Abstract
Breast cancer metastasis suppressor 1 (BRMS1) is a metastasis suppressor gene in several solid tumors. The role of BRMS1 in non-small cell lung cancer (NSCLC) is not well established. To assess in vitro and in vivo metastatic behavior H1299 NSCLC cells stably expressing BRMS1 or a vector control were created. BRMS1 expression significantly decreases both migration and invasion of NSCLC cells in vitro. Importantly, in flank xenografts, BRMS1 suppresses the formation of pulmonary and hepatic metastases but does not significantly affect primary tumor growth. To evaluate whether BRMS1 is related to the progression of NSCLC, we examined BRMS1 expression in human NSCLC. Both BRMS1 mRNA and protein levels are diminished in NSCLC compared to adjacent non-cancerous lung. BRMS1 expression is also lower in squamous cell carcinoma compared to adenocarcinoma. Moreover, preservation of tumor BRMS1 expression is associated with improved patient survival. Thus, BRMS1 functions as a metastasis suppressor and may be a prognostic indicator for human NSCLC.
Keywords: BRMS1, Lung cancer, Metastasis, Prognosis
1. Introduction
Lung cancer remains the most common cause of cancer death in the United States in both men and women. Lung cancer has an overall 15% 5-year survival, largely secondary to the majority of patients having loco-regionally advanced or distant metastatic disease at initial presentation [1]. Approximately, 80% of lung cancer is non-small cell lung cancer (NSCLC). A better understanding of the biology contributing to the development and metastatic movement of NSCLC is important to improving clinical outcomes.
Metastasis is a multistep process, and interruption of this process at any individual step can arrest the metastatic cascade [2]. Welch and colleagues initially identified breast cancer metastasis suppressor 1 (BRMS1), a gene subsequently shown to suppress metastasis in multiple malignancies [3]. BRMS1 localizes predominantly (>90%) to the nucleus, is expressed in all normal human tissues, and is conserved across species with orthologs existing in mouse, rat, and dog as well as homologues in Drosophila [3,4].
BRMS1 has been distinctly classified as a metastasis suppressor gene; a class of genes defined by their ability to suppress metastatic movement while having little or no affect on primary tumor growth [5]. Metastasis suppressor genes are considered distinct from tumor suppressor genes whose hallmark is that their mutation predisposes the host to primary tumorigenesis [6]. BRMS1 is considered a true metastasis suppressor gene and was originally identified as such in breast cancer and melanoma cell lines where stable overexpression of BRMS1 resulted in suppression of pulmonary metastasis, but did not inhibit primary tumor growth [3,7]. Reduced BRMS1 protein expression has been correlated with reduced disease-free survival and poor prognosis in breast cancer [8,9] with breast cancer brain metastases having reduced BRMS1 expression [10]. While several genes act as metastasis suppressor genes in some models and as a tumor suppressor gene in other models [11], BRMS1 has only been reported to suppress metastases, not primary tumorigenesis or progression.
Limited data exists regarding BRMS1 in NSCLC. We, and others, recently have shown that BRMS1 mRNA and protein expression is decreased in small samples of NSCLC tumor compared to adjacent lung, and in NSCLC cells lines compared to bronchial epithelial cell lines [12,13]. It also has been demonstrated in NSCLC cells that transient over-expression of BRMS1 reduces CXCR4-induced chemotaxis while selective knockdown of BRMS1 promotes migration [14]. To date no animal models have been reported evaluating the role of BRMS1 in NSCLC and no data exists on the putative role of BRMS1 as a prognostic marker in NSCLC.
In this study, we identified that BRMS1 significantly decreases both NSCLC cell migration and invasion in our 2D and 3D cell culture models of invasion. Further studies in our murine NSCLC xenografts that stably express BRMS1 demonstrate a dramatic reduction in the number of pulmonary and hepatic metastases. Protein and mRNA analysis of human NSCLC specimens demonstrates a robust decrease in tumor BRMS1 levels which correlates with a worse clinical prognosis. Taken together this data suggests that BRMS1 functions as a potent suppressor of metastases in human NSCLC; furthermore, the loss of BRMS1 is associated with a decreased survival in patients with NSCLC.
2. Materials and methods
2.1. Cell culture, surgical specimens, reagents, and plasmid constructs
NCI-H1299 cells were obtained from American Type Culture Collection (Manassas, VA) and grown as described previously [15]. Human NSCLC specimens and adjacent non-cancerous lung were preserved in liquid nitrogen following standard surgical resection from patients at the Division of Thoracic Surgery, University of Virginia with individual informed consent and Human Investigations Committee approval. The antibodies used in this study were BRMS1 (Abnova Corporation, Taiwan); His-probe and α-tubulin (Sigma Aldrich, St. Louis, MO). The BRMS1 antibody used in this study has been used in previously published reports by our group [12] as well as others [16]. Recombinant TNF and TGFβ were purchased from Sigma Aldrich (St. Louis, MO).
