Summary
Posttranslational histone modifications participate in modulating the structure and function of chromatin. Promoters of transcribed genes are enriched with K4 trimethylation and hyperacetylation on the N-terminal tail of histone H3. Recently, PHD finger proteins, like Yng1 in the NuA3 HAT complex, were shown to interact with H3K4me3, indicating a biochemical link between K4 methylation and hyperacetylation. By using a combination of mass spectrometry, biochemistry, and NMR, we detail the Yng1 PHD-H3K4me3 interaction and the importance of NuA3-dependent acetylation at K14. Furthermore, genome-wide ChIP-Chip analysis demonstrates colocalization of Yng1 and H3K4me3 in vivo. Disrupting the K4me3 binding of Yng1 altered K14ac and transcription at certain genes, thereby demonstrating direct in vivo evidence of sequential trimethyl binding, acetyltransferase activity, and gene regulation by NuA3. Our data support a general mechanism of transcriptional control through which histone acetylation upstream of gene activation is promoted partially through availability of H3K4me3, “read” by binding modules in select subunits.
Introduction
In eukaryotes, DNA is complexed with histone proteins to form the nucleosomal subunits of chromatin, the context in which nuclear factors differentially interpret the genome. A wealth of histone posttranslational modifications (PTMs), including methylation, acetylation, phosphorylation, and ubiquitination, have been identified whose functional effects are under active investigation. Specific histone PTMs may contribute to a “histone/epigenetic code” that dictates distinct biological outputs such as transcription, silencing, and DNA repair (Jenuwein and Allis, 2001; Strahl and Allis, 2000; Turner, 2000). For example, methylation of K9 and K27 on histone H3 is often associated with heterochromatin, whereas K4 methylation is largely associated with euchromatin.
Many protein motifs characteristically associated with chromatin have recently been shown to have affinity for modified histone tails, acting as “effectors” for histone PTMs, notably lysine methylation. Chromodomains, for example, are often found in subunits of silencing complexes, providing a mechanism to bind to H3 methylated at K9 and/or K27. In keeping, modules such as chromodomains transduce specific PTM signals into changes in local chromatin structure, thereby limiting the accessibility to the underlying DNA (Khorasanizadeh, 2004; Seet et al., 2006). This chromodomain recruitment served to establish a useful paradigm wherein modules bind PTMs on histones, allowing distinct chromatin-associated enzymatic machineries to properly engage the chromatin fiber at discrete regions.
In contrast to the silencing paradigm with K9 and K27 methylation, chromatin immunoprecipitation (ChIP) experiments have consistently localized histone H3K4 trimethylation (hereafter H3K4me3) and H3/H4 hyperacetylation to promoter and 5′ regions of transcriptionally active genes (Ng et al., 2003; Santos-Rosa et al., 2002; Schneider et al., 2004). Furthermore, “ChIP-Chip” approaches that combine immunoprecipitation of chromatin-associated proteins with DNA microarray analysis have permitted generation of histone PTM “maps” along vast stretches of the S. cerevisiae genome that reinforce the correlation between K4 methylation, H3 hyperacetylation, and transcription as a global phenomenon (Pokholok et al. [2005] and references within). The spatial and temporal confinement of H3K4me3 and hyperacetylation along the genome strongly suggests that these PTMs participate in pathways involved in recruitment of general transcription factors or other elongation machinery (Ng et al., 2003). No direct role for H3K4 targeting an H3-specific histone acetyltransferase (HAT) complex has been established. Such a mechanism has, however, been suggested for SAGA and SLIK via interaction of the tandem chromodomains of yeast Chd1 with H3K4me2 (Pray-Grant et al., 2005), and the crystal structure of the human CHD1 bound to H3K4me3 (Flanagan et al., 2005), although human CHD1 has not been directly linked to H3 HAT activity and the preference for K4me2 and K4me3 has not been rigorously established (Sims et al., 2005). It remains unclear if there are other mechanisms contributing to the codistribution of H3K4me3 and histone H3 hyperacetylation, especially given recent identification of several H3K4me3 effectors (reviewed in Mellor [2006]).
The yeast orthologs of the PHD finger (plant homeodomain)-containing ING tumor suppressor family, Yng1, Yng2, and Pho23 (Loewith et al., 2000; Bienz 2006), have been shown to bind K4-trimethylated peptides with low μM affinity (Pena et al., 2006; Shi et al., 2006; Martin et al., 2006a). Interestingly, Yng1 is a member of the NuA3 complex, one of four known multiprotein H3 HAT complexes (NuA3, ADA, SAGA, and SLIK/SALSA) that have been isolated from yeast (Eberharter et al., 1998; Grant et al., 1997; Sterner et al., 2002; Howe et al., 2002). Although NuA3 has been implicated in transcriptional elongation through the interaction of Sas3, the MYST family member of NuA3 that serves as the HAT activity, with the FACT component Spt16 (John et al., 2000), the genome-wide binding pattern of NuA3 along chromatin has not been reported.
Removal of Yng1 from NuA3 reduces NuA3 H3 HAT activity on chromatin (Howe et al., 2002), and a recent report details a genetic link to Yng1 PHD function using toxicity of Yng1 overexpression (Martin et al., 2006a). However, the Yng1 PHD finger has not been directly linked to HAT activity in in vitro or in vivo assays (Howe et al., 2002; Loewith et al., 2000). Thus, the functional contribution of the Yng1 PHD finger to H3K4me3 remains uncharacterized. Furthermore, since prior NuA3 functional assays were performed without regard for the methylation status of H3K4, any connections between K4 methylation, NuA3-dependent H3 acetylation, and the events surrounding transcription remain ambiguous. To better understand if H3K4me3 and hyperacetylation at the promoter (and 5′ regions) are coupled by NuA3 HAT targeting through the Yng1 PHD finger, we undertook studies aimed at characterizing the Yng1 PHD-H3K4me3 interaction and describing the functional significance and genomic localization of the associated NuA3 HAT complex. Here, we provide genetic, biochemical, and biophysical evidence (including ChIP-Chip analysis) that Yng1 mediates NuA3-dependent H3K14 acetylation through a specific interaction between the Yng1 PHD finger and H3K4me3. Our studies are consistent with the general view that a hierarchical sequence of posttranslational histone modifications occurs at a promoter during the events that lead to transcriptional activation.
Results
Isolation and Characterization of the Yng1-Containing NuA3 HAT Complex
To verify that Sas3 and Taf30 are present in the purified NuA3 HAT complex and to identify additional stable components, we employed the recently described isotopic differentiation of interactions as random or targeted (I-DIRT) technology (Tackett et al., 2005a). Yng1 was affinity tagged to specifically isolate NuA3, keeping in mind that Sas3, its catalytic HAT subunit, has recently been associated with complexes found at boundary elements (Tackett et al., 2005b), and Taf30, another NuA3 component, has been shown to interact with TFIID, TFIIF, INO80, and RSC (Kabani et al., 2005). We isolated genomically TAP-tagged Yng1 under conditions that preserve in vivo protein interactions (Tackett et al., 2005a, 2005b); in all cases, only the protein A (PrA) component of the TAP tag was utilized for purification. We believe that our extraction conditions maintained the integrity of the Yng1-containing complex because gel filtration analysis revealed a protein complex of similar size as previously reported for NuA3 (see Figure S1 in the Supplemental Data available with this article online) (John et al., 2000).
