Abstract
The T cell receptor (TCR) is unique in that its affinity for ligand is unknown prior to encounter and can vary by orders of magnitude. How the immune system regulates individual T cells that display highly different reactivity to antigen remains unclear. Here we identified that activated CD4+ T cells, at the peak of clonal expansion, persistently downregulate TCR expression in proportion to the strength of initial antigen recognition. This programmed response increases the threshold for cytokine production and recall proliferation in a clone-specific manner, ultimately excluding clones with the highest antigen reactivities. Thus, programmed TCR downregulation represents a negative feedback mechanism to constrain T cell effector function with a suitable time delay, thereby allowing pathogen control while avoiding excess inflammatory damage.
INTRODUCTION
Efficient recognition and control of microbial infections relies on the availability of a diverse TCR repertoire1,2. In developing T cells, TCR diversity is generated via V(D)J recombination, during which different gene segments are combined with nucleotide addition and removal at recombination junctions. This process results in TCRs with highly variable sequences, and therefore generates individual T cells that recognize peptide–MHC with a wide range of binding strengths, or avidities. This has important functional consequences, as high avidity T cells are generally more sensitive to antigen, undergo more proliferation and produce more cytokines3–9. However, T cells at the upper end of the avidity spectrum can also display substantial limitations in biological activity10–13, suggesting the existence of regulatory mechanisms that prevent deleterious consequences of very strong antigen reactivity. Hence, the optimal avidity associated with long-term T cell functionality remains elusive, particularly in the context of immunity against microbial infections.
Mycobacterium tuberculosis remains one of the most detrimental human pathogens and has been challenging to combat due to lack of an effective vaccine14,15. Although CD4+ T cells are essential to control M. tuberculosis16,17, bacterial eradication is rarely achieved. One explanation is that M. tuberculosis is capable of inhibiting MHC class II expression18,19, resulting in limited antigen presentation in mycobacterial granulomas20,21. Therefore, one way to improve vaccination against tuberculosis might be to engage T cells of an overall very high avidity, because these would require less antigen to activate effector mechanisms.
To investigate how T cell avidity influences immunity against M. tuberculosis, we generated a TCR transgenic mouse in which CD4+ T cells have very high avidity for the mycobacterial antigen ESAT6(1–20). By comparing this mouse to a previously described TCR transgenic with intermediate avidity for ESAT622,23, we found that the higher avidity T cells were less capable of controlling bacterial growth; a phenotype that was associated with a drastic loss in TCR expression. Based on this, we found that clonally expanded T cells undergo a program of persistent TCR downregulation that is proportional to the strength of initial antigen recognition. This previously unappreciated feedback mechanism increases the threshold for cytokine production and recall proliferation in an avidity-dependent manner, resulting in intermediate avidity T cells having the greatest contribution to anti-microbial defense.
RESULTS
Avidity of C7 and C24 CD4+ T cells
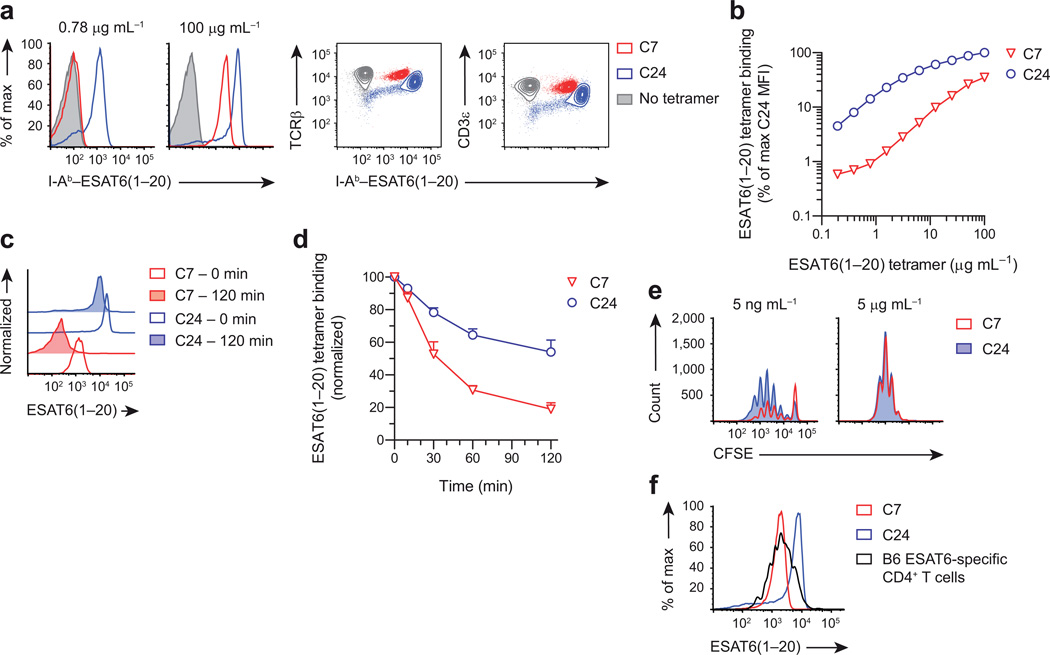
To investigate CD4+ T cell immunity against tuberculosis, we previously generated a TCR transgenic mouse, called C7, with all CD4+ T cells specific for the mycobacterial antigen ESAT6(1–20). Using this mouse, we demonstrated that TH1-differentiated C7 cells are capable of greatly reducing, but not eliminating mycobacterial growth22,23. To examine whether this failure of bacterial eradication is a general feature of T cell immunity, we generated a second TCR transgenic using an ESAT6(1–20)-specific T cell hybridoma, called C24, which was obtained from an M. tuberculosis-infected mouse. We chose the C24 TCR because it uses different α and β chains compared to the C7 TCR, creating the possibility that it recognizes ESAT6(1–20) with different affinity. To test this, we incubated naïve C7 and C24 T cells with different concentrations of ESAT6(1–20) tetramers, to evaluate their antigen binding strength24. At saturating concentrations, C24 T cells bound 3-fold more tetramer compared to C7 T cells, and this difference increased to 15-fold at non-saturating concentrations (Fig. 1a,b). Since TCR expression of naïve C7 and C24 T cells was comparable (Fig. 1a), this suggested that C24 has substantially higher structural avidity for ESAT6(1–20); which was confirmed by tetramer dissociation studies (Fig. 1c,d). To assess whether C7 and C24 T cells also differ in functional avidity, we activated these cells with different concentrations of ESAT6(1–20) peptide. At low antigen dose, 45% of C24 T cells had proliferated after 4 days compared to only 18% of C7 T cells, whereas at high antigen dose there was no difference (Fig. 1e), indicating that TCR stimulation of C24 was stronger. Consistent with this, activated C24 T cells also displayed increased CTLA-4 expression (Supplementary Fig. 1)25.
Figure 1.
C7 and C24 are TCR transgenic CD4+ T cells with intermediate and very high avidity for ESAT6(1–20). (a) Flow cytometry showing I-Ab–ESAT6(1–20) tetramer binding of naïve C7 and C24 T cells stained with different tetramer concentrations. The level of TCRβ and CD3ε expression is shown as a control. (b) Quantification of ESAT6(1–20) tetramer binding as a function of tetramer concentration. Tetramer binding was quantified using the median fluorescence intensity (MFI) and normalized to C24. (c) Flow cytometry showing ESAT6(1–20) tetramer dissociation of naïve C7 and C24 T cells stained with 50 µg mL−1 tetramer. (d) Quantification of ESAT6(1–20) tetramer dissociation as a function of time. (e) Flow cytometry showing CFSE dilution of C7 and C24 T cells activated in vitro with different ESAT6(1–20) peptide concentrations for 4 days. (f) Flow cytometry showing ESAT6(1–20) tetramer binding of C7 and C24 T cells, as well as endogenous CD4+ T cells stained with 50 µg mL−1 tetramer. The latter cells were pulled-down from the lungs of a representative C57BL/6J (B6) mouse, 28 days after infection with M. tuberculosis. Data are from one experiment representative of two independent experiments (mean + s.d. of n = 3 male mice per group (d) or n = 5 male mice per group (f)).