2.2. Total RNA isolation, quantitative real-time RT-PCR
Total RNA was extracted using TRIzol (Invitrogen, Carlsbad, CA) according to manufacturer's protocol. cDNAs were synthesized using the Advantage RT for PCR enzyme kit (Clontech, Palo Alto, CA). BRMS1 expression was determined by quantitative real time RT-PCR as described previously [12]. Human HPRT gene was amplified as a reference gene.
2.3. Creation of NSCLC cells stably expressing BRMS1
The H1299 NSCLC cell line was stably transfected with a plasmid encoding the His(6)-tagged BRMS1 as described previously [15]. Briefly, parental H1299 cells were transfected with either 2 μg of the pcDNA-His(6)-tagged BRMS1 expression plasmid or a vector control plasmid utilizing Polyfect reagent (Qiagen, Valencia, CA). Cells were allowed to grow under neomycin (600 μg/ml) selection, and individual clones were subsequently harvested and expanded into cell lines. Western blots were analyzed for the expression of His(6)-tagged BRMS1 using a His-specific primary antibody (H-3). Clones expressing the His(6)-tagged BRMS1 were pooled and renamed H1299B while the vector control cells were renamed H1299V.
2.4. Western blot
Whole cell lysates were obtained and western blots were performed as previously described [12]. The primary antibodies were used at 1:1000 dilution and secondary antibodies (Promega, Madison, WI) were used at 1:5000 dilution.
2.5. Transwell migration and invasion assays
Migration and invasion of H1299 B/V cells was assessed using commercially available chambers (BD Biosciences, San Jose, CA) according to manufacturer protocol. Briefly, cells were harvested and 2.5 × 104 cells suspension was placed in the migration chamber. Chambers were incubated at 37 °C for 20 h. After fixing and drying, cells were photographed and counted. Parallel experiments were performed in triplicate to assess migration in control chambers and invasion.
2.6. 3-D matrix gel invasion assays
H1299 V/B cells were diluted to 8 × 105 cells/ml and drops of the cell suspension (25 μl) were placed onto the lids of 100 mm dishes, which were inverted over dishes containing 10 ml RPMI. Hanging drop cultures were incubated, and after 5 days the spheroids of both cell lines were transferred and implanted into a three-dimensional collagen I gels including 2.5 mg/ml rat collagen I (R&D systems, Inc, Minneapolis, MN), and overlaid with 200 νl RPMI with or without TNF (20 ng/ml) and TGFβ (2 ng/ml). The maximal invasion distances were measured from the center of the spheroids to the population of invading cells most distant from the spheroids 18 h post-implantation. This method has been demonstrated to be a reproducible approach to determining invasive potential in implantable spheroids [17].
2.7. NSCLC murine xenograft models
NSCLC xenografts were created as previously described [18]. The flanks of 4 week-old athymic nude mice (Taconic, Germantown, NY) were inoculated with 0.1 ml of serum free and antibiotic free media containing 2 × 106 cells (H1299 parental, H1299B, and H1299V, N = 6/group). While orthotopic models of lung cancer xenografts are well described [19], we chose a heterotopic flank model to allow us to more readily to determine any potential effect of BRMS1 on primary tumor growth. Mice were then maintained under standard animal husbandry conditions. Tumors were measured twice per week using digital calipers and volumes were calculated (volume = long axis × short axis2). Mice were euthanized at the end of the study period. Necropsy was performed including weighing the whole mouse, the primary tumor, the lungs, the liver, and examining the lungs and the livers under a dissecting microscope to count visible lung surface tumor metastatic deposits. “Tumor weight index” is defined as the weight of the primary flank tumor divided by whole body weight of the mouse.
2.8. NSCLC tissue microarrays (TMA)
We created a TMA containing 80 NSCLC and adjacent non-cancerous tissue. Representative pathologic NSCLC stages and histologies from patients undergoing attempted curative resection for NSCLC at our institution were used. All patients had an R0 resection. Patients with small cell cancer, carcinoid tumors, or those with preoperative chemotherapy or radiotherapy were excluded.
Each NSCLC specimen block was reviewed microscopically by a pathologist. Non-necrotic tumor and surrounding areas of immediately adjacent, non-cancerous, parenchyma were marked. A map of the receiver blocks was prepared with coordinates for each sample to correctly identify the tumour samples. Using a tissue micro arrayer (Beecher Instruments, Woodland, USA), 0.6 mm diameter cores were punched from the donor blocks and positioned in a recipient paraffin array block in 0.4 mm holes on 0.8 mm centers. For each patient sample, three tumor cores were taken from different areas of the marked tumor. In blocks which contained non-cancerous adjacent stroma, a single core of non-cancerous stroma was also taken. Cores of spleen and kidney were placed at the edge of the block for orientation and control specimens. The array blocks were then incubated for 30 min at 37 °C. The blocks were cut with a standard microtome (Microm, Heidelberg, Germany).