Briefly, tagged YNG1 cells, grown in isotopically light media, and wild-type (untagged) cells, grown in heavy isotopic (d4-lysine) media, were mixed in a 1:1 ratio by cell weight and colysed under cryogenic conditions. Proteins purifying with tagged Yng1 were resolved by SDS-PAGE, visualized by Coomassie staining, and subjected to mass spectrometric protein identification (Figure 1A). This analysis allowed identification of proteins that were either stably associated (peptides yielding an exclusively light labeled signature) or nonstably associated (1:1 ratio of light to heavy labeled peptides) with tagged Yng1. No significant enrichment of yeast proteins was identified in a mock purification from a yeast strain without an affinity tag (Figure 1A).
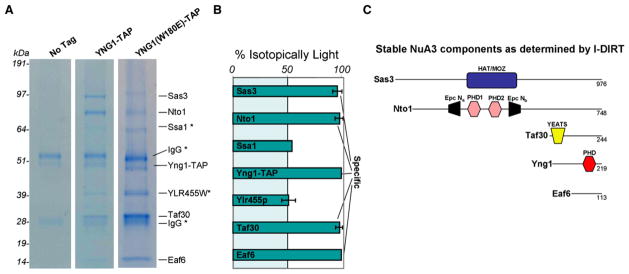
Figure 1. Isolation of an Yng1-Containing NuA3 Protein Complex.
(A) Isolation and identification of an Yng1-TAP-containing NuA3 protein complex. IgG-coated Dynabeads were incubated with S. cerevisiae lysate from either a strain containing no affinity tag or a strain containing TAP-tagged Yng1. The Yng1-TAP lysate contained an equal amount of d4-lysine-labeled cell lysate (untagged) for I-DIRT analysis. Yng1-TAP and associating proteins were resolved by 4%–12% denaturing gel electrophoresis, visualized by Coomassie staining, and excised for mass spectrometric protein identification. Proteins associated with an in-genome mutated version of Yng1 (W180E) were also identified (see text for details).
(B) I-DIRT analysis of the Yng1-TAP-associated proteins identified in (A). Proteins identified as containing near 100% h4-lysine (isotopically light) are true protein complex components, while protein identifications containing 50% h4-lysine (and therefore 50% d4-lysine) are contaminants. Error bars show the standard deviation for lysine-containing peptides.
(C) Schematic representation of proteins identified in (B) as stable components of the Yng1-TAP-containing protein complex.
We inferred from the I-DIRT analysis that the stable NuA3 complex contains five proteins: Sas3 (98 kDa), Nto1 (86 kDa), Taf30 (27 kDa), Yng1 (25 kDa), and Eaf6 (13 kDa) (Figure 1B); a schematic cartoon of these subunits and their known domains is represented in Figure 1C. Nto1 and Eaf6 have not been previously published as members of the yeast NuA3 complex, and Eaf6 is also a stable member of the H2A/H4 NuA4 HAT complex (Doyon et al., 2004). The mammalian homologs of Nto1 (Jade1/2/3 and BRPF 1/2/3) and Eaf6 (hEaf6) were recently shown to be in two HAT complexes containing ING 4/5 (H4-specific HBO1 HAT) and ING5 (H3-specific MOZ/MORF HAT) (Doyon et al., 2006). Interestingly, Nto1 has two PHD fingers surrounded by an interrupted Epc-N domain that is predicted to fold into a bromodomain-like α-helical structure with potential acetyllysine binding properties (Doyon et al., 2006; Perry, 2006). For reasons that remain unclear, we did not detect Spt16, a mammalian FACT (facilitates chromatin transcription) complex member, shown previously to interact with the carboxyl terminus of Sas3, nor did we detect any of the other ribonuclear polypeptides isolated in the initial NuA3 characterization (John et al., 2000). We also purified NuA3 from a genomically tagged Yng1 point mutant (W180E), predicted by our alignment between the Yng1 PHD and the second PHD finger in BPTF to ablate interaction with H3K4me3 (Li et al., 2006; Wysocka et al., 2006). Importantly for the interpretation of studies presented below, this point mutant yielded an intact NuA3 complex, with a highly similar, if not identical, subunit composition to wild-type Yng1 (Figure 1A).
Intact PHD Finger of Yng1 Is Required for NuA3 Association with H3 K4me3
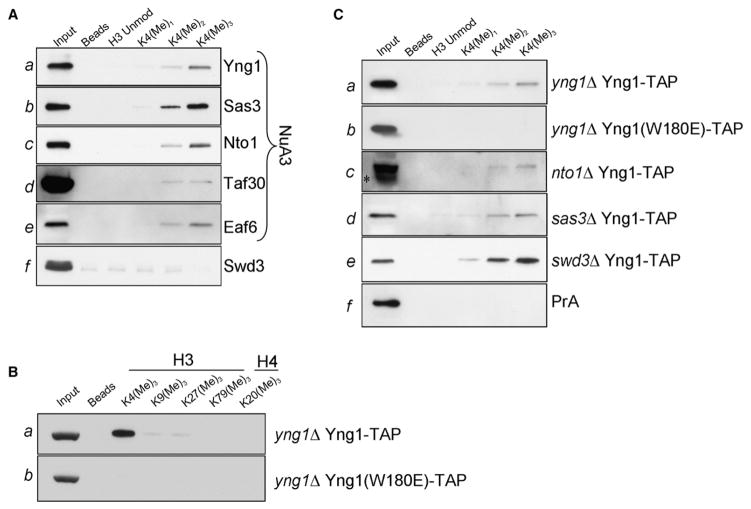
Multisubunit complexes in which HATs typically exist in vivo serve to enhance the specificity of the HATs for their histone or chromatin substrates (Eberharter et al., 1998; Grant et al., 1997; Sendra et al., 2000). To address whether the NuA3 complex can specifically bind methylated K4, we tested the ability of H3 histone peptides with different methyl states to pull down genomically tagged NuA3 members from cellular extracts. Members of the NuA3 complex were TAP tagged and extracted as above. These extracts were then incubated with biotinylated H3 peptides bearing unmodified, mono-, di-, or trimethylated K4. Associated proteins were pulled down with streptavidin resin, resolved by SDS-PAGE, and visualized with antibodies against the PrA epitope. We observed that the NuA3 members Yng1, Sas3, Nto1, and Eaf6 were enriched in the trimethylated K4 peptide pull-down, as compared to unmodified, monomethyl, and dimethyl K4 peptides (Figures 2Aa–2Ac and 2Ae), suggesting that NuA3 displays a preference for H3K4me3. Swd3, a homolog of WDR5, was tested in this assay as a potential H3K4me2 effector (Flanagan et al., 2005; Wysocka et al., 2005) but did not detectably interact with H3 peptides (Figure 2Af). To determine if endogenous H3 was pulled down with NuA3 complex members, membranes were probed with antibodies recognizing the various states of K4 methylation (Figure S2). Since little to no H3 was detected in the immunopurification lanes, it is likely that the Yng1/NuA3 complex is interacting directly with the histone peptides.
Figure 2. NuA3 Is Targeted to H3 K4me3 by Yng1.
(A) Peptide pull-down assays were performed by using yeast lysates from tagged strains and biotinylated H3 peptides (either unmodified, monomethylated, dimethylated, or trimethylated at K4) bound to streptavidin-linked Dynabeads. In all cases, pull-downs were analyzed with antibodies recognizing the PrA epitope in the TAP tag.
(B) NuA3 binding to H3K4me3 is context specific. Lysates from wild-type or Yng1 W180E strains were processed as in (A), and peptide pull-downs were performed with the H3K4me3peptides indicated.
(C) The binding of NuA3 to H3K4me3 peptide is directed solely through the PHD finger of Yng1. Lysates from knockout strains indicated were processed and pull-downs were performed as in (A). The asterisk on (Cc) represents a breakdown product of the tagged Yng1.