Because both C7 and C24 T cells were generated using TCRs of M. tuberculosis-infected mice, we sought to compare their relative avidity to endogenous ESAT6(1–20)-specific CD4+ T cells. Therefore, we infected mice with M. tuberculosis and pulled-down ESAT6(1–20) tetramer binding cells at the peak of the response. We observed a normal distribution of tetramer binding by endogenous CD4+ T cells, spanning two orders of magnitude, likely reflecting cells with different avidities for ESAT6(1–20) (Fig. 1f). Notably, ~50% of tetramer positive endogenous CD4+ T cells bound tetramer at equal or higher levels than C7 T cells, whereas only ~5% bound tetramer equally or higher than C24 T cells. This suggested that, relative to endogenous CD4+ T cells, C7 and C24 represent T cells with intermediate and very high avidity, respectively.
Control of M. tuberculosis by C7 and C24 T cells
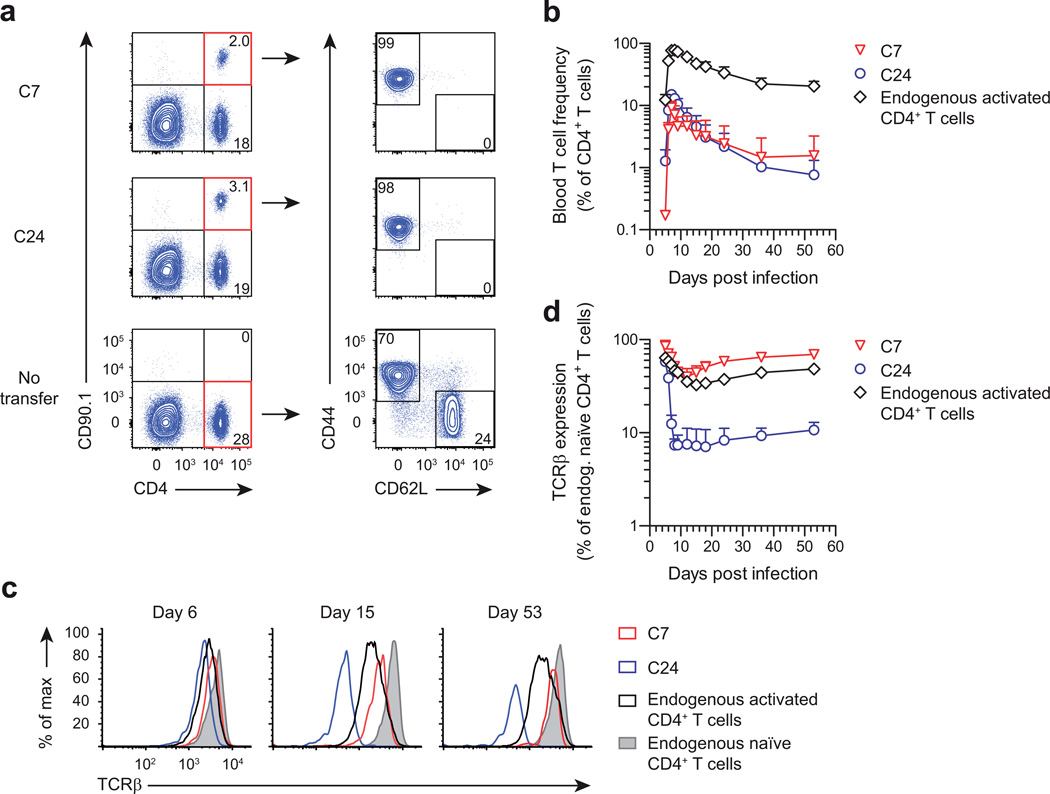
Given the very high avidity of C24 T cells, we anticipated that these cells might be very efficient at controlling M. tuberculosis infection, which is associated with limited antigen presentation20,21. Since activated TH1 cells confer greater protection than naïve T cells23, we polarized C7 and C24 T cells in vitro using TH1 conditions, transferred these cells into mice, and infected them with M. tuberculosis. Analysis of peripheral blood revealed that C7 and C24 TH1 cells continued to expand for 10 days after initial activation (Fig. 2a). The expansion of both clones peaked at a similar frequency, indicating that their capacity for proliferation is not inherently different. To determine bacterial control, we harvested lungs on day 16 post infection, when protection by transferred TH1 cells is known to be maximal22. Importantly, although both TH1 clones had inhibited bacterial growth, the degree of protection by C24 TH1 cells was significantly lower than by C7 TH1 cells (Fig. 2b), indicating impaired functionality associated with very high antigen avidity.
Figure 2.
Differential capacity of C7 and C24 TH1 cells to control M. tuberculosis infection. (a) Blood frequency of mice that received 5 × 106 CD90.1+ C7 or C24 TH1 cells, which had been activated in vitro for 4 days, following which the mice were infected with M. tuberculosis (b) Lung bacterial burden of T cell recipients described in a, 16 days after infection. ‘No transfer’ mice did not receive any T cells. Horizontal bars depict geometric mean. (c,d) Flow cytometry showing TCRβ and CD44 expression of lung C7 and C24 TH1 cells from the recipient mice described in b. Individual flow plots are shown in c and aggregate data are shown in d. Endogenous CD4+ T cells were identified as CD90.1− CD4+ T cells. Antibody binding was quantified using the MFI and normalized to the signal obtained for endogenous CD4+ T cells. Horizontal bars depict geometric mean. * P < 0.001 (unpaired two-tailed Student’s t test). Data are from one experiment representative of three independent experiments (mean + s.d. of n = 5 female mice per group (a)).
To investigate this apparent failure of C24 T cells, we analyzed the expression of several activation markers. Although C24 TH1 cells were uniformly CD44hi, they displayed dramatic downregulation of TCR expression (Fig. 2c). Specifically, TCR expression of C24 TH1 cells was only 6% of that of endogenous CD4+ T cells, whereas expression of C7 TH1 cells was 78% (Fig. 2d). Thus, the unexpected weaker performance of C24 TH1 cells could be due to loss of TCR expression.
Programmed TCR downregulation of C7 and C24 T cells
In mice, M. tuberculosis causes a chronic infection during which ESAT6 is consistently expressed26. Hence, we reasoned that TCR downregulation of C24 TH1 cells could be due to chronic antigen exposure. To test this, we transferred C7 and C24 TH1 cells into mice that were then either infected with M. tuberculosis, or left uninfected. As before, C7 and C24 TH1 cells continued to proliferate for several days after adoptive transfer before undergoing contraction in cell numbers (Fig. 3a). The extent of TH1 cell expansion and contraction was highly similar for both M. tuberculosis-infected and uninfected mice, indicating that these properties were programmed during initial T cell activation in vitro27–31. Importantly, both C7 and C24 TH1 cells exhibited high TCR expression during the expansion phase (Fig. 3b,c). However, at the peak of clonal expansion, both clones had downregulated TCR expression and this downregulation persisted throughout the contraction phase. By day 16 post activation, TCR expression of blood C7 and C24 TH1 cells was only 28% and 4% of the level of endogenous CD4+ T cells, respectively. Therefore, both C7 and C24 TH1 cells had downregulated TCR expression, but to different degrees, with the higher avidity cell displaying more TCR downregulation. Surprisingly, the extent of TCR downregulation was the same between infected and uninfected mice (Fig. 3c), suggesting that this process is not driven by chronic antigen exposure, but rather is programmed during initial T cell activation.