After immunostaining, the TMA was reviewed by a pathologist. Since BRMS1 is principally a nuclear protein [4], the level of nuclear expression was scored for percent of cells with positive nuclear staining for BRMS1 which was graded from 1 to 4 and intensity of nuclear staining which was graded from 1 to 3. The product of the percentage and intensity is expressed as the scoring index (range 1–12) [20]. Clinical and pathological variables regarding the tumor and the patient, including smoking status as measured by pack-years (duration × intensity of smoking) were collected from medical records following approval from our institutional review board.
2.9. Statistical analysis
Results of all experiments represent the mean ± S.D. of three separate experiments performed in triplicate, unless otherwise noted. Chi square, Fisher's exact, and Wilcoxon two sample non-parametric tests were used as appropriate. Kaplan–Meier survival estimates were used to evaluate BRMS1 expression and patient survival and difference between groups determined by log-rank test. A p-value of <0.05 was considered significant for all calculations. Statistical analysis was performed using SAS version 9.1 for Windows (SAS Institute, Cary, North Carolina).
3. Results
3.1. BRMS1 expression inhibits migration and invasion in NSCLC cells
To explore the effect of BRMS1 on lung cancer cell invasion we created H1299B/V NSCLC cell lines that stably express BRMS1 or an empty vector. Western blot of whole cell lysates confirmed the absence of BRMS1 protein in H1299 parental and H1299V cells compared to strong expression in H1299B cells (Fig. 1A). In vitro models of migration and invasion were performed using H1299 B/V cells lines. As shown in Fig. 1A, cells with ectopic expression of BRMS1 demonstrated significantly less migration and invasion compared to H1299V cells. These results are further quantified by calculating the migration ratio and invasion ratio per the manufacturer's protocol. The H1299B/H1299V migration ratio was 0.65 and the invasion ratio was 0.53 (both p < 0.001 by t-test), indicating significantly less migration and invasion of NSCLC cells in the presence of BRMS1.
Fig. 1.
BRMS1 expression inhibits migration and invasion in NSCLC cells. (A) pcDNA-BRMS1 or pcDNA empty vector (control) was stably transfected into H1299 cells. Western blot, with α-tubulin as loading control, shows strong staining for BRMS1 in H1299 NSCLC cells stably expressing BRMS1 (H1299B or B), compared to H1299 cells stably expressing empty vector (H1299V or V) and parental H1299 cells. Migration and invasion were assessed using commercially available chambers (BD Biosciences). Results are expressed as mean ± standard deviation of parallel experiments performed in triplicate. Photomicrographs taken under 20× magnification. (†p<0.001 compared to H1299 V). (B) H1299 B/V 3D cell spheroids were prepared by hanging drop culture for 5 days and implanted into 3D Culture Matrix Collagen I gel in the presence or absence TNF (20 ng/ml)/TGFβ (2 ng/ml). Maximal invasion distances were measured 18 h later. Values are expressed as mean ± SD, and N = 6. Photomicrographs of indicated spheroids show individual cells invading through the collagen I matrix in a sun-burst pattern 18 h post-implantation. (*p < 0.01 compared to H1299V No add; § p < 0.01 compared to H1299B No add).
To further investigate the role of BRMS1 in regulating cellular movement, the same cell lines were grown in a hanging drop model and implanted into a 3D culture matrix collagen gel. We treated the cells with TNF and TGFβ as these cytokines are needed for epithelial-mesenchymal transition (EMT) and closely mimic the natural peri-tumoral stromal environment. As seen in Fig. 1B, in the absence of stimulus, H1299B cells demonstrated very little movement, compared to H1299V cells which demonstrated much more spread. Movement was stimulated by TNF and TGFβ compared to no-add in both cell lines, but remained significantly inhibited in the presence of BRMS1 expression, compared to the absence of BRMS1. These findings, together with the results seen in the transwell assays, strongly suggest that BRMS1 is a metastasis suppressor in NSCLC.