To confirm that the interaction detected in Figure 2A was specific for H3K4me3, we performed pull-downs with additional trimethylated histone peptides. Since we were unable to detect significant enrichment with trimethylated peptides other than H3K4me3 (Figure 2Ba), we conclude that NuA3 interaction with H3K4me3 also required the K4 proximal sequence. In contrast, Yng1 W180E protein was not pulled down with any trimethylated histone peptide (Figure 2Bb), including H3K4me3, suggesting that the interaction of Yng1 with H3K4me3 is solely directed through the PHD finger.
Given that NuA3 contains several proteins besides Yng1 with potential chromatin-interacting modules, the association of NuA3 with trimethylated K4 peptide could be partially mediated by another complex member (i.e., Nto1 contains two PHD fingers in tandem). Therefore, we performed pull-downs in Yng1-tagged strains deleted for various complex members. As expected, a positive control, consisting of tagged Yng1 reintroduced into an YNG1 knockout, was enriched in the trimethyl K4 pull-down (Figure 2Ca). Furthermore the W180E mutant was not enriched in any pull-down (Figure 2Cb), suggesting the complete ablation of H3K4me3 binding in this mutant. A pull-down with a strain expressing a genomic copy of PrA alone served as a negative control (Figure 2Cf). As shown in (Figures 2Cc and 2Cd), the interaction between Yng1 and H3K4me3 remains robust in preparations from strains missing the Nto1 and Sas3 NuA3 components, suggesting that Yng1 binding to H3K4me3 is direct. Moreover, the binding of Yng1 to H3K4me3 in extracts from strains deleted for Swd3 (Figure 2Ce) also remained intact, suggesting that this protein linked to H3K4me2 is not contributing to the NuA3-H3K4me3 interaction. These data suggest that the NuA3 complex binds to H3K4 directly through an interaction between Yng1 and H3K4me3 and that this specificity is likely dictated through the Yng1 PHD finger.
Yng1 PHD Binding to H3 K4me3 Is Direct and Specific
While the data above strongly implicate the Yng1 PHD finger as the mechanistic link between NuA3 and H3K4me3, they do not exclude the formal possibility that the Yng1 PHD finger binds other histones or other histone modifications. To explore this possibility, a recombinant GST-Yng1 PHD finger fusion was incubated with acid-extracted histones from wild-type yeast and glutathione beads used to pull down Yng1-bound histones followed by western analyses with histone-specific antibodies. Importantly, histone-modification-independent antibodies detected an enrichment of H3 compared to the other histones in pull-downs from the wild-type GST-Yng1 PHD fusion (Figure 3Ac). This H3 contained a relatively high percentage of K4me3 (Figure 3Aa) and, to a lesser extent, K4me2 (Figure 3Ab), suggesting that the PHD finger of Yng1 preferentially interacts with H3K4me3 (H3K9me and K27me were not tested, as these PTMs have not been shown to exist in S. cerevisiae). Conversely, we did not detect enrichment of H2B, H2A, or H4 (Figures 3Ad, 3Ae, and 3Af, respectively). The absence of significant enrichment of any histone, or histone modification, in GST-W180E mutant pull-downs reinforced our earlier observations that the aromatic cage tryptophan in the Yng1-PHD finger is required for the interaction with H3K4me3 (see below).
Figure 3. Yng1 Binds Directly to H3 K4me3 through Its Single PHD Finger.
(A) Peptide pull-down assays were performed using cryogenically processed yeast, and the pull-down lane was compared to the input lane to gauge enrichment. As expected, anti-GST antibodies show that GST fusions were enriched in glutathione pull-downs (see Ag).
(B) A titration plot based on the chemical shift changes of the W180 side chain imino resonance from PHD domain of YNG1 protein. Chemical shift changes were measured by 1H15N-HSQC spectra upon addition of monomethylated, dimethylated, trimethylated, and unmodified lysine 4 from histone H3 peptide as a function of the molar ratio. The concentrated peptide (20 mM) was added at the molar ratio of 0.5, 1.0, 1.5, 2.2, 3.0, and 5.0 peptide to protein. Δδ was calculated based on the method described in Supplemental Experimental Procedures.
(C) Fluorescence anisotropy was used to determine the dissociation constants for the interaction between the Yng1 PHD finger and different methylation states of H3K4. Error bars are the standard deviation from triplicate analyses.
(D) Tabulated values for the dissociation constants measured in Figure 3C.
Characterization of Yng1 PHD Interaction with H3 Trimethylated at K4
Using tryptophan fluorescence, the PHD finger of Yng1 was recently reported to have a low μM affinity for H3K4me3 (Pena et al., 2006); however, the preference between different K4 modification states was not determined. Although our peptide pull-down experiments with recombinant protein demonstrated a preference of the Yng1 PHD finger for H3K4me3 (data not shown), we sought to biophysically define the parameters of this interaction with solution-based binding assays. As shown in Figure 3B in a series of NMR titrations based on binding-dependent chemical shift changes of W180 (attributed to the K4me3 peptide binding in ING2 and BPTF) (Li et al., 2006; Pena et al., 2006), the 15N-labeled YNG1 PHD finger binds preferentially to H31–9K4me3 peptide, less so to H31–9K4me2, and significantly less to the K4me1 and unmethylated H31–9 peptides. Binding assays that used fluorescence anisotropy (Figure 3C) were consistent with NMR titration assays. We found that the Yng1 PHD-H3K4me3 interaction has a KD of 9.1 ± 1.6 μM, with monotonic 2-fold decreases in binding affinity from K4me3 to K4me2 to K4me1 and with binding to unmodified H3 reduced to a KD of >400 μM (Figure 3D). Mutation of the W180E aromatic cage residue dramatically reduces binding affinity to levels similar to unmodified H3 peptides (Figure 3D), reinforcing the importance of this residue in the interaction with H3K4me3.
