. Am. J. Hum. Genet. 1993;53:488–496. [PMC free article] [PubMed] [Google Scholar]
- Mourier A, Ruzzenente B, Brandt T, Kühlbrandt W, Larsson NG. Loss of LRPPRC causes ATP synthase deficiency. Hum. Mol. Genet. 2014;23:2580–2592. doi: 10.1093/hmg/ddt652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnich A, Rustin P. Clinical spectrum and diagnosis of mitochondrial disorders. Am. J. Med. Genet. 2001;106:4–17. doi: 10.1002/ajmg.1391. [DOI] [PubMed] [Google Scholar]
- Newgard CB. Interplay between lipids and branched-chain amino acids in development of insulin resistance. Cell Metab. 2012;15:606–614. doi: 10.1016/j.cmet.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsucci D, Filosto M, Siciliano G, Mancuso M. Electron transfer mediators and other metabolites and cofactors in the treatment of mitochondrial dysfunction. Nutr. Rev. 2009;67:427–438. doi: 10.1111/j.1753-4887.2009.00221.x. [DOI] [PubMed] [Google Scholar]
- Pajares S, Arias A, García-Villoria J, Briones P, Ribes A. Role of creatine as biomarker of mitochondrial diseases. Mol. Genet. Metab. 2013;108:119–124. doi: 10.1016/j.ymgme.2012.11.283. [DOI] [PubMed] [Google Scholar]
- Pastore A, Petrillo S, Tozzi G, Carrozzo R, Martinelli D, Dionisi-Vici C, Di Giovamberardino G, Ceravolo F, Klein MB, Miller G, et al. Glutathione: a redox signature in monitoring EPI-743 therapy in children with mitochondrial encephalomyopathies. Mol. Genet. Metab. 2013;109:208–214. doi: 10.1016/j.ymgme.2013.03.011. [DOI] [PubMed] [Google Scholar]
- Paxton R, Scislowski PW, Davis EJ, Harris RA. Role of branched-chain 2-oxo acid dehydrogenase and pyruvate dehydrogenase in 2-oxobutyrate metabolism. Biochem. J. 1986;234:295–303. doi: 10.1042/bj2340295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen JE, Landaas S, Eldjarn L. The occurrence of 2-hydroxybutyric acid in urine from patients with lactic acidosis. Clin. Chim. Acta. 1973;48:213–219. doi: 10.1016/0009-8981(73)90367-7. [DOI] [PubMed] [Google Scholar]
- Pfeffer G, Horvath R, Klopstock T, Mootha VK, Suomalainen A, Koene S, Hirano M, Zeviani M, Bindoff LA, Yu-Wai-Man P, et al. New treatments for mitochondrial disease-no time to drop our standards. Nat. Rev. Neurol. 2013;9:474–481. doi: 10.1038/nrneurol.2013.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts LD, Gerszten RE. Toward new biomarkers of cardiometabolic diseases. Cell Metab. 2013;18:43–50. doi: 10.1016/j.cmet.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe DS, Yang BZ, Vianey-Saban C, Struys E, Sweetman L, Roe CR. Differentiation of long-chain fatty acid oxidation disorders using alternative precursors and acylcarnitine profiling in fibroblasts. Mol. Genet. Metab. 2006;87:40–47. doi: 10.1016/j.ymgme.2005.09.018. [DOI] [PubMed] [Google Scholar]
- Rosalki SB, Wilkinson JH. Reduction of alpha-ketobutyrate by human serum. Nature. 1960;188:1110–1111. doi: 10.1038/1881110a0. [DOI] [PubMed] [Google Scholar]
- Ruzzenente B, Metodiev MD, Wredenberg A, Bratic A, Park CB, Cámara Y, Milenkovic D, Zickermann V, Wibom R, Hultenby K, et al. LRPPRC is necessary for polyadenylation and coordination of translation of mitochondrial mRNAs. EMBO J. 2012;31:443–456. doi: 10.1038/emboj.2011.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sas K, Robotka H, Toldi J, Vécsei L. Mitochondria, metabolic disturbances, oxidative stress and the kynurenine system, with focus on neurodegenerative disorders. J. Neurol. Sci. 2007;257:221–239. doi: 10.1016/j.jns.2007.01.033. [DOI] [PubMed] [Google Scholar]
- Sasarman F, Brunel-Guitton C, Antonicka H, Wai T, Shoubridge EA LSFC Consortium. LRPPRC and SLIRP interact in a ribonucleoprotein complex that regulates posttranscriptional gene expression in mitochondria. Mol. Biol. Cell. 2010;21:1315–1323. doi: 10.1091/mbc.E10-01-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasarman F, Nishimura T, Antonicka H, Weraarpachai W, Shoubridge EA LSFC Consortium. Tissue-specific responses to the LRPPRC founder mutation in French Canadian Leigh Syndrome. Hum. Mol. Genet. 2015;24:480–491. doi: 10.1093/hmg/ddu468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schapira AH. Mitochondrial diseases. Lancet. 2012;379:1825–1834. doi: 10.1016/S0140-6736(11)61305-6. [DOI] [PubMed] [Google Scholar]