3.2. BRMS1 inhibits metastases in NSCLC xenografts
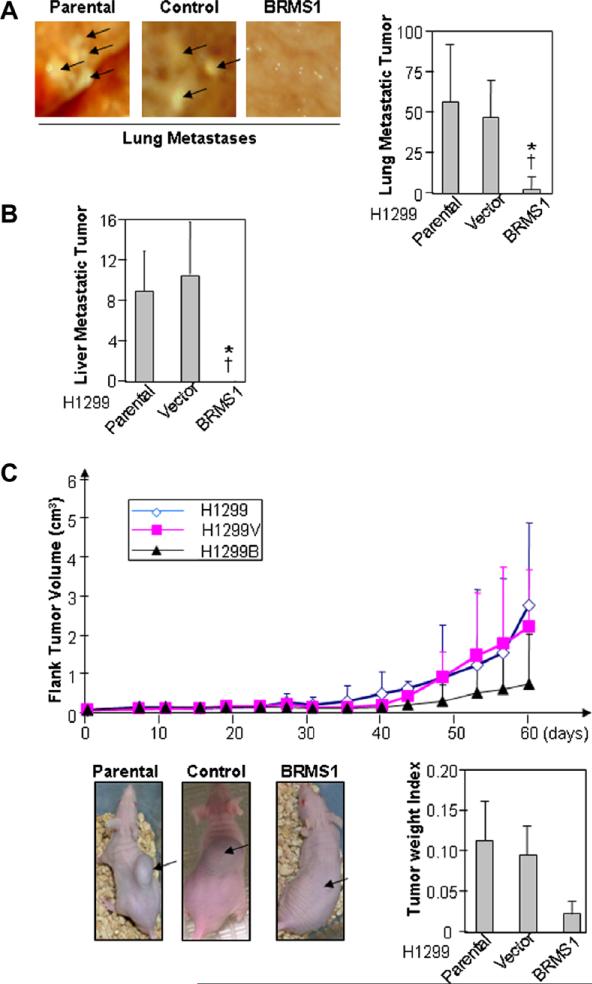
Given our in vitro findings, we next sought to determine if similar observations would be found in an animal model of NSCLC. Using the H1299 B/V and parental cells, flank tumors were created in athymic nude mice. Fig. 2A demonstrates the effect of BRMS1 on the development of pulmonary metastases in our flank xenograft model. Representative images of the visceral pleural surface of the murine lung are shown and demonstrate significantly lower metastatic burden to the lungs in the H1299B xenografts compared to the control or parental tumors. This observation is quantified in the associated bar graphs, demonstrating significantly lower metastatic burden to the lung in the presence of BRMS1 (p<0.01). Decreased metastatic spread in this model is further confirmed by the lack of liver metastases in the H1299B xenografts compared to parental and vector control, which is graphically represented in Fig. 2B.
Fig. 2.
BRMS1 expression inhibits metastasis in a mouse model of NSCLC. (A) Mouse flank tumor xenografts were created using the H1299, H1299B, and H1299V cell lines, N = 6 animals per group. Pulmonary metastases were measured as described in Section 2 at necropsy. Graphic data are expressed as the mean ± SEM (*p < 0.01 vs. H1299 parental, †p < 0.01 vs. H1299V). (B) Liver metastases were measured as described in Section 2 at necropsy. Graphic data are expressed as the mean ± SEM. (*p < 0.01 vs. H1299 parental, †p < 0.01 vs. H1299V). (C) Primary tumor growth was measured volumetrically over time and primary tumor weight measured at necropsy. Photographs demonstrate representative flank tumors for the three cell lines, all taken on the same day post inoculation.
As seen in Fig. 2C, in representative photographs as well as graphically, ectopic expression of BRMS1 resulted in slower absolute growth of the primary tumor, although this was not statistically significant (p = 0.37). This is true based both on volume calculations as well as the tumor weight index at necropsy (Fig. 2).
3.3. BRMS1 transcription is diminished in human NSCLC
We previously have shown that BRMS1 expression is decreased in all five NSCLC cell lines tested compared to NL20 immortalized bronchial epithelial cells and HEK293T cells by real-time RT-PCR [12]. Expanding this type of analysis to human lung cancer specimens we investigated the expression of BRMS1 mRNA in surgically resected human NSCLC. Samples of NSCLC and matched adjacent lung from the same patient were selected from our institutionally-approved tissue bank. Patients (N = 12) were selected to represent a distribution of histology and pathologic stage. QRT-PCR identified that BRMS1 mRNA expression was decreased compared to patient-matched adjacent lung for each sample tested (Fig. 3A). The patient with the stage IIIB adenocarcinoma who had a higher than expected BRMS1 mRNA level had a T4N0M0 tumor with the T4 classification based on multiple tumors in the same lobe and absence of lymphatic spread.
Fig. 3.
BRMS1 mRNA expression is diminished in human NSCLC. (A) BRMS1 gene expression was measured by QRT-PCR using patient tumor samples and matched adjacent, non-cancerous lung. Adjacent lung was normalized to one. Tumor histologic type and pathologic TNM stage [29] are given for each sample. (B) BRMS1 mRNA expression grouped by pathologic node status (*p = 0.002 compared to node-negative). (C) BRMS1 mRNA expression grouped by tumor histologic classification (†p < 0.001 compared to adenocarcinoma).