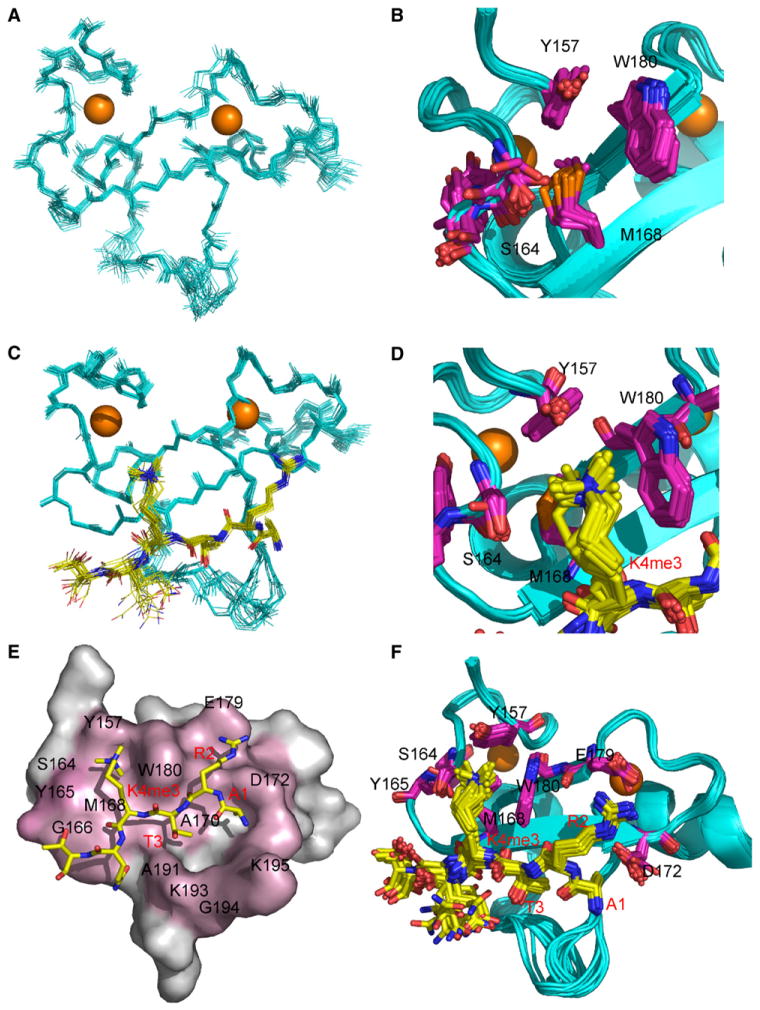
Structural Basis for the Interaction between Yng1 PHD and Histone H3 K4me3
The structure of YNG1 PHD finger (152–212) was determined by high-resolution NMR in two states: (1) free and (2) in the complex with H31–9K4me3 peptide. The overall fold of PHD finger in the complex is very similar to its apo state and undergoes only minor adjustments to accommodate the peptide binding (rms 0.61). The structure consists of three loops stabilized by two zinc clusters, a distinct feature of the PHD finger (Cys4-His-Cys3 zinc fingers), and a double-stranded antiparallel β sheet (Figures 4A and 4C). As shown in Figure 4C, the H31–9K4me3 peptide interacts extensively with the surface opposite the zinc clusters forming a third antiparallel β strand, while its side chains sit snugly in two connected grooves (Figures 4E and 4F). The first pocket formed by Y157, S164, M168, and W180 is occupied by the methylated side chain of K4, which forms a cation-π interaction with the aromatic moieties of Y157 and W180 (Figures 4B and 4D). The second groove, formed by D172, E179, and W180, is occupied by the H3R2 side chain, which establishes hydrogen bonds with the carboxylate bearing side chains of D172 and E179. The grooves are separated by the indole moiety of W180, which is sandwiched between H3R2 and H3K4me3. This recognition motif has also been observed in two additional PHD fingers from BPTF and ING2 (Li et al., 2006; Pena et al., 2006). In all three cases, tryptophan plays a central role in the recognition of the modified H3 tail.
Figure 4. NMR Structure of the Yng1 PHD Interaction with Trimethylated H3 K4.
NMR-derived structure of the PHD finger (152–212) in the free form. For clarity, residues 152–154 and 208–212 in the unstructured regions were omitted.
(A) Backbone superposition of 20 energy-minimized structures of the PHD finger.
(B) Aromatically rich surface that shows extensive chemical shift changes upon peptide binding (Y157 and W180).
(C) Backbone superposition of 20 energy-minimized structures of the PHD finger in complex with H31–9K4me3 peptide.
(D) Aromatically rich surface that shows extensive interactions with trimethylated lysine K4 (Y157 and W180).
(E) Surface representation of the YNG1 PHD complex with H31–9K4me3 peptide. Surface residues that undergo the largest chemical shift upon binding are highlighted in pink.
(F) An ensemble of 20 structures with side chains involved in complex formation colored in purple (D172 and E179 for H3 R2, Y157 and W180 for H3K4).
Our structural and mutational studies demonstrate that H3R2 and K4me3, as well as the W180 from the PHD finger that interdigitates these residues, are important for binding of the Yng1 PHD finger to the H31–9K4me3 peptide. Unlike PHD fingers that bind the H3 “activation mark” AR2TK4me3, chromodomains recognize the “silencing marks” of H3K9 and K27 in the context of AR8/26K9/27me. Since H3T3 is directly opposite W180 of the Yng1 PHD finger (Figures 4E and 4F), we wanted to determine if the presence or absence of H3T3 could alter specificity of the PHD finger for the N terminus of H3 trimethylated at K4. Interestingly, elimination of T3 from H3K4me3 peptide reduces the binding of wild-type YNG1 PHD finger to levels resembling those of unmethylated H3 peptide (Figure 3D and data not shown). This suggests a requirement for H3T3 as a molecular spacer, maintaining appropriate distance between H3R2 and K4me3 as dictated by W180 of the Yng1 PHD finger. Together, these data detail the molecular recognition of the Yng1 PHD finger for H3K4me3 and indicate that the methylation state of K4 is a substantial determinant of binding affinity, whereas the location of the lysine methylation relative to a proximal arginine determines specificity.
The Interaction of the Yng1 PHD Finger with K4me3 Enhances NuA3 HAT Activity on Histone H3 Substrates
Our demonstration that the Yng1 PHD finger preferentially binds K4me3, along with our structural basis for this binding preference, raised the intriguing possibility that NuA3 HAT activity may be increased on K4me3 peptides substrates. If correct, this finding would serve to provide a molecular explanation for why both hyperacetylation and K4me3 have been linked on the same H3 tail (Zhang et al., 2004). The HAT activity of purified NuA3 was therefore assayed using H3 histone peptides methylated to different degrees at K4. As shown in Figure 5A, K4me3 peptide was consistently a better substrate for acetylation by NuA3 relative to di-, mono-, and unmethylated peptides. Since we demonstrated that the W180E point mutant abrogates Yng1 binding to K4me3, we reasoned that the NuA3 purified from W180E strains would no longer show higher activity in trimethylated peptides. Indeed, the activity of the W180E NuA3 was significantly reduced in K4 trimethylated peptides versus the unmodified controls.
Figure 5. HAT Activity of the Yng1-TAP-Containing Protein Complex.
(A) NuA3 exhibits enhanced HAT activity upon trimethylation of H3K4. Peptide-eluted Yng1-TAP-containing protein complex was incubated with differentially modified versions of histone H31–20 peptide and with radiolabeled acetyl CoA. Data were normalized to the amount of acetylation observed for the unmodified peptide.
(B) Yng1-TAP-containing NuA3 protein complex incorporated one acetyl group per peptide. Peptide-eluted Yng1-TAP containing NuA3 protein complex was incubated with H3K4me3 peptide and acetyl CoA. Following the HAT reaction, unacetylated lysines were chemically acetylated with d6-acetic anhydride. The mass spectrum from this reaction showed that only one acetylation was detectable on any given peptide. The shaded area shows the theoretical isotopic distribution from the singly acetylated H3 peptide. Peak area excluded from the shaded area is due to the unacetylated version of the H3 peptide.
(C) The peptide-eluted Yng1-TAP-containing NuA3 protein complex preferentially acetylated the input H3K4me3 peptide on K14. Shown is a plot of the fraction of acetylation at each modifiable lysine on the peptide. The fraction acetylated was determined by mass spectrometric fragmentation of the singly acetylated peptide from (B). Error bars are the standard deviation from triplicate analyses.