- Schiff M, Bénit P, El-Khoury R, Schlemmer D, Benoist JF, Rustin P. Mouse studies to shape clinical trials for mitochondrial diseases: high fat diet in Harlequin mice. PLoS ONE. 2011;6:e28823. doi: 10.1371/journal.pone.0028823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SH, Kraus WE, Newgard CB. Metabolomic profiling for the identification of novel biomarkers and mechanisms related to common cardiovascular diseases: form and function. Circulation. 2012;126:1110–1120. doi: 10.1161/CIRCULATIONAHA.111.060368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaham O, Slate NG, Goldberger O, Xu Q, Ramanathan A, Souza AL, Clish CB, Sims KB, Mootha VK. A plasma signature of human mitochondrial disease revealed through metabolic profiling of spent media from cultured muscle cells. Proc. Natl. Acad. Sci. USA. 2010;107:1571–1575. doi: 10.1073/pnas.0906039107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeitink JA, Zeviani M, Turnbull DM, Jacobs HT. Mitochondrial medicine: a metabolic perspective on the pathology of oxidative phosphorylation disorders. Cell Metab. 2006;3:9–13. doi: 10.1016/j.cmet.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Smith AJ, Strang LB. An inborn error of metabolism with the urinary excretion of alpha-hydroxy-butyric acid and phenylpyruvic acid. Arch. Dis. Child. 1958;33:109–113. doi: 10.1136/adc.33.168.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele RD, Weber H, Patterson JI. Characterization of alpha-ketobutyrate metabolism in rat tissues: effects of dietary protein and fasting. J. Nutr. 1984;114:701–710. doi: 10.1093/jn/114.4.701. [DOI] [PubMed] [Google Scholar]
- Tan L, Yu JT, Tan L. The kynurenine pathway in neurodegenerative diseases: mechanistic and therapeutic considerations. J. Neurol. Sci. 2012;323:1–8. doi: 10.1016/j.jns.2012.08.005. [DOI] [PubMed] [Google Scholar]
- Torraco A, Diaz F, Vempati UD, Moraes CT. Mouse models of oxidative phosphorylation defects: powerful tools to study the pathobiology of mitochondrial diseases. Biochim. Biophys. Acta. 2009;1793:171–180. doi: 10.1016/j.bbamcr.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vafai SB, Mootha VK. Mitochondrial disorders as windows into an ancient organelle. Nature. 2012;491:374–383. doi: 10.1038/nature11707. [DOI] [PubMed] [Google Scholar]
- Vafai SB, Mootha VK. Medicine. A common pathway for a rare disease? Science. 2013;342:1453–1454. doi: 10.1126/science.1248449. [DOI] [PubMed] [Google Scholar]
- Verhoeven NM, Roe DS, Kok RM, Wanders RJ, Jakobs C, Roe CR. Phytanic acid and pristanic acid are oxidized by sequential peroxisomal and mitochondrial reactions in cultured fibroblasts. J. Lipid Res. 1998;39:66–74. [PubMed] [Google Scholar]
- Xu F, Morin C, Mitchell G, Ackerley C, Robinson BH. The role of the LRPPRC (leucine-rich pentatricopeptide repeat cassette) gene in cytochrome oxidase assembly: mutation causes lowered levels of COX (cytochrome c oxidase) I and COX III mRNA. Biochem. J. 2004;382:331–336. doi: 10.1042/BJ20040469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Tavintharan S, Sum CF, Woon K, Lim SC, Ong CN. Metabolic signature shift in type 2 diabetes mellitus revealed by mass spectrometry-based metabolomics. J. Clin. Endocrinol. Metab. 2013;98:E1060–E1065. doi: 10.1210/jc.2012-4132. [DOI] [PubMed] [Google Scholar]
- Zhu W, Stevens AP, Dettmer K, Gottfried E, Hoves S, Kreutz M, Holler E, Canelas AB, Kema I, Oefner PJ. Quantitative profiling of tryptophan metabolites in serum, urine, and cell culture supernatants by liquid chromatography-tandem mass spectrometry. Anal. Bioanal. Chem. 2011;401:3249–3261. doi: 10.1007/s00216-011-5436-y. [DOI] [PubMed] [Google Scholar]
- Zulyniak MA, Ralston JC, Tucker AJ, MacKay KA, Hillyer LM, McNicholas PD, Graham TE, Robinson LE, Duncan AM, Ma DW, Mutch DM. Vaccenic acid in serum triglycerides is associated with markers of insulin resistance in men. Appl. Physiol. Nutr. Metab. 2012;37:1003–1007. doi: 10.1139/h2012-081. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.