Additionally, significantly higher levels of BRMS1 transcription were seen in tumors without pathologic evidence of lymphatic spread (Fig. 3B). Interestingly, NSCLC with squamous cell histology demonstrated significantly less BRMS1 transcription than that seen in tumors with adenocarcinoma histology (Fig. 3C).
3.4. BRMS1 protein expression is decreased in human NSCLC and correlates with patient smoking history
To determine whether meaningful clinical correlations exist with BRMS1 expression, we used our NSCLC TMA. As seen in the representative photomicrographs (Fig. 4A), there was markedly less BRMS1 immuno-staining in the tumor compared to the patient matched, non-cancerous lung tissue. The mean scoring index for non-cancerous lung was 10 ± 0.7, compared to 4.7 ± 0.6 for NSCLC. Fig. 4B graphically represents the lower BRMS1 expression in NSCLC compared to matched non-cancerous lung for these 80 patients. The mean scoring index difference between patient matched tumor and adjacent lung was 5.3 ± 0.9.
Fig. 4.
BRMS1 protein is down-regulated in human NSCLC and loss of BRMS1 expression correlates with decreased patient survival. (A) Representative 100× photomicrographs showing nuclear staining for BRMS1 in non-cancerous lung and NSCLC. (B) Scoring index for BRMS1 nuclear staining for 80 matched samples of NSCLC and adjacent, non-cancerous lung. Box extends from 25th to 75th percentile with line at median (no line is seen for “non-tumor” because the median is 12, the upper limit of the range). Whiskers cover full range of data (*p < 0.0001 between NSCLC and patient-matched adjacent, non-cancerous lung). (C) Kaplan–Meier actuarial survival plot with time 0 being the date of lung cancer resection. Tick marks represent either patient death or censor (†p = 0.02, log-rank test).
To determine if BRMS1 levels were associated with cigarette smoking we compared BRMS1 expression levels as measured by our scoring index. Interestingly, patient smoking history as measured by pack-years had a positive correlation with decreases in BRMS1 protein scores (Spearman rank correlation 0.23, p < 0.037, 95%CI (0.01, 0.43). This suggests that heavy tobacco use may be a potential etiology of BRMS1 expression levels and this observation will require further investigation.
3.5. Preservation of BRMS1 expression in NSCLC correlates with improved patient survival
The BRMS1 scoring index was lower in 85% of tumors than in patient-matched adjacent lung (Fig. 4B). As a measure of preservation or loss of BRMS1 expression in tumor compared to adjacent lung, we calculated a gradient of BRMS1 expression for each matched pair of NSCLC compared to adjacent lung. “BRMS1 lost” was defined as a scoring index that was >4 points lower in the tumor compared to matched non-cancerous lung. The corollary “BRMS1 preserved” was defined as a scoring index that was ≤4 points lower in the tumor than in the matched non-cancerous lung. The clinical and pathologic variables for the patients are shown in Table 1.
Table 1.
Clinicopathologic variables and BRMS1 expression in human NSCLC.
| All | BRMS1 lost | BRMS1 preserved | p Value | |
|---|---|---|---|---|
| N | 80 | 61 | 19 | |
| Female | 39 (49%) | 27 (44%) | 12 (63%) | 0.15a |
| Non-caucasian | 10 (l3%) | 7 (12%) | 3 (16%) | 0.69b |
| Median age | 66 (58-73) | 66 (57-72) | 66 (58-75) | 0.86c |
| Histologic type | ||||
| Adeno | 45 (56%) | 31 (51%) | 14 (74%) | 0.009b |
| Squamous | 29 (36%) | 27 (44%) | 2 (11%) | |
| Other | 6 (8%) | 3 (5%) | 3 (16%) | |
| Histologic grade | ||||
| G1 | 8 (10.0%) | 6 (9.8%) | 2 (10.5%) | 0.30b |
| G2 | 35 (43.8%) | 25 (41.0%) | 10 (52.6%) | |
| G3 | 33 (41.3%) | 28 (45.9%) | 5 (26.3%) | |
| G4 | 4 (5.0%) | 2 (3.3%) | 2 (10.5%) | |
| Median size (cm) | 2.7 (2.0-4.5) | 3.0 ± (2.0-5.0) | 2.5 (2.0-3.0) | 0.15c |
| Node status | ||||
| N0 | 59 (74%) | 44 (72%) | 15 (79%) | >0.99b |
| N1 | 10 (13%) | 8 (13%) | 2 (11%) | |
| N2 | 11 (14%) | 9 (15%) | 2 (11%) | |
| TNM stage | ||||
| I | 51 (63.8%) | 36 (59.0%) | 15 (78.9%) | 0.61b |
| II | 9 (11.3%) | 7 (11.5%) | 2 (10.5%) | |
| III | 15 (18.8%) | 12 (19.7%) | 3 (15.8%) | |
| IV | 5 (6.3%) | 5 (8.2%) | 0 (0.0%) |
Chi square.