To further test the substrate preference of our NuA3 preparation, H3 peptides preacetylated either on K14 alone or dually on K9/K14 (Figure 5A) were used in NuA3 acetylation assays. Consistent with previous characterizations for NuA3 enriched fractions (Eberharter et al., 1998), NuA3 could not appreciably acetylate peptides already preacetylated on K14. These data suggested that K14 was the preferred site of NuA3 activity on the N-terminal tail of histone H3, an observation reinforced by a previous study in which H3K14 was identified as a major site of acetylation on a nucleosomal substrate (Howe et al., 2001). In order to further determine the amount and position of acetylation detected in Figure 5A for the trimethylated K4 peptide, Yng1-TAPcontaining NuA3 protein complex was incubated with trimethylated K4 peptide and acetyl coenzyme A, and reaction products were analyzed by mass spectrometry (Tackett et al., 2005b). The mass spectrum of this reaction showed that only a single acetylation was detectable for the H3K4me3 N-terminal peptide (Figure 5B). Tandem mass spectrometric analysis of this sample revealed that this single acetylation is primarily on K14, with acetylations at K9 and K18 detected at levels barely above background (Figure 5C). The demonstration that our tagged Yng1 containing NuA3 complex catalyzes preferential acetylation of K14 when K4 is trimethylated supports our HAT assays performed with preacetylated H3K14ac peptide in Figure 5A. Therefore, our data are most consistent with the idea that K4me3 increases H3K14 acetylation activity of NuA3, likely through increased affinity of the HAT complex via the PHD finger on Yng1.
Yng1 Genomic Localization Overlaps Significantly with K4me3 by High-Resolution ChIP-Chip
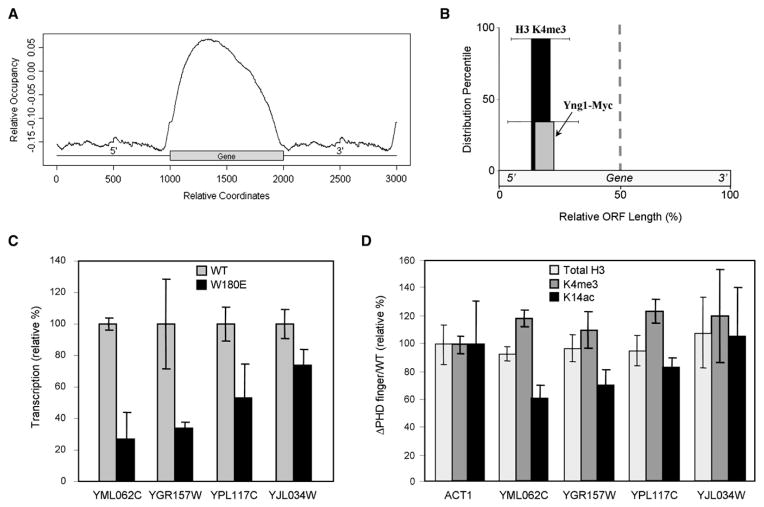
After determining that the PHD finger of Yng1 binds to H3K4me3 in vitro (Figures 2, 3B–3D, and 4), we sought to determine if Yng1 was targeted to chromosome regions enriched with the H3K4me3 mark in vivo. To perform these analyses, we utilized a ChIP-Chip technique with readout on high resolution, tiled microarrays (NimbleGen Systems Inc.) that covered the entire S. cerevisiae genome (Ren et al., 2000). Use of this technique, Pokholok et al. (2005) showed enrichment of H3K4me3 at the 5′ end of transcriptionally active ORFs, while H3K4me2 was enriched at the median and H3K4me1 was enriched at the 3′ end. Although HAT complexes are not well characterized by genome-wide high-resolution ChIP-Chip studies, we hypothesized that Yng1 would be enriched in the 5′ half of targeted ORFs due to the preferential association with H3K4me3 that we detected in our in vitro assays (Figures 2 and 3).
We show a composite profile of the Yng1-associated DNA occupancy (relative occupancy) plotted as a function of relative coordinates on an average gene flanked with intergenic regions in a manner similar to Pokholok et al. (2005) (Figure 6A). Relative occupancy is a value that represents the binding intensity for a protein (i.e., Yng1) on a hypothetical gene. From the graph, one can ascertain that Yng1 is enriched at coding sequences (relative to intergenic regions) and, with an obvious peak enrichment within the 5′ half of the ORF, consistent with our above mentioned hypothesis. The lower level of Yng1 association throughout the non-5′ region of the ORF could be due to (1) the trailing levels of H3K4me3 extending throughout ORFs on average (Pokholok et al., 2005) and/or (2) a weaker interaction of NuA3 with an as yet-uncharacterized chromatin component. Indeed, it was recently reported that Sas3 retention on chromatin is partially mediated by the activity of Set2, an H3K36 HMT, as well as Set1 (Martin et al., 2006b).
Figure 6. Genome-Wide Localization of Yng1-Myc, Its Transcriptional Impact, and Its Epigenetic Effect.
(A) YNG1 was genomically MYC tagged and subjected to ChIP-Chip analysis on high-resolution microarrays that covered the entire S. cerevisiae genome. Yng1-Myc binding sites are plotted in accordance to their relative position within or surrounding an ORF.
(B) The top 50 ORFs bound by Yng1-Myc (identified from [A]) were compared to the H3K4me3 data reported by (Pokholok et al., 2005), and 92% of the Yng1-Myc-associated ORFs contained H3K4me3 enriched in the 5′ half of the ORF. Of these ORFs containing bound Yng1-Myc and 5′ enriched H3K4 trimethylation, Yng1-Myc was enriched at the 5′ half of the ORF in 35% of the cases. The distribution percentiles are plotted in accordance to the point of enrichment in the 5′ half of the ORF, and the standard deviation of this point of enrichment is shown.
(C) Quantitative rtPCR was used to determine differences in transcription levels of Yng1-targeted ORFs. These overall cDNA levels were normalized to the expression levels of actin in wild-type and W180E mutant strains. The transcription level was normalized to wild-type as 100%. Error bars show the standard deviation of triplicate analyses. Figure S5 shows all tested ORFs.
(D) ChIP with real-time PCR readout was used to determine the relative levels of H3K4me3 and H3K14ac at the Yng1-bound ORFs from (C) (total H3 signal serves as a nucleosome occupancy control). Levels of H3K4me3, H3K14ac, and total H3 are reported as the ratio of signal from the strain lacking the PHD finger of Yng1 versus that from a wild-type strain. Each ratio was normalized to the ratio observed at ACT1. Error bars show the standard deviation from triplicate analyses.
Of the top 50 highest affinity sites bound by Yng1, 92% of the ORFs had an enrichment of H3K4me3 in the 5′ half of the gene (Figure 6B). As would be expected from the overall distribution of Yng1 observed in Figure 6A, the highest-affinity sites were also enriched for Yng1 binding at the 5′ end of the ORF (Figure 6B), although minor amounts of Yng1 were distributed elsewhere within the ORF. Overall, the 5′ localization of Yng1 in our ChIP-Chip data is consistent with the notion that H3K4me3 may serve to recruit or retain Yng1 and the attendant NuA3 complex to 5′ regions of ORFs in order to effect acetylation of H3K14 that, in turn, would be anticipated to stimulate transcription.
Mutation of the Yng1 PHD Results in Decreased Transcription and Loss of K14ac at a Subset of NuA3-Targeted Genes
While H3K4me3 has been linked with transcription and silencing (Shi et al., 2006; Santos-Rosa et al., 2002), H3 acetylation is well known to positively impact transcriptional competency (Agalioti et al., 2002; Pokholok et al., 2005). Since the NuA3 H3 HAT member Yng1 binds H3K4me3, and the genomic localization of Yng1 overlaps to a high degree with K4me3 at transcriptionally active ORFs (Figure 6), we reasoned that NuA3 may contribute to positive regulation of transcription at targeted loci. The Yng1 ChIP-Chip data permit us to examine genes potentially regulated by NuA3 and to address the in vivo significance of the Yng1 PHD finger-H3K4me3 interaction.