Figure 3.
C7 and C24 TH1 cells undergo graded TCR downregulation that is programmed during initial activation. (a) Blood frequency of mice that received 106 CD90.1+ C7 or C24 TH1 cells, which had been activated in vitro for 3 days, following which the mice were either infected with M. tuberculosis, or left uninfected. (b,c) Flow cytometry showing TCRβ expression kinetics of blood C7 and C24 TH1 cells from the recipient mice described in a. Individual flow plots are shown in b and aggregate data are shown in c. Endogenous CD4+ T cells were identified as CD90.1− CD4+ T cells. Note that in the blood, C7 T cells appear to display greater TCR downregulation compared to in the lung (as shown in Fig. 2d). Data are from one experiment representative of two independent experiments (mean + s.d. of n = 3 male mice per group (a,c)).
Although the substantial TCR downregulation of C24 TH1 cells did not require continuous exposure to foreign antigen, it could still have resulted from greater self-reactivity. Notably, naïve C24 T cells displayed 50% lower CD5 expression, but 70% higher basal TCRζ phosphorylation compared to naïve C7 T cells, suggesting that, of the two, C24 has somewhat higher self-reactivity (Supplementary Fig. 2). However, transfer of C7 and C24 TH1 cells into wild-type or MHC class II-deficient mice revealed indistinguishable TCR downregulation (Supplementary Fig. 3), implying that no further TCR signaling is required to induce downregulation beyond initial T cell activation, and thus the response appears to be truly programmed.
Programmed TCR downregulation of endogenous CD4+ T cells
The finding that both C7 and C24 TH1 cells displayed TCR downregulation raised the question whether this represented a general behavior of activated CD4+ T cells. To investigate this in vivo, we generated a recombinant Listeria monocytogenes strain that expresses ESAT632. L. monocytogenes induces potent T cell responses against multiple antigens, and therefore use of the ESAT6-expressing strain should allow activation of naïve C7 and C24 T cells, as well as endogenous L. monocytogenes-specific CD4+ T cells in vivo. To allow unbiased assessment of TCR downregulation on endogenous L. monocytogenes-specific T cells, we reasoned that flow cytometry gating on activation markers might enable identification of such cells without requiring TCR-based methodology. To test this, we transferred naïve CD90.1+ C7 and C24 T cells into mice, and infected them with L. monocytogenes-ESAT6. A third group of infected mice did not receive any cells (no transfer), to allow natural development of the endogenous L. monocytogenes-specific response. On day 7 post infection, blood C7 and C24 T cells were readily identified by the expression of CD90.1, and further gating revealed these cells uniformly expressed an activated phenotype, being CD44hi CD62L− (Fig. 4a). In contrast, gating on endogenous CD4+ T cells in ‘no transfer’ recipients revealed two subpopulations, with cells expressing either an activated or a naïve (CD44lo CD62L+) phenotype.
Figure 4.
Programmed TCR downregulation is a general feature of activated CD4+ T cells. (a) Flow cytometry showing blood frequency of mice that received 104 naïve CD90.1+ C7 or C24 CD4+ T cells, or did not receive any cells (no transfer), and were infected with recombinant L. monocytogenes-ESAT6. Blood samples were analyzed on day 7 post infection. Gating on CD90.1+ C7 or C24 CD4+ T cells revealed that these cells uniformly expressed an activated phenotype (CD44hi CD62L−). In contrast, gating on CD90.1− endogenous CD4+ T cells in ‘no transfer’ recipients revealed two subpopulations, with cells expressing either an activated or a naïve (CD44lo CD62L+) phenotype. Numbers depict the percentage of gated cells. (b) Blood frequency of C7 and C24 CD4+ T cells, as well as endogenous activated CD4+ T cells from the recipient mice described in a. Endogenous activated CD4+ T cells were defined as CD44hi CD62L− CD4+ T cells in ‘no transfer’ recipients. (c,d) Flow cytometry showing TCRβ expression kinetics of blood C7 and C24 CD4+ T cells, as well as endogenous activated and naïve CD4+ T cells from the recipient mice described in a. Individual flow plots are shown in c and aggregate data are shown in d. Endogenous naïve CD4+ T cells were defined as CD44lo CD62L+ CD4+ T cells in ‘no transfer’ recipients. Data are from one experiment representative of two independent experiments (mean + s.d. of n = 5 male mice per group (b,d)).
We then examined the response kinetics of activated C7 and C24 T cells, as well as endogenous CD44hi CD62L− CD4+ T cells. As expected, C7 and C24 T cells proliferated until day 7 post infection, after which the response contracted leaving a memory T cell population by day 35 (Fig. 4b). Notably, the expansion and contraction pattern of endogenous CD44hi CD62L− CD4+ T cells was highly similar to that of C7 and C24 T cells, suggesting that this gating strategy identified activated L. monocytogenes-specific T cells.
Being able to detect endogenous activated T cells independent of their TCR, we investigated the kinetics of TCR expression. Similar to our observations with in vitro-activated TH1 cells, in vivo-activated C7 and C24 T cells exhibited high TCR expression during initial proliferation, with expression rapidly downregulated at the peak of clonal expansion (Fig. 4c,d). TCR downregulation reached its lowest amount 15 days post infection, after which expression increased somewhat. However, as late as 53 days post infection, TCR expression of C24 T cells was still only 11% of the expression of endogenous naïve CD4+ T cells, suggesting that once the adjusted TCR expression level is set, it remains highly stable. Despite having almost no TCR, expanded C24 T cells had normal expression of costimulatory molecules, activation markers, cytokine receptors and inhibitory receptors, and did not express Foxp3 (Supplementary Fig. 4), indicating they are ‘ordinary’ effector cells. Importantly, endogenous activated CD4+ T cells also displayed persistent TCR downregulation, with similar kinetics to C7 and C24 T cells (Fig. 4c,d). Although the median TCR downregulation of endogenous T cells resembled C7, there were clearly cells present in this population that had TCR expression equivalent to C24.
To investigate whether programmed TCR downregulation is also observed during chronic bacterial infection, we analyzed the response of naïve C7 and C24 T cells activated by M. tuberculosis in vivo, which revealed the same pattern of TCR downregulation as seen during acute L. monocytogenes infection (Supplementary Fig. 5). Furthermore, endogenous M. tuberculosis-specific CD4+ T cells also displayed TCR downregulation, and low ESAT6(1–20) tetramer binding cells displayed reduced TCR expression compared to high tetramer binding cells (Supplementary Fig. 5), indicating that tetramer analyses can be confounded by TCR downregulation. Together, these data establish that activated CD4+ T cells display a program of persistent TCR downregulation that is initiated at the peak of clonal expansion and results in a repertoire with graded TCR expression, of which C7 and C24 T cells represent an average and an extreme example, respectively.
Programmed TCR downregulation linked to TCRζ degradation
Given that the TCRα and β chains of C7 and C24 T cells are driven by a constitutive promoter, we hypothesized that programmed TCR downregulation was regulated by other components of the TCR complex. Notably, activated C24 T cells with near-absent surface TCR expression had unaltered transcription of CD3γ, δ, ε and TCRζ (Supplementary Fig. 6), suggesting that TCR downregulation is mediated via post-transcriptional mechanisms. TCR complex assembly is generally limited by the availability of TCRζ33, and transient TCR downregulation can be mediated via TCRζ degradation34–36. This prompted us to investigate TCRζ protein expression in CD4+ T cells activated by L. monocytogenes. As before, C7 and C24 T cells, as well as endogenous activated CD4+ T cells displayed graded and persistent TCR downregulation starting at the peak of clonal expansion (Fig. 5a–d). Importantly, total TCRζ protein expression demonstrated a markedly similar pattern of downregulation compared to surface TCRβ expression (Fig. 5e), suggesting that programmed TCR downregulation is mediated via TCRζ degradation.