Fisher's exact.
Wilcoxon two sample non-parametric test.
Patients with preserved tumor BRMS1 expression more frequently had adenocarcinoma histology tumors (as also was seen at the mRNA level above), and demonstrated trends towards being female and having smaller tumors. No correlation was seen between preservation of BRMS1 tumor expression and race, age, histologic grade, presence of lymphatic spread, and TNM staging.
Kaplan–Meier survival estimates were performed based on whether they had preserved vs. lost BRMS1 expression, as defined above. As shown in Fig. 4C, preserved expression of BRMS1 in NSCLC is significantly associated with improved survival compared to those patients with tumors who have lost BRMS1 expression (p = 0.02). The 5-year survival for patients with tumors in which BRMS1 expression was preserved was 75%, while those with a loss of BRMS1 had an actuarial 5-year survival of only 33% (Fig. 4C).
4. Discussion
BRMS1 has previously been demonstrated to function as a metastasis suppressor gene in in vivo models of breast cancer [3,21,22], melanoma [7], and ovarian cancer [23]. In this study we have shown that BRMS1 acts as a metastasis suppressor in NSCLC in cell culture models of metastatic movement, and in flank xenograft models. Consistent with reports of BRMS1 in other malignancies, we observed no significant effect of BRMS1 on growth of the primary tumor. This supports that BRMS1 functions as a metastasis suppressor gene in NSCLC. It is interesting to note that there was a statistically non-significant trend towards lower volume of the primary tumor in cells expressing BRMS1 compared to controls. Further study is needed to determine if any tumor suppressor function of BRMS1 exists in NSCLC.
Prior studies have revealed lower levels of BRMS1 in cancer cell lines compared to non-malignant cells, as well as lower levels of BRMS1 in patient tumors in malignancies including breast [3,8,9], melanoma [7], bladder [24], and ovarian cancer [25]. We, and others, recently have shown limited data regarding expression of both BRMS1 protein and mRNA in NSCLC cell lines [12,13]. Data in this study extends and confirms those findings of decreased BRMS1 expression in human NSCLC specimens, compared to patient matched adjacent lung.
Interestingly, we found a marked difference in BRMS1 mRNA and protein in different NSCLC histologies, with squamous histology tumors having much lower levels of BRMS1 than adenocarcinomas. This, coupled with our observation that BRMS1 has prognostic value, suggests that it may be candidate for inclusion in future NSCLC prognostic models.
We also found that patients with pathologically confirmed nodal metastasis had lower levels of BRMS1 mRNA than patients with node-negative disease. However, when we examined BRMS1 protein levels by immunohistochemistry in our larger TMA sample, we found no correlation between nodal spread and BRMS1. Similar seemingly discordant findings have been reported for BRMS1 in breast cancer [8]. The difference between our mRNA and protein data may be due to sample size, or could be a result of post-translational modifications that result in BRMS1 protein stabilization and increased immunopositivity. Finally, it is well recognized that standard histopathologic techniques lack sensitivity and can underestimate the true incidence of node-positive disease [25].
An important, and potentially clinically relevant, observation of this study is that loss of BRMS1 protein in the primary tumor negatively correlated with patient survival. These observations were independent of tumor stage. Our findings are similar to those observed in breast cancer where expression of BRMS1 in the primary breast cancer tumor did not correlate with axillary nodal metastases, but does correlate significantly with overall survival [8,26].
The mechanisms of BRMS1 as a metastasis suppressor gene are not clearly elucidated. While genomic deletion has not been found to be a major cause for loss of BRMS1 expression [8], BRMS1 is known to be a target of epigenetic silencing where a CpG island in the BRMS1 promoter is hypermethylated across breast cancer cell lines and in patient samples in 45% of primary tumors, and 60% of matched lymph node metastases [27]. We have reported similar BRMS1 promoter methylation in our lung cancer cells and human tumors [28]. Future studies of the mechanism of BRMS1 function will be important, particularly if restoration of intratumoral BRMS1 levels were to become a desirable therapeutic strategy.
In conclusion, using both in vitro and in vivo model systems we have shown that BRMS1 is a metastasis suppressor gene in NSCLC. We have provided the initial observations that expression of BRMS1 in NSCLC has prognostic relevance as a predictor of survival with tumor loss of BRMS1 expression portending a worse survival. Given the increasing appreciation of the importance of BRMS1 in NSCLC, as well as other tumors, future studies will need to focus on how BRMS1 is regulated and the mechanisms of its anti-metastatic functions, with the goal being to critically examine the possibilities of exploiting BRMS1 as a therapeutic target.