Quantitative real-time RT-PCR analysis was performed for genes identified as genomic targets for Yng1 that also highly correlated with genomic localization of H3K4me3 as determined in previous work (Pokholok et al., 2005). Transcription levels from many of the highest scoring genes in Yng1 and H3K4me3 colocalization were examined in either wild-type or Yng1 W180E strains; in all cases, gene expression was normalized to expression level in wild-type cells. Actin, which was not detected as a genomic target for Yng1, did not differ in expression between strains, as expected. Interestingly, the transcript levels of two Yng1/NuA3 target genes (YGR157W and YML062C) were significantly downregulated by approximately 70%, while YPL117C was decreased approximately 50% (Figure 6C). Other transcripts remained either at wild-type levels or with <20% reduction in expression (Figure 6C and Figure S5). This analysis implies that transcription of a subset of genes targeted by NuA3 is dependent on the interaction between the Yng1 PHD finger and H3K4me3.
Our binding data suggest that NuA3 HAT activity preferentially targeted H3K4me3 peptides (Figure 5). We sought then to determine if mutation of the Yng1 PHD would also result in loss of H3K14 acetylation at targeted genes in vivo. To examine changes in acetylation levels, ChIP experiments were performed at targeted genes in either wild-type or strains deleted for the Yng1 PHD. H3K4me3 levels remained virtually unchanged between wild-type and mutant strains across all tested loci, as did α-H3, serving as a control for nucleosome occupancy (Figure 6D). However, those genes most significantly affected in transcription levels, YGR157W, YML062C, and YPL117C, were found to be deficient for K14ac as well in strains lacking the Yng1 PHD (Figure 6D). This trend strongly implies an in vivo role for K4me3 in targeting K14Ac by NuA3 to promote transcription as modeled in Figure 7.
Figure 7. Model for H3K4me3-Directed Activity of NuA3.

(A) Set1, the H3K4 HMT, is recruited to promoter-proximal nucleosomes at the ORF to be activated, resulting in H3K4me3.
(B) NuA3 is targeted to and/or retained at sites of H3K4me3 through interactions with the PHD finger of Yng1, promoting H3K14ac via the Sas3 HAT, positively regulating downstream transcription events.
Discussion
An increasing body of literature indicates that histone H3 methylation participates in an epigenetic marking system, distinguishing regions of the genome as either transcriptionally activating or repressive through the binding of histone effector modules. In euchromatin, for example, H3K4me3 and H3 hyperacetylation are commonly localized to the promoter and 5′ regions of actively transcribing genes. While bromodomains and PHD fingers have been identified as effectors for acetylated and methylated histone lysines, respectively, it has remained unclear how these modules function together to promote transcriptional activation of target genes. We propose that a coordinated sequence of histone modifications occurs in a subset of target genes—namely, that Set1 complex-mediated H3K4me3 (Figure 7A) functions to target NuA3 binding and/or retention at distinct genomic regions through the PHD finger of Yng1. This localization of NuA3 HAT activity at promoter proximal nucleosomes culminates in the positive transcriptional regulation of these genes (Figure 7B).
Yng1 PHD Finger: A Yeast H3K4me3 Binding Module
Our structural and biophysical studies characterizing the Yng1-H3K4me3 interaction are consistent with recent reports identifying ING2 and BPTF PHD fingers as H3K4me3 binding modules (Li et al., 2006; Pena et al., 2006). Indeed, these data support a model implicating the tryptophan W180 of the Yng1 PHD finger that interleaves the H3R2 and K4 histone tail residues, as a major component of binding. Furthermore, the loss of binding observed with the T3-deleted trimethyl H3K4 peptide supports the structural interpretation that T3 is required as a molecular spacer to appropriately direct the R2 and K4 of histone H3 into their respective binding grooves. In this way, T3 in the context of the extreme H3 N terminus (ART3K) participates in the ability of the PHD finger of Yng1 and its associated machinery to distinguish nucleosomes trimethylated on H3K4 from regions of chromatin enriched in other lysine methylation, like methylated H3K9 or K27, for example.
An intriguing possibility is the prospect of PTM of residues in either the Yng1 PHD or the histone tail contributing to a molecular “switch,” abrogating binding of the Yng1 PHD finger, as has been proposed with other histone binding modules such as HP1 (Fischle et al., 2003; 2005). While H3T3 phosphorylation is known to occur during mitosis in metazoans (Dai et al., 2005), this modification has yet to be documented in yeast. Methylation of R2 (if co-occurring with H3K4me3) may also impact binding to the PHD finger as well.
The Role of Yng1 in Transcription
Trimethylation of H3K4 and increased acetylation on H3 are strongly associated with promoter and 5′ regions of actively transcribing genes (Pokholok et al. [2005] and references within). For example, a strong correlation has been observed between H3K4me and H3 hyperacetylation at MLL target genes (Milne et al., 2005) and at c-fos and c-jun (Hazzalin and Mahadevan, 2005). Here, we have demonstrated that an intact Yng1 PHD finger enhances K14-specific NuA3 HAT activity on H3K4me3 peptides. Furthermore, our ChIP-Chip experiments tracked Yng1 to the 5′ end of actively transcribing genes, with significant overlap in regions enriched in H3K4me3 and H3K14ac, suggesting that Set1-dependent K4me3 precedes NuA3-dependent K14ac on promoter-proximal nucleosomes (see model in Figure 7).
Despite the fact that only a subset of the Yng1-targeted genes tested showed decreased transcription levels in H3K4me3 binding-defective Yng1 point mutants, we found no evidence of increased transcription at any tested loci in the mutant strain (Figure 6). While these results suggest a function for NuA3 in promoting transcription, they contrast with a reported role of NuA3 in gene silencing (Nourani et al., 2003). Since Sas3, the catalytic subunit of NuA3, is also found in the Dpb4-chromatin remodeling complex and the Yta7 boundary chromatin complex and impacts epigenetic regulation of the HMR locus (Tackett et al., 2005b), mutational analyses of Sas3 may lack a clear functional interpretation.
H3K4me3 as a General Complex Targeting Signal
In our genome-wide array, the majority of Yng1 is localized to sites enriched in H3K4me3. Yet, the genomic localization of Yng1 only represents a subpopulation of the total sites occupied by H3K4me3 in a previous publication (Pokholok et al., 2005). These results are also consistent with previous reports that ablation of NuA3 function does not result in a detectable decrease of bulk acetylation levels (Martin et al., 2006b). However, even in genes in which Yng1 is enriched, decreased transcription rates and decreased H3K14ac were not ubiquitous; further studies will be required to determine site occupancy of NuA3 complex members other than Yng1. Interestingly, Taf30 was equally enriched in K4me2 and K4me3 pull-downs (Figure 2Ad), possibly representing K4me2 binding character contributed by a module in a different Taf30-containing complex (Kabani et al., 2005).
Since our observations suggest that NuA3 participates in a specialized transcriptional control mechanism over a distinct subset of genes in part through the H3K4me3/Yng1 PHD interaction, the recent identification of many H3K4me3 binding modules raises the intriguing question of how diverse sets of complexes with H3K4me3 binding modules can be preferentially targeted to one particular K4me3 region over another. While H3K4me3 could target NuA3 to distinct regions of the genome through interactions with the Yng1 PHD finger, it seems likely that additional modules in a complex or histone PTMs serve to “fine tune” this localization. For example, Nto1 contains a PZPM (PHD/Zn2+-knuckle/PHD motif) surrounded by an Epc-N domain (Figure 1C) that together may bind unknown combinations of histone PTMs (Perry, 2006). In this way, chromatin-templated activities may integrate PTMs on histone tails, adjusting the molecular readout of chromatin, based on the modularity of the complexes that evolved to read them in a combinatorial fashion (Mellor, 2006). Collectively, these observations strengthen an emerging paradigm wherein effectors for H3K4me3 function to facilitate chromatin-associated activities at distinct regions along the genome with clear developmental ramifications (Bernstein et al., 2006).