Figure 5.
Programmed TCR downregulation is associated with TCRζ degradation. (a) Blood frequency of mice that received 104 naïve CD90.1+ C7 or C24 CD4+ T cells, or did not receive any cells (no transfer), and were infected with recombinant L. monocytogenes-ESAT6. Endogenous activated CD4+ T cells were defined as CD44hi CD62L− CD4+ T cells in ‘no transfer’ recipients. (b–d) Flow cytometry showing surface TCRβ and total TCRζ expression kinetics of blood C7 and C24 CD4+ T cells, as well as endogenous activated and naïve CD4+ T cells from the recipient mice described in a. Individual flow plots are shown in b and aggregate data are shown in c and d. Endogenous naive CD4+ T cells were defined as CD44lo CD62L+ CD4+ T cells in ‘no transfer’ recipients. (e) Pearson correlation of surface TCRβ and total TCRζ expression of C7 and C24 CD4+ T cells, as well as endogenous activated CD4+ T cells (r = 0.86; P < 0.001). Data are from one experiment representative of two independent experiments (mean + s.d. of n = 5 male mice per group (a,c,d) or mean of n = 5 per group (e)).
Programmed TCR downregulation controls antigen reactivity
TCR expression is critical to efficiently respond the antigenic stimulation37–39. To investigate to what extent programmed TCR downregulation influences the antigen reactivity of activated T cells, we transferred C7 and C24 TH1 cells into uninfected mice. This generated a setting where both TH1 clones had high TCR expression initially (day 7 post activation), whereas later on (day 13), expression was intermediate on C7 and low on C24 (Fig. 6a,b). To functionally test these cells, we restimulated them with ESAT6(1–20) peptide, to measure the TCR-dependent capacity for cytokine production. As a control, we incubated the same cells with PMA + Ionomycin, which measures the TCR-independent capacity for cytokine production. On day 7, TCR-dependent IFN-γ production of C7 and C24 TH1 cells was equal to TCR-independent IFN-γ production, indicating that when TCR expression is high, the antigen reactivity of both clones is fully intact (Fig. 6c,d). In contrast, on day 13, the TCR-dependent IFN-γ production of C7 TH1 cells was intermediate and that of C24 TH1 cells was low, resulting in a very strong correlation between TCR expression and TCR-dependent IFN-γ production (r = 0.98; Fig. 6c–e). Notably, TCR-independent IFN-γ production of both clones was very high on day 13, indicating that following TCR downregulation T cells continue to have a high intrinsic capacity to produce cytokine, however their cytokine production in response to antigen is drastically altered.
Figure 6.
Programmed TCR downregulation controls CD4+ T cell cytokine production. (a,b) Flow cytometry showing splenocyte TCRβ expression of mice that received 106 CD90.1+ C7 or C24 TH1 cells, which had been activated in vitro for 4 days. Individual flow plots are shown in a and aggregate data are shown in b. Endogenous CD4+ T cells were identified as CD90.1− CD4+ T cells. Horizontal bars depict geometric mean. (c,d) Flow cytometry showing IFN-γ production of splenic C7 and C24 TH1 cells from the recipient mice described in a. Cells were restimulated with either ESAT6(1–20) peptide or PMA + Ionomycin to determine both the TCR-dependent and TCR-independent capacity for cytokine production. Individual flow plots are shown in c and aggregate data are shown in d. Data are expressed as the percentage of peptide-induced IFN-γ+ T cells normalized to the percentage of PMA + Ionomycin-induced IFN-γ+ T cells. Horizontal bars depict geometric mean. (e) Pearson correlation of TCRβ expression and TCR-dependent IFN-γ production of C7 and C24 TH1 cells (r = 0.98; P < 0.001). n.s., not significant. * P = 0.003 and ** P < 0.001 (unpaired two-tailed Student’s t test). Data are from one experiment representative of two independent experiments (mean of n = 3 male mice per group (e)).
Since programmed TCR downregulation is initiated at the peak of clonal expansion and persists for at least 50 days, we reasoned this could have substantial impact on the participation in secondary immune responses. To address this, we infected recipients of naïve C7 and C24 T cells with L. monocytogenes-ESAT6 and reinfected these mice one month later. As before, primary expansion of C7 and C24 T cells was similar, and TCR expression on both clones was initially high, whereas by day 27 it was intermediate on C7 and very low on C24 (Fig. 7a–c). Strikingly, rechallenge on day 28 resulted in a > 500% increase in the frequency of C7 T cells by day 33, whereas there was a 50% decrease in the frequency of C24 T cells, indicating that these could not participate in the secondary response.
Figure 7.
Programmed TCR downregulation controls CD4+ T cell recall proliferation. (a) Blood frequency of mice that received 104 naïve CD90.1+ C7 or C24 CD4+ T cells, and were infected with recombinant L. monocytogenes-ESAT6 on day 0 and reinfected with the same bacteria on day 28. (b,c) Flow cytometry showing TCRβ expression kinetics of blood C7 and C24 CD4+ T cells from the recipient mice described in a. Individual flow plots are shown in b and aggregate data are shown in c. Endogenous naïve CD4+ T cells were identified as CD44lo CD62L+ CD4+ T cells. Data are from one experiment representative of three independent experiments (mean + s.d. of n = 5 male mice per group (a,c)).
As L. monocytogenes induces both CD4+ and CD8+ T cell expansion, we questioned whether CD8+ T cells would also undergo TCR downregulation. To this end, we gated on endogenous activated CD8+ T cells (CD44hi CD62L−) and found that these cells displayed persistent TCR downregulation with similar kinetics to activated CD4+ T cells (Supplementary Fig. 7). Therefore, programmed TCR downregulation appears to be a feature of both T cell lineages to regulate antigen reactivity after clonal expansion.