Supplementary Material
Acknowledgments
Supported, in part, through grants from the University of Virginia Cancer Center (to DRJ) and from the National Institutes of Health T32 HL007849 (to PWS).
Footnotes
Conflicts of interest statement
None declared.
Appendix A. Supplementary material
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.canlet. 2008.11.024.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA: A Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Fidler IJ. Critical factors in the biology of human cancer metastasis. Am Surg. 1995;61:1065–1066. [PubMed] [Google Scholar]
- 3.Seraj MJ, Samant RS, Verderame MF, Welch DR. Functional evidence for a novel human breast carcinoma metastasis suppressor, BRMS1, encoded at chromosome 11q13. Cancer Res. 2000;60:2764–2769. [PubMed] [Google Scholar]
- 4.Samant RS, Seraj MJ, Saunders MM, Sakamaki TS, Shevde LA, Harms JF, Leonard TO, Goldberg SF, Budgeon L, Meehan WJ, Winter CR, Christensen ND, Verderame MF, Donahue HJ, Welch DR. Analysis of mechanisms underlying BRMS1 suppression of metastasis. Clin Exp Metastasis. 2000;18:683–693. doi: 10.1023/a:1013124725690. [DOI] [PubMed] [Google Scholar]
- 5.Shevde LA, Welch DR. Metastasis suppressor pathways –an evolving paradigm. Cancer Lett. 2003;198:1–20. doi: 10.1016/s0304-3835(03)00304-5. [DOI] [PubMed] [Google Scholar]
- 6.Hasty P. Is NHEJ a tumor suppressor or an aging suppressor? Cell Cycle. 2008;7:1139–1145. doi: 10.4161/cc.7.9.5807. [DOI] [PubMed] [Google Scholar]
- 7.Shevde LA, Samant RS, Goldberg SF, Sikaneta T, Alessandrini A, Donahue HJ, Mauger DT, Welch DR. Suppression of human melanoma metastasis by the metastasis suppressor gene, BRMS1. Exp Cell Res. 2002;273:229–239. doi: 10.1006/excr.2001.5452. [DOI] [PubMed] [Google Scholar]
- 8.Hicks DG, Yoder BJ, Short S, Tarr S, Prescott N, Crowe JP, Dawson AE, Budd GT, Sizemore S, Cicek M, Choueiri TK, Tubbs RR, Gaile D, Nowak N, Accavitti-Loper MA, Frost AR, Welch DR, Casey G. Loss of breast cancer metastasis suppressor 1 protein expression predicts reduced disease-free survival in subsets of breast cancer patients. Clin Cancer Res. 2006;12:6702–6708. doi: 10.1158/1078-0432.CCR-06-0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Z, Yamashita H, Toyama T, Yamamoto Y, Kawasoe T, Iwase H. Reduced expression of the breast cancer metastasis suppressor 1 mRNA is correlated with poor progress in breast cancer. Clin Cancer Res. 2006;12:6410–6414. doi: 10.1158/1078-0432.CCR-06-1347. [DOI] [PubMed] [Google Scholar]
- 10.Stark AM, Tongers K, Maass N, Mehdorn HM, Held-Feindt J. Reduced metastasis-suppressor gene mRNA-expression in breast cancer brain metastases. J Cancer Res Clin Oncol. 2005;131:191–198. doi: 10.1007/s00432-004-0629-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stafford LJ, Vaidya KS, Welch DR. Metastasis suppressors genes in cancer. Int J Biochem Cell Biol. 2008;40:874–891. doi: 10.1016/j.biocel.2007.12.016. [DOI] [PubMed] [Google Scholar]
- 12.Liu Y, Smith PW, Jones DR. Breast cancer metastasis suppressor 1 functions as a corepressor by enhancing histone deacetylase 1-mediated deacetylation of RelA/p65 and promoting apoptosis. Mol Cell Biol. 2006;26:8683–8696. doi: 10.1128/MCB.00940-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang J, Zhang B, Lin Y, Yang Y, Liu X, Lu F. Breast cancer metastasis suppressor 1 inhibits SDF-1alpha-induced migration of non-small cell lung cancer by decreasing CXCR4 expression. Cancer Lett. 2008 doi: 10.1016/j.canlet.2008.04.016. [DOI] [PubMed] [Google Scholar]
- 14.Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, Mayo MW. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. Embo J. 2004;23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones DR, Broad RM, Madrid LV, Baldwin AS, Jr., Mayo MW. Inhibition of NF-kappaB sensitizes non-small cell lung cancer cells to chemotherapy-induced apoptosis. Ann Thorac Surg. 2000;70:930–936. doi: 10.1016/s0003-4975(00)01635-0. [DOI] [PubMed] [Google Scholar]