Experimental Procedures
S. cerevisiae Strains
All strains are listed in Table S1.
Mass Spectrometric Protein Identification
m/z values were determined on a MALDI-prOTOF (PerkinElmer Sciex) mass spectrometer. Tandem MS analysis of detectable peptides was performed with aMALDI-LTQ MS (Thermo). Proteins were identified with the program XProteo.
Yng1-TAP Immunopurifications with I-DIRT Analysis
Yng1-TAP protein complex purification and I-DIRT were performed essentially as described (Tackett et al., 2005a, 2005b). For enzymatic assays, the Yng1-TAP and interacting proteins were released from IgG-coated Dynabeads under nondenaturing conditions with a PrA elution peptide (Tackett et al., 2005b).
In Vivo Binding Assay
Extracts were made as described for the Yng1-TAP IP with changes noted in the Supplemental Experimental Procedures. Total represented 0.5% of input. Unmodified, K14ac, K9/14ac, and K4me1 and K4me3 H3 peptides were from Upstate Biotech (UBI). K4me2, K9me3, K27me3, and K79me3 H3 peptides and trimethylated K20me3 H4 peptide were synthesized at the Proteomics Resource Center of The Rockefeller University.
Protein Preparation of Yng1 PHD Finger
The Yng1 PHD finger constructs were made with an N-terminal glutathione S-transferase (GST) tag. The expression and purification conditions are detailed in Supplemental Experimental Procedures. The W180E mutant was made using the QuikChange Site-Directed Mutagenesis Kit (Stratagene).
Yng1 PHD Binding Assays
Binding assays were performed by mixing 1 μg YNG1-GST or YNG1-W180E-GST PHD fingers (141–219 of full-length Yng1) with acid extracts from wild-type yeast (5.5 × 108 cell equivalents) that were resuspended in 500 μL of binding buffer (150 mM NaCl, 20 mM HEPES [pH 7.9], 25% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 1 mM PMSF, 0.2% Triton X-100) for 1 hr at room temperature. Glutathione Sepharose 4B beads (Amersham Pharmacia) (25 μL) were added to the binding reaction and nutated for 1 hr at room temperature. Beads were washed three times in 300 mM KCl wash buffer (300 mM KCl, 20mMHEPES [pH 7.9], 0.2% Triton X-100) one time in final wash buffer (4 mM HEPES [pH 7.5] and 10 mM NaCl). Antibodies used to probe membranes included the following: general H3 (ab1791, Abcam); H3K4(Me)2 (ab7766, Abcam); H3K4(Me)3 (ab8580, Abcam); general H2B (07-371, UBI), general H2A (07-146, UBI), and H4 (ab10158, Abcam); and GST (RPN1236, Amersham).
NMR Binding Study
The binding of the peptides H31–9K4me3, H31–9K4me2, H31–9K4me1, and H31–9K4 to the YNG1 PHD domain was monitored by recording 1H15N HSQC spectra at the peptide to protein molar ratios of 0.5, 1.0, 1.5, 3.0, and 5.0.
Fluorescence Anisotropy
Fluorescence polarization assays were performed essentially as described in Taverna et al. (2002) in buffer containing 20 mM HEPES (pH 7.5), 150 mM NaCl, 0.1% Tween-20. Reactions (60 μl) were incubated with 100 nM C-terminally fluorescein-labeled peptide for 1 hr at room temperature; fluorescence polarization was then measured in a Hidex Chameleon plate reader, also at room temperature.
NMR Spectroscopy
Collection of NMR spectra and structural calculations are detailed in Supplemental Experimental Procedures.
HAT Assay
HAT assays with NuA3 were performed as described previously in Lo et al. (2004) with the exception that 0.5 μg biotinylated histone peptide was used per reaction. Unmodified, K14 acetylated, K9/14 acetylated, and K4 mono- and trimethylated H3 peptides were from UBI. Dimethylated H3 was synthesized at the Proteomics Resource Center of The Rockefeller University.
Chromatin Immunoprecipitation and Genome-Wide ChIP-Chip
ChIP experiments were performed using the anti-H3K14ac antibody (UBI, #07-353) and anti-H3K4me3 (Abcam, # ab8580-50) according to established protocols (Tackett et al., 2005b).
For the ChIP-Chip array, the IP of Yng1-Myc was performed as in Ren et al. (2000). The PCR amplification of the immunoprecipitated DNA was carried out using the protocol from NimbleGen Systems Inc. DNA was labeled and hybridized by NimbleGen Systems Inc. to S. cerevisiae whole-genome tiled arrays. Analyses were done in triplicate with dye swapping.
Measurement of Transcription Levels by Q-PCR
Total RNA was prepared from yng1::YNG1-TAP and yng1:: YNG1(W180E)-TAP strains via hot phenol extraction. cDNA was synthesized with the Superscript First Strand Reagents Kit (Invitrogen). Differences in transcription levels between wild-type and mutant strains of Yng1 target genes were measured with Q-PCR on the Stratagene MX 3000 using Platinum SYBR Green (Invitrogen).
Supplementary Material
Acknowledgments
All FITC-labeled and most biotin-labeled peptides were generated by N. Chandramouli and S. Powell at the Proteomics Resource Center of The Rockefeller University. Histone antibodies from UBI/Serologicals were a gift from the company. A.J.T. thanks C. Warthen for MS support from AR INBRE (P20RR016460), and AR Biosciences Inst. We thank Allis laboratory members for helpful discussions, especially E. Goneska and Drs. A. Ruthenburg, Y. Dou, and T. Milne for critical reading of this manuscript. A.J.T. acknowledges insightful discussions with Drs. W. Wahls and K. Raney. This work was supported by NIH grants to S.D.T. (GM53512), L.B., (CA09673), C.D.A. (GM63959), A.J.T. (P20RR015569), J.D.A. (RR022220 and GM076547), and B.T.C. (RR00862 and RR022220) as well as Rockefeller University. D.J.P. is supported by funds from the Abby Rockefeller Mauze Trust, Dewitt Wallace and Maloris Foundations, and the NIH and acknowledges generous access to the New York Structural Biology Center’s NMR facilities.
Footnotes
Supplemental Data include five figures, two tables, Supplemental Experimental Procedures, and Supplemental References and can be found with this article online at http://www.molecule.org/cgi/content/full/24/5/785/DC1/.
Accession Numbers
The structural coordinates of YNG1 PHD protein free and in the complex with H31–9K4me3 peptide have been deposited in the Protein Data Base under accession codes 2JMI and 2JMJ, respectively.