Programmed TCR downregulation is driven by T cell avidity
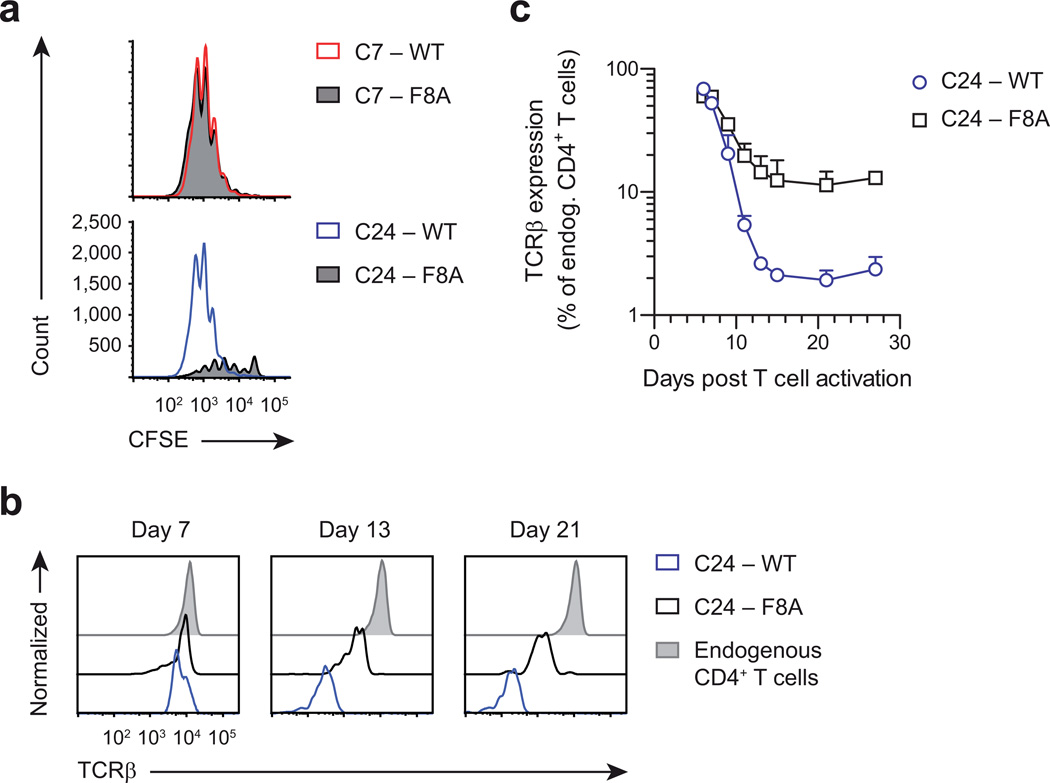
Given that activated C7 and C24 T cells display TCR downregulation to a different degree, we wondered whether this was a reflection of their differential avidity for antigen. Notably, activation of C24 T cells with a 99% lower peptide dose, which resulted in greatly diminished clonal expansion, had only minimal effect on the extent of TCR downregulation (Supplementary Fig. 8), in agreement with previous studies that low peptide quantities can still engage many TCRs via serial triggering40,41. Therefore, to truly manipulate T cell avidity, we screened ESAT6(1–20) peptide mutants and identified a peptide, F8A, that was a full agonist for C7 T cells, but only a partial agonist for C24 T cells (Fig. 8a). We then generated C24 TH1 cells in the presence of ESAT6(1–20) wild-type or F8A peptide and adoptively transferred these cells into mice. Importantly, following clonal expansion, wild-type-activated TH1 cells displayed 80% greater TCR downregulation compared to F8A-activated cells, and this difference was maintained up to a month (Fig. 8b,c). Together, these data strongly argue that T cell avidity is a driving factor behind programmed TCR downregulation, which implies that clonally expanded T cells adjust their reactivity according to the strength of initial antigen recognition.
Figure 8.
Programmed TCR downregulation is driven by the strength of initial antigen recognition. (a) Flow cytometry showing CFSE dilution of C7 and C24 T cells activated with 5 µg mL−1 ESAT6(1–20) wild-type (WT) or F8A peptide for 4 days. (b,c) Flow cytometry showing blood TCRβ expression of mice that received 106 CD90.1+ C24 TH1 cells, which had been activated in vitro with either ESAT6(1–20) WT or F8A peptide for 4 days. Individual flow plots are shown in b and aggregate data are shown in c. Endogenous CD4+ T cells were identified as CD90.1− CD4+ T cells. Data are from one experiment representative of two independent experiments (mean + s.d. of n = 5 male mice per group (c)).
DISCUSSION
Among cell surface receptors, the TCR is peculiar in that its affinity for ligand is not fixed, but highly variable. Therefore, how strongly individual T cells will react to antigen is essentially unknown prior to its encounter. Given that higher avidity interactions generally lead to greater biological output, this raises the question whether negative feedback mechanisms exist that regulate T cell effector function in an avidity-dependent manner. In this study, we have identified such a mechanism, and show that it operates via programmed downregulation of TCR expression following clonal expansion, thereby effectively lowering the functional avidity of the TCR–ligand interaction.
What is the rationale for programmed TCR downregulation and why does it lead to greater attenuation of high avidity compared to intermediate avidity T cells? One explanation is that greater suppression of high avidity cells preserves the polyclonality of the response and thereby counteracts pathogen immune escape13,25. In general, the aim of any response is to control pathogen replication with the least amount of collateral damage. Programmed TCR downregulation in that sense represents a typical example of ‘reactive regulation’, meaning a negative feedback response that is executed proportional to the strength of signaling input, but with a suitable time delay42. This enables pathogen control, while automatically shutting down the response to avoid excess tissue damage. Given the reactive nature of programmed TCR downregulation, this implies that at the end of the avidity spectrum cells will be excluded from further participation in the response. In this context, it is important to stress that programmed TCR downregulation does not commence until clonal expansion has peaked, implying there can be millions of very high avidity T cells distributed over the host. It is conceivable that a few million of such cells with full TCR expression would be difficult to restrain, especially given that at the peak of clonal expansion antigen may still be abundant27. Therefore, these cells could generate substantial inflammatory damage if left unchecked43. As such, programmed TCR downregulation appears an elegant solution, where the biological activity of a given T cell clone is tuned according to its intrinsic antigen reactivity, ultimately favoring cells of moderate avidity.
It remains puzzling how T cells use the strength of initial antigen recognition to adjust TCR expression following days of clonal expansion. Previous studies have shown that transient TCR downregulation during initial T cell activation also correlates with antigen avidity, meaning that functionally more potent ligands induce greater TCR downregulation40,41. In these experiments, TCR loss paralleled the extent of full TCRζ phosphorylation and activation of ZAP-70. Ultimately, such biochemical changes would have to induce chromatin remodeling, such that graded expression of a 'TCR targeting factor', like a ubiquitin ligase34–36, is achieved once T cells reach full expansion. As such, it might be expected that other molecules that influence TCR signaling would also contribute to programmed TCR downregulation39,44. Indeed, CD4+ T cells with very low CD5 expression also display TCR downregulation following clonal expansion45. In our case, C24 T cells displayed lower CD5 expression, but higher basal TCRζ phosphorylation compared to C7 T cells, indicating that both these factors could have added to the extensive TCR loss of C24. These results also emphasize that while CD5 expression and basal TCRζ phosphorylation often correlate46,47, there are exceptions. Since CD5 can also be upregulated on activated T cells45, some care must be used when interpreting this marker.
Our findings have important implications for the design of T cell-based immunization strategies, as it is now clear that T cells with the highest long-term functionality are those that recognize antigen with an intermediate rather than a very high avidity10,48–50. Given the impact of programmed TCR downregulation on T cell biological activity, monitoring of TCR expression should prove a useful biomarker to help new therapies select the optimal repertoire of responding T cells.
ONLINE METHODS
Generation of C24 TCR transgenic mice
C24 TCR transgenic mice specific for the I-Ab–ESAT6(1–20) antigen of M. tuberculosis were generated on the C57BL/6J (B6) background as previously described for the C7 TCR transgenic mice22. The C24 transgene is driven by the human CD2 promoter and uses Vα11.3 – Jα32 and Vβ4 – Dβ2.1 – Jβ2.1 TCRα and β chains, respectively.
Mouse models
B6, B6.PL-Thy1a/CyJ (B6.PL), B6.129S7-Rag1tm1Mom/J (Rag1−/−) and B6.129S2-H2dlAb1-Ea/J (MHC II−/−) mice were purchased from the Jackson Laboratory. All animal procedures were performed in agreement with approved protocols by the Memorial Sloan-Kettering Cancer Center Institutional Animal Care and Use Committee. C7 and C24 TCR transgenic mice were bred to B6.PL and Rag1−/− mice to generate Rag1−/− C7.PL and Rag1−/− C24.PL mice, which were used as CD4+ T cell donors in all experiments. C7 and C24 mice were maintained in the heterozygous state for the TCR-encoding transgenes. All animals were used between 6 and 12 weeks of age, were maintained under SPF conditions in individually ventilated cages, were kept on a 12 h light/dark cycle and had access to food and water ad libitum. The sizes of experimental groups were determined by the investigators, without the use of statistical methods. All age- and sex-matched mice of a given strain were considered genetically identical and were randomly assigned to treatment groups, without blinding of the investigator to mouse identity. Both male and female mice were used throughout the study, however T cell donors and recipients were always sex-matched. In case T cell recipients showed clear signs of graft rejection (>100-fold lower T cell frequency compared to any other mouse in the same experimental group), these samples were excluded from further analysis.