- 16.Rivera J, Megias D, Bravo J. Proteomics-based strategy to delineate the molecular mechanisms of the metastasis suppressor gene BRMS1. J Proteome Res. 2007;6:4006–4018. doi: 10.1021/pr0703167. [DOI] [PubMed] [Google Scholar]
- 17.Del Duca D, Werbowetski T, Del Maestro RF. Spheroid preparation from hanging drops: characterization of a model of brain tumor invasion. J Neurooncol. 2004;67:295–303. doi: 10.1023/b:neon.0000024220.07063.70. [DOI] [PubMed] [Google Scholar]
- 18.Denlinger CE, Rundall BK, Jones DR. Inhibition of phosphatidylinositol 3-kinase/Akt and histone deacetylase activity induces apoptosis in non-small cell lung cancer in vitro and in vivo. J Thorac Cardiovasc Surg. 2005;130:1422–1429. doi: 10.1016/j.jtcvs.2005.06.051. [DOI] [PubMed] [Google Scholar]
- 19.Ueda K, Kawashima H, Ohtani S, Deng WG, Ravoori M, Bankson J, Gao B, Girard L, Minna JD, Roth JA, Kundra V, Ji L. The 3p21.3 tumor suppressor NPRL2 plays an important role in cisplatin- induced resistance in human non-small-cell lung cancer cells. Cancer Res. 2006;66:9682–9690. doi: 10.1158/0008-5472.CAN-06-1483. [DOI] [PubMed] [Google Scholar]
- 20.Maaser K, Daubler P, Barthel B, Heine B, von Lampe B, Stein H, Hoffmeister B, Scherer H, Scherubl H. Oesophageal squamous cell neoplasia in head and neck cancer patients: upregulation of COX-2 during carcinogenesis. Br J Cancer. 2003;88:1217–1222. doi: 10.1038/sj.bjc.6600865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hedley BD, Welch DR, Allan AL, Al-Katib W, Dales DW, Postenka CO, Casey G, Macdonald IC, Chambers AF. Downregulation of osteopontin contributes to metastasis suppression by breast cancer metastasis suppressor 1. Int J Cancer. 2008;123:526–534. doi: 10.1002/ijc.23542. [DOI] [PubMed] [Google Scholar]
- 22.Samant RS, Debies MT, Hurst DR, Moore BP, Shevde LA, Welch DR. Suppression of murine mammary carcinoma metastasis by the murine ortholog of breast cancer metastasis suppressor 1 (Brms1) Cancer Lett. 2006;235:260–265. doi: 10.1016/j.canlet.2005.04.032. [DOI] [PubMed] [Google Scholar]
- 23.Zhang S, Lin QD, Di W. Suppression of human ovarian carcinoma metastasis by the metastasis-suppressor gene. BRMS1, Int J Gynecol Cancer. 2006;16:522–531. doi: 10.1111/j.1525-1438.2006.00547.x. [DOI] [PubMed] [Google Scholar]
- 24.Seraj MJ, Harding MA, Gildea JJ, Welch DR, Theodorescu D. The relationship of BRMS1 and RhoGDI2 gene expression to metastatic potential in lineage related human bladder cancer cell lines. Clin Exp Metastasis. 2000;18:519–525. doi: 10.1023/a:1011819621859. [DOI] [PubMed] [Google Scholar]
- 25.Ito M, Minamiya Y, Kawai H, Saito S, Saito H, Imai K, Ogawa J. Intraoperative detection of lymph node micrometastasis with flow cytometry in non-small cell lung cancer. J Thorac Cardiovasc Surg. 2005;130:753–758. doi: 10.1016/j.jtcvs.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 26.Kelly LM, Buggy Y, Hill A, O'Donovan N, Duggan C, McDermott EW, O'Higgins NJ, Young L, Duffy MJ. Expression of the breast cancer metastasis suppressor gene, BRMS1, in human breast carcinoma: lack of correlation with metastasis to axillary lymph nodes. Tumour Biol. 2005;26:213–216. doi: 10.1159/000086955. [DOI] [PubMed] [Google Scholar]
- 27.Metge BJ, Frost AR, King JA, Dyess DL, Welch DR, Samant RS, Shevde LA. Epigenetic silencing contributes to the loss of BRMS1 expression in breast cancer. Clin Exp Metastasis. 2008 doi: 10.1007/s10585-008-9187-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y, Smith PW, Taylor MD, Jones DR. BRMS1 transcription is repressed by DNA methylation and functions as a both a metastasis and tumor suppressor gene in non-small cell lung cancer. Proc Am Assn Cancer Res. 2008;326 [Google Scholar]
- 29.Mountain CF. Revisions in the international system for staging lung cancer. Chest. 1997;111:1710–1717. doi: 10.1378/chest.111.6.1710. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.