References
- Agalioti T, Chen G, Thanos D. Deciphering the transcriptional histone acetylation code for a human gene. Cell. 2002;111:381–392. doi: 10.1016/s0092-8674(02)01077-2. [DOI] [PubMed] [Google Scholar]
- Bienz M. The PHD finger, a nuclear protein-interaction domain. Trends Biochem Sci. 2006;31:35–40. doi: 10.1016/j.tibs.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Dai J, Sultan S, Taylor SS, Higgins JM. The kinase haspin is required for mitotic histone H3 Thr 3 phosphorylation and normal metaphase chromosome alignment. Genes Dev. 2005;19:472–488. doi: 10.1101/gad.1267105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon Y, Selleck W, Lane WS, Tan S, Cote J. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol Cell Biol. 2004;24:1884–1896. doi: 10.1128/MCB.24.5.1884-1896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon Y, Cayrou C, Ullah M, Landry AJ, Cote V, Selleck W, Lane WS, Tan S, Yang XJ, Cote J. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol Cell. 2006;21:51–64. doi: 10.1016/j.molcel.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Eberharter A, John S, Grant PA, Utley RT, Workman JL. Identification and analysis of yeast nucleosomal histone acetyltransferase complexes. Methods. 1998;15:315–321. doi: 10.1006/meth.1998.0635. [DOI] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Allis CD. Binary switches and modification cassettes in histone biology and beyond. Nature. 2003;425:475–479. doi: 10.1038/nature02017. [DOI] [PubMed] [Google Scholar]
- Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438:1116–1122. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- Flanagan JF, Mi LZ, Chruszcz M, Cymborowski M, Clines KL, Kim Y, Minor W, Rastinejad F, Khorasanizadeh S. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- Grant PA, Duggan L, Cote J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, et al. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- Hazzalin CA, Mahadevan LC. Dynamic acetylation of all lysine 4-methylated histone H3 in the mouse nucleus: analysis at c-fos and c-jun. PLoS Biol. 2005;3:e393. doi: 10.1371/journal.pbio.0030393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe L, Auston D, Grant P, John S, Cook RG, Workman JL, Pillus L. Histone H3 specific acetyltransferases are essential for cell cycle progression. Genes Dev. 2001;15:3144–3154. doi: 10.1101/gad.931401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe L, Kusch T, Muster N, Chaterji R, Yates JR, 3rd, Workman JL. Yng1p modulates the activity of Sas3p as a component of the yeast NuA3 histone acetyltransferase complex. Mol Cell Biol. 2002;22:5047–5053. doi: 10.1128/MCB.22.14.5047-5053.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- John S, Howe L, Tafrov ST, Grant PA, Sternglanz R, Workman JL. The something about silencing protein, Sas3, is the catalytic subunit of NuA3, a yTAF(II)30-containing HAT Role of Yng1 in H3 Acetylation and Transcription 795 complex that interacts with the Spt16 subunit of the yeast CP (Cdc68/Pob3)-FACT complex. Genes Dev. 2000;14:1196–1208. [PMC free article] [PubMed] [Google Scholar]
- Kabani M, Michot K, Boschiero C, Werner M. Anc1 interacts with the catalytic subunits of the general transcription factors TFIID and TFIIF, the chromatin remodeling complexes RSC and INO80, and the histone acetyltransferase complex NuA3. Biochem Biophys Res Commun. 2005;332:398–403. doi: 10.1016/j.bbrc.2005.04.158. [DOI] [PubMed] [Google Scholar]
- Khorasanizadeh S. The nucleosome: from genomic organization to genomic regulation. Cell. 2004;116:259–272. doi: 10.1016/s0092-8674(04)00044-3. [DOI] [PubMed] [Google Scholar]
- Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo WS, Henry KW, Schwartz MF, Berger SL. Histone modification patterns during gene activation. Methods Enzymol. 2004;377:130–153. doi: 10.1016/S0076-6879(03)77007-4. [DOI] [PubMed] [Google Scholar]
- Loewith R, Meijer M, Lees-Miller SP, Riabowol K, Young D. Three yeast proteins related to the human candidate tumor suppressor p33ING1 are associated with histone acetyltransferase activities. Mol Cell Biol. 2000;20:3807–3816. doi: 10.1128/mcb.20.11.3807-3816.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DG, Baetz K, Shi X, Walter KL, Macdonald VE, Wlodarski MJ, Gozani O, Hieter P, Howe L. The yng1p plant homeodomain finger is a methyl-histone binding module that recognizes lysine 4-methylated histone H3. Mol Cell Biol. 2006a;26:7871–7879. doi: 10.1128/MCB.00573-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DG, Grimes DE, Baetz K, Howe L. Methylation of histone H3 mediates the association of the NuA3 histone acetyltransferase with chromatin. Mol Cell Biol. 2006b;26:3018–3028. doi: 10.1128/MCB.26.8.3018-3028.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor J. It takes a PHD to read the histone code. Cell. 2006;126:22–24. doi: 10.1016/j.cell.2006.06.028. [DOI] [PubMed] [Google Scholar]
- Milne TA, Dou Y, Martin ME, Brock HW, Roeder RG, Hess JL. MLL associates specifically with a subset of transcriptionally active target genes. Proc Natl Acad Sci USA. 2005;102:14765–14770. doi: 10.1073/pnas.0503630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- Nourani A, Howe L, Pray-Grant MG, Workman JL, Grant PA, Cote J. Opposite role of yeast ING family members in p53-dependent transcriptional activation. J Biol Chem. 2003;278:19171–19175. doi: 10.1074/jbc.C300036200. [DOI] [PubMed] [Google Scholar]
- Pena PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006;442:100–103. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry J. The Epc-N domain: a predicted protein-protein interaction domain found in select chromatin associated proteins. BMC Genomics. 2006;7:6. doi: 10.1186/1471-2164-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Pray-Grant MG, Daniel JA, Schieltz D, Yates JR, 3rd, Grant PA. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature. 2005;433:434–438. doi: 10.1038/nature03242. [DOI] [PubMed] [Google Scholar]
- Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Schneider R, Bannister AJ, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat Cell Biol. 2004;6:73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- Seet BT, Dikic I, Zhou MM, Pawson T. Reading protein modifications with interaction domains. Nat Rev Mol Cell Biol. 2006;7:473–483. doi: 10.1038/nrm1960. [DOI] [PubMed] [Google Scholar]
- Sendra R, Tse C, Hansen JC. The yeast histone acetyltransferase A2 complex, but not free Gcn5p, binds stably to nucleosomal arrays. J Biol Chem. 2000;275:24928–24934. doi: 10.1074/jbc.M003783200. [DOI] [PubMed] [Google Scholar]
- Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, III, Chen CF, Santos-Rosa H, Kouzarides T, Patel SS, Reinberg D. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J Biol Chem. 2005;280:41789–41792. doi: 10.1074/jbc.C500395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner DE, Belotserkovskaya R, Berger SL. SALSA, a variant of yeast SAGA, contains truncated Spt7, which correlates with activated transcription. Proc Natl Acad Sci USA. 2002;99:11622–11627. doi: 10.1073/pnas.182021199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Tackett AJ, DeGrasse JA, Sekedat MD, Oeffinger M, Rout MP, Chait BT. I-DIRT, a general method for distinguishing between specific and nonspecific protein interactions. J Proteome Res. 2005a;4:1752–1756. doi: 10.1021/pr050225e. [DOI] [PubMed] [Google Scholar]
- Tackett AJ, Dilworth DJ, Davey MJ, O’Donnell M, Aitchison JD, Rout MP, Chait BT. Proteomic and genomic characterization of chromatin complexes at a boundary. J Cell Biol. 2005b;169:35–47. doi: 10.1083/jcb.200502104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna SD, Coyne RS, Allis CD. Methylation of histone H3 at lysine 9 targets programmed DNA elimination in Tetrahymena. Cell. 2002;110:701–711. doi: 10.1016/s0092-8674(02)00941-8. [DOI] [PubMed] [Google Scholar]
- Turner BM. Histone acetylation and an epigenetic code. Bioessays. 2000;22:836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- Zhang K, Siino JS, Jones PR, Yau PM, Bradbury EM. A mass spectrometric “Western blot” to evaluate the correlations between histone methylation and histone acetylation. Proteomics. 2004;4:3765–3775. doi: 10.1002/pmic.200400819. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.