MHC class II tetramer and antibody staining
For MHC class II tetramer staining, cells were incubated with various concentrations of PE-labeled I-Ab–ESAT6(1–20) tetramers (NIH Tetramer Core Facility), in RPMI medium containing L-glutamine, 25 mM HEPES, 10% fetal bovine serum (FBS) and 200 µM 2-Mercaptoethanol (all Invitrogen) for 1 h at 37 °C. For tetramer dissociation, cells were incubated as above but with the addition of 0.1% NaN3 (Sigma-Aldrich), to prevent tetramer internalization, and dissociation was followed after washing free tetramer away. For tetramer pull-down, tetramer stained cells were purified using anti-PE MicroBeads and LS columns (both Miltenyi Biotec). For restimulation, cells were incubated in RPMI medium containing GolgiPlug (BD Biosciences) and 5 µg mL−1 ESAT6(1–20) peptide or 50 ng mL−1 phorbol 12-myristate 13-acetate (PMA; Calbiochem) and 1 µg mL−1 Ionomycin (Sigma-Aldrich) for 5 h at 37 °C. For cell surface staining, cells were incubated in PBS containing 0.5% bovine serum albumin (BSA; Sigma-Aldrich) and 0.1% NaN3 for 15 min at 4 °C. For intracellular staining, cells were fixed and permeabilized after cell surface staining in Cytofix/Cytoperm solution (BD Biosciences) for 15 min at 4 °C, followed by blocking in Perm/Wash solution (BD Biosciences) for 15 min at 4 °C and staining for intracellular proteins in Perm/Wash solution for 15 min at 4 °C. For nuclear staining, cells were stained using a Foxp3 Staining Buffer Set (eBioscience). Antibodies to the following proteins were used (with indicated clone numbers and dilutions; Supplementary Table 1): CD3ε (145-2C11; 1:100), CD4 (RM4-5; 1:100), CD5 (53-7.3; 1:100), CD8α (53-6.7; 1:50), CD25 (PC61; 1:100), CD27 (LG.3A10; 1:100), CD28 (37.51; 1:50), CD44 (IM7; 1:100), CD45 (30-F11; 1:200), CD62L (MEL-14; 1:100), CD69 (H1.2F3; 1:50), CD90.1 (OX-7; 1:1,600), CD122 (TM-β1; 1:50), CTLA-4 (UC10-4F10-11; 1:50), IFN-γ (XMG1.2; 1:200), PD-1 (J43; 1:50), TCRβ (H57-597; 1:100), Hamster IgG1 isotype control (A19-3; 1:50) and Rat IgG2a isotype control (R35-95; 1:100) (all BD Biosciences), CD127 (A7R34; 1:50) and Foxp3 (FJK-16s; 1:100) (both eBioscience) and TCRζ (H146-968; 1:100) (Thermo Scientific). Dead cells were excluded using 7-AAD Viability Staining Solution (eBioscience). Cells were measured on an LSRII flow cytometer (BD Biosciences) and data were analyzed using FlowJo software (TreeStar).
In vitro TH1 differentiation and adoptive cell transfer
For in vitro TH1 differentiation of C7 and C24 T cells, CD4+ T cells from pooled spleen and lymph nodes of donor mice were purified using CD4 (L3T4) MicroBeads and LS columns (both Miltenyi Biotec). To prepare antigen-presenting cells (APCs), B6 spleens were depleted of endogenous T cells using CD90.2 MicroBeads and LS columns, after which remaining splenocytes were irradiated with 25 Gy. 4 × 106 CD4+ T cells were co-cultured with 12 × 106 irradiated APCs in RPMI medium containing 5 µg mL−1 ESAT6(1–20) peptide, 10 ng mL−1 recombinant mouse IL-12 (R&D systems) and 5 µg mL−1 neutralizing antibody to mouse IL-4 (11B11 hybridoma). Cells were cultured in 6-well plates at 37 °C and 5% CO2. On days 2 and 3 of culture, cells were split in half and 12.5 ng mL−1 recombinant mouse IL-2 (R&D systems) was added. On day 4 of culture, C7 and C24 TH1 cells were harvested, washed in PBS and transferred intravenously into sex-matched B6 recipients.
For in vitro cell proliferation assays, purified C7 and C24 T cells were resuspended in PBS containing 0.1% BSA and labeled with 5 µM CFSE (Invitrogen) for 10 min at 37 °C. After washing in RPMI medium, cells were activated using APCs as described above. The percentage divided cells was calculated using the Proliferation tool of FlowJo software.
For adoptive transfer of naïve C7 and C24 T cells, CD4+ T cells were purified from pooled spleen and lymph nodes of donor mice using the CD4+ T cell Isolation Kit II (Miltenyi Biotec) and LS columns, to generate untouched CD4+ T cells. Following washing in PBS, 104 CD4+ T cells were transferred intravenously into sex-matched B6 recipients.
M. tuberculosis infection and T cell analysis
M. tuberculosis Erdman was grown in Middlebrook 7H9 broth (BD Biosciences) until early log phase (OD600 of ~0.2). Cultures were washed in PBS containing 0.05% Tween 80 (Fischer Scientific), sonicated and resuspended in water. B6 mice were infected with aerosolized bacteria using an inhalation exposure system (Glas-Col), which was calibrated to deliver ~100 colony-forming units (CFU) per animal.
To monitor C7 and C24 T cell responses over time, blood samples were harvested in PBS containing 5 mM EDTA (US Biological), followed by red blood cell lysis using NH4Cl buffer. To determine lung T cell responses, left lungs were minced using scissors and digested in PBS containing 2% FBS, 500 U mL−1 Collagenase Type 4 (Worthington) and 20 µg mL−1 DNase I (Roche) for 30 min at 37 °C. Cell suspensions were homogenized by passage through 18G needles followed by 100 µM cell strainers (BD Biosciences). Blood and lung samples were stained as described above, fixed in PBS containing 2% paraformaldehyde (Electron Microscopy Sciences) and analyzed by flow cytometry.
To determine lung bacterial burdens, right lungs were homogenized in PBS containing 0.05% Tween 80, after which serial dilutions were made in PBS containing 0.05% Tween 80. Serial dilutions were spread on Middlebrook 7H10 agar (BD Biosciences) and incubated for 21 days at 37 °C and 5% CO2, after which the number of colonies was determined.
Immunoblot analysis
Naïve C7 and C24 T cells were purified from spleens of donor mice using CD4 MicroBeads and LS columns. 106 cells were lysed in 100 µL RIPA buffer (Sigma-Aldrich) containing protease and phosphatase inhibitors (Thermo Scientific). 15 µL of lysates was mixed with 2× loading buffer, separated on 10% SDS-PAGE and transferred to PVDF membrane. Membranes were incubated with rabbit anti-phospho-TCRζ (pTyr142) and rabbit anti β-actin primary antibodies (both Sigma-Aldrich) O/N at 4 °C, with a dilution factor of 1:1,000 in blocking buffer (PBS with 5% skim milk and 0.1% Tween 20). Following incubation with HRP-linked anti-rabbit secondary antibody (Cell Signaling; diluted 1:5,000 in blocking buffer), detection was performed using ECL reagent (GE Healthcare). To allow fair comparison, blots containing phospho-TCRζ and β-actin staining were imaged simultaneously. Densitometry was assessed using ImageJ software (NIH).
