Summary
The Flaviviridae are a family of viruses that cause severe human diseases. For example, dengue virus (DENV) is a rapidly emerging pathogen causing an estimated 100 million symptomatic infections annually worldwide1. No approved antivirals are available to date and clinical trials with a tetravalent dengue vaccine showed disappointingly low protection rates2. Also hepatitis C virus (HCV) remains a major medical problem with 160 million chronically infected patients and only expensive treatment on the market3. Despite distinct differences in pathogenesis and mode of transmission, the two viruses share common replication strategies4. A detailed understanding of the host functions that determine viral infection is lacking. Here, we utilized a pooled CRISPR genetic screening strategy5,6 to comprehensively dissect host factors required for these two highly important Flaviviridae members. For DENV, we identified ER-associated multi-protein complexes involved in signal sequence recognition, N-linked glycosylation and ER associated degradation. Dengue virus replication was nearly completely abrogated in cells deficient in the oligosaccharyltransferase (OST) complex. Mechanistic studies pinpointed viral RNA replication and not entry or translation as critical step requiring the OST complex. Moreover, we showed that viral non-structural proteins bind to the OST complex. The identified ER-associated protein complexes were also important for other mosquito-borne flaviviruses including Zika virus, an emerging pathogen causing severe birth defects7. In contrast, the most significant genes identified in the HCV screen were distinct and included viral receptors, RNA binding proteins and