Recombinant L. monocytogenes-ESAT6 infection
To generate a recombinant L. monocytogenes strain that stably expresses and secretes ESAT6, the entire ESAT6 coding region of M. tuberculosis, except the first ATG (ESAT6(2–95)), was fused in-frame behind the hly signal sequence of the L. monocytogenes secreted protein listeriolysin O. The hly-ESAT6(2–95) fusion product was cloned behind the hly promoter and the entire construct was inserted into the pPL2 shuttle integration vector32. The pPL2-ESAT6 vector was then introduced as a single copy into the genome of the L. monocytogenes strain 10403S via conjugation. Following selection by chloramphenicol, successful transconjugants were confirmed by PCR and passaged through B6 mice to ensure virulence.
For recombinant L. monocytogenes-ESAT6 infection, bacteria were grown in Brain Heart Infusion broth (BD Biosciences) until early log phase (OD600 of 0.1). Cultures were washed in PBS and injected intravenously into B6 mice at a dose of 5×103 CFU for primary infections and 5×105 CFU for secondary infections.
Real-time PCR analysis
To analyze gene expression of adoptively transferred CD90.1+ C7 and C24 T cells, these cells were purified from the spleens of CD90.2+ recipient mice using CD90.1 MicroBeads (Miltenyi Biotec) and separation over two consecutive MS columns (Miltenyi Biotec). Eluted cells were lysed in RLT buffer (QIAGEN), homogenized using QIAshredder columns (QIAGEN) and stored at –80 °C. Total RNA from frozen homogenates was extracted using an RNeasy mini kit (QIAGEN), followed by cDNA generation using a QuantiTect Reverse Transcription kit (QIAGEN). TaqMan probe-based real-time PCR analysis was performed on a StepOnePlus Real-Time PCR System (Applied Biosystems), using a DyNAmo probe qPCR kit (Thermo Scientific) and specific TaqMan probes (Invitrogen). Actb was used as endogenous control. Relative gene expression was determined using the Comparative CT method.
Statistical analysis
Statistical analysis was performed using an unpaired two-tailed Student’s t test, assuming that data was normally distributed and there was equal variance between the groups under comparison. P < 0.05 was considered statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Samstein, H. Yan and S. Reddy for technical assistance. This work was supported by the US National Institutes of Health (NIH) (F32 grant AI074248 to A.M.G. and grant AI080619 to M.S.G. and E.G.P., and P30 CA008748) and the Netherlands Organisation for Scientific Research (NWO) (Veni grant 91614038 to J.W.J.v.H.).
Footnotes
AUTHOR CONTRIBUTIONS
A.M.G., M.S.G., E.G.P. and J.W.J.v.H. designed the study; A.M.G., H.X. and J.W.J.v.H. performed experiments and analyzed data. E.G.P. and J.W.J.v.H. wrote the manuscript. A.M.G. generated the C7 and C24 TCR transgenic mice, and the L. monocytogenes-ESAT6 strain. I.M.L. and B.S. bred and genotyped the C7 and C24 mice, and provided technical assistance. J.W.J.v.H. first identified programmed TCR downregulation.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Nikolich-Zugich J, Slifka MK, Messaoudi I. The many important facets of T-cell repertoire diversity. Nat Rev Immunol. 2004;4:123–132. doi: 10.1038/nri1292. [DOI] [PubMed] [Google Scholar]
- 2.van Heijst JW, et al. Quantitative assessment of T cell repertoire recovery after hematopoietic stem cell transplantation. Nat Med. 2013;19:372–377. doi: 10.1038/nm.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Busch DH, Pamer EG. T cell affinity maturation by selective expansion during infection. J Exp Med. 1999;189:701–710. doi: 10.1084/jem.189.4.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corse E, Gottschalk RA, Allison JP. Strength of TCR-peptide/MHC interactions and in vivo T cell responses. J Immunol. 2011;186:5039–5045. doi: 10.4049/jimmunol.1003650. [DOI] [PubMed] [Google Scholar]
- 5.King CG, et al. T cell affinity regulates asymmetric division, effector cell differentiation, and tissue pathology. Immunity. 2012;37:709–720. doi: 10.1016/j.immuni.2012.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rees W, et al. An inverse relationship between T cell receptor affinity and antigen dose during CD4(+) T cell responses in vivo and in vitro. Proc Natl Acad Sci U S A. 1999;96:9781–9786. doi: 10.1073/pnas.96.17.9781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Savage PA, Boniface JJ, Davis MM. A kinetic basis for T cell receptor repertoire selection during an immune response. Immunity. 1999;10:485–492. doi: 10.1016/s1074-7613(00)80048-5. [DOI] [PubMed] [Google Scholar]
- 8.Varela-Rohena A, et al. Control of HIV-1 immune escape by CD8 T cells expressing enhanced T-cell receptor. Nat Med. 2008;14:1390–1395. doi: 10.1038/nm.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zehn D, Lee SY, Bevan MJ. Complete but curtailed T-cell response to very low-affinity antigen. Nature. 2009;458:211–214. doi: 10.1038/nature07657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corse E, Gottschalk RA, Krogsgaard M, Allison JP. Attenuated T cell responses to a high-potency ligand in vivo. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hebeisen M, et al. SHP-1 phosphatase activity counteracts increased T cell receptor affinity. J Clin Invest. 2013;123:1044–1056. doi: 10.1172/JCI65325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalergis AM, et al. Efficient T cell activation requires an optimal dwell-time of interaction between the TCR and the pMHC complex. Nat Immunol. 2001;2:229–234. doi: 10.1038/85286. [DOI] [PubMed] [Google Scholar]
- 13.Vigano S, et al. Functional avidity: a measure to predict the efficacy of effector T cells? Clinical & developmental immunology. 2012;2012:153863. doi: 10.1155/2012/153863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Garra A, et al. The immune response in tuberculosis. Annu Rev Immunol. 2013;31:475–527. doi: 10.1146/annurev-immunol-032712-095939. [DOI] [PubMed] [Google Scholar]
- 15.Ottenhoff TH, Kaufmann SH. Vaccines against tuberculosis: where are we and where do we need to go? PLoS Pathog. 2012;8:e1002607. doi: 10.1371/journal.ppat.1002607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lawn SD, Myer L, Edwards D, Bekker LG, Wood R. Short-term and long-term risk of tuberculosis associated with CD4 cell recovery during antiretroviral therapy in South Africa. Aids. 2009;23:1717–1725. doi: 10.1097/QAD.0b013e32832d3b6d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mogues T, Goodrich ME, Ryan L, LaCourse R, North RJ. The relative importance of T cell subsets in immunity and immunopathology of airborne Mycobacterium tuberculosis infection in mice. J Exp Med. 2001;193:271–280. doi: 10.1084/jem.193.3.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hmama Z, Gabathuler R, Jefferies WA, de Jong G, Reiner NE. Attenuation of HLA-DR expression by mononuclear phagocytes infected with Mycobacterium tuberculosis is related to intracellular sequestration of immature class II heterodimers. J Immunol. 1998;161:4882–4893. [PubMed] [Google Scholar]
- 19.Pai RK, Convery M, Hamilton TA, Boom WH, Harding CV. Inhibition of IFN-gamma-induced class II transactivator expression by a 19-kDa lipoprotein from Mycobacterium tuberculosis: a potential mechanism for immune evasion. J Immunol. 2003;171:175–184. doi: 10.4049/jimmunol.171.1.175. [DOI] [PubMed] [Google Scholar]
- 20.Bold TD, Banaei N, Wolf AJ, Ernst JD. Suboptimal activation of antigen-specific CD4+ effector cells enables persistence of M. tuberculosis in vivo. PLoS Pathog. 2011;7:e1002063. doi: 10.1371/journal.ppat.1002063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Egen JG, et al. Intravital imaging reveals limited antigen presentation and T cell effector function in mycobacterial granulomas. Immunity. 2011;34:807–819. doi: 10.1016/j.immuni.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gallegos AM, Pamer EG, Glickman MS. Delayed protection by ESAT-6-specific effector CD4+ T cells after airborne M. tuberculosis infection. J Exp Med. 2008;205:2359–2368. doi: 10.1084/jem.20080353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gallegos AM, et al. A gamma interferon independent mechanism of CD4 T cell mediated control of M. tuberculosis infection in vivo. PLoS Pathog. 2011;7:e1002052. doi: 10.1371/journal.ppat.1002052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crawford F, Kozono H, White J, Marrack P, Kappler J. Detection of antigen-specific T cells with multivalent soluble class II MHC covalent peptide complexes. Immunity. 1998;8:675–682. doi: 10.1016/s1074-7613(00)80572-5. [DOI] [PubMed] [Google Scholar]
- 25.Egen JG, Allison JP. Cytotoxic T lymphocyte antigen-4 accumulation in the immunological synapse is regulated by TCR signal strength. Immunity. 2002;16:23–35. doi: 10.1016/s1074-7613(01)00259-x. [DOI] [PubMed] [Google Scholar]
- 26.Winslow GM, Roberts AD, Blackman MA, Woodland DL. Persistence and turnover of antigen-specific CD4 T cells during chronic tuberculosis infection in the mouse. J Immunol. 2003;170:2046–2052. doi: 10.4049/jimmunol.170.4.2046. [DOI] [PubMed] [Google Scholar]
- 27.Badovinac VP, Porter BB, Harty JT. Programmed contraction of CD8(+) T cells after infection. Nat Immunol. 2002;3:619–626. doi: 10.1038/ni804. [DOI] [PubMed] [Google Scholar]
- 28.Kaech SM, Ahmed R. Memory CD8+ T cell differentiation: initial antigen encounter triggers a developmental program in naive cells. Nat Immunol. 2001;2:415–422. doi: 10.1038/87720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee WT, Pasos G, Cecchini L, Mittler JN. Continued antigen stimulation is not required during CD4(+) T cell clonal expansion. J Immunol. 2002;168:1682–1689. doi: 10.4049/jimmunol.168.4.1682. [DOI] [PubMed] [Google Scholar]
- 30.Mercado R, et al. Early programming of T cell populations responding to bacterial infection. J Immunol. 2000;165:6833–6839. doi: 10.4049/jimmunol.165.12.6833. [DOI] [PubMed] [Google Scholar]
- 31.van Stipdonk MJ, Lemmens EE, Schoenberger SP. Naive CTLs require a single brief period of antigenic stimulation for clonal expansion and differentiation. Nat Immunol. 2001;2:423–429. doi: 10.1038/87730. [DOI] [PubMed] [Google Scholar]
- 32.Lauer P, Chow MY, Loessner MJ, Portnoy DA, Calendar R. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J Bacteriol. 2002;184:4177–4186. doi: 10.1128/JB.184.15.4177-4186.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sussman JJ, et al. Failure to synthesize the T cell CD3-zeta chain: structure and function of a partial T cell receptor complex. Cell. 1988;52:85–95. doi: 10.1016/0092-8674(88)90533-8. [DOI] [PubMed] [Google Scholar]
- 34.Cenciarelli C, et al. Activation-induced ubiquitination of the T cell antigen receptor. Science. 1992;257:795–797. doi: 10.1126/science.1323144. [DOI] [PubMed] [Google Scholar]
- 35.Naramura M, et al. c-Cbl and Cbl-b regulate T cell responsiveness by promoting ligand-induced TCR down-modulation. Nat Immunol. 2002;3:1192–1199. doi: 10.1038/ni855. [DOI] [PubMed] [Google Scholar]
- 36.Valitutti S, Muller S, Salio M, Lanzavecchia A. Degradation of T cell receptor (TCR)-CD3-zeta complexes after antigenic stimulation. J Exp Med. 1997;185:1859–1864. doi: 10.1084/jem.185.10.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Labrecque N, et al. How much TCR does a T cell need? Immunity. 2001;15:71–82. doi: 10.1016/s1074-7613(01)00170-4. [DOI] [PubMed] [Google Scholar]
- 38.Schrum AG, Turka LA, Palmer E. Surface T-cell antigen receptor expression and availability for long-term antigenic signaling. Immunol Rev. 2003;196:7–24. doi: 10.1046/j.1600-065x.2003.00083.x. [DOI] [PubMed] [Google Scholar]
- 39.Viola A, Lanzavecchia A. T cell activation determined by T cell receptor number and tunable thresholds. Science. 1996;273:104–106. doi: 10.1126/science.273.5271.104. [DOI] [PubMed] [Google Scholar]
- 40.Itoh Y, Hemmer B, Martin R, Germain RN. Serial TCR engagement and down-modulation by peptide:MHC molecule ligands: relationship to the quality of individual TCR signaling events. J Immunol. 1999;162:2073–2080. [PubMed] [Google Scholar]
- 41.Valitutti S, Muller S, Cella M, Padovan E, Lanzavecchia A. Serial triggering of many T-cell receptors by a few peptide-MHC complexes. Nature. 1995;375:148–151. doi: 10.1038/375148a0. [DOI] [PubMed] [Google Scholar]
- 42.Germain RN. Maintaining system homeostasis: the third law of Newtonian immunology. Nat Immunol. 2012;13:902–906. doi: 10.1038/ni.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baniyash M. TCR zeta-chain downregulation: curtailing an excessive inflammatory immune response. Nat Rev Immunol. 2004;4:675–687. doi: 10.1038/nri1434. [DOI] [PubMed] [Google Scholar]
- 44.Anderton SM, Wraith DC. Selection and fine-tuning of the autoimmune T-cell repertoire. Nat Rev Immunol. 2002;2:487–498. doi: 10.1038/nri842. [DOI] [PubMed] [Google Scholar]
- 45.Weber KS, et al. Distinct CD4+ helper T cells involved in primary and secondary responses to infection. Proc Natl Acad Sci U S A. 2012;109:9511–9516. doi: 10.1073/pnas.1202408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mandl JN, Monteiro JP, Vrisekoop N, Germain RN. T cell-positive selection uses self-ligand binding strength to optimize repertoire recognition of foreign antigens. Immunity. 2013;38:263–274. doi: 10.1016/j.immuni.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Persaud SP, Parker CR, Lo WL, Weber KS, Allen PM. Intrinsic CD4+ T cell sensitivity and response to a pathogen are set and sustained by avidity for thymic and peripheral complexes of self peptide and MHC. Nat Immunol. 2014;15:266–274. doi: 10.1038/ni.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fasso M, et al. T cell receptor (TCR)-mediated repertoire selection and loss of TCR vbeta diversity during the initiation of a CD4(+) T cell response in vivo. J Exp Med. 2000;192:1719–1730. doi: 10.1084/jem.192.12.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McCue D, Ryan KR, Wraith DC, Anderton SM. Activation thresholds determine susceptibility to peptide-induced tolerance in a heterogeneous myelin-reactive T cell repertoire. Journal of neuroimmunology. 2004;156:96–106. doi: 10.1016/j.jneuroim.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 50.McMahan RH, et al. Relating TCR-peptide-MHC affinity to immunogenicity for the design of tumor vaccines. J Clin Invest. 2006;116:2543–2551. doi: 10.1172/JCI26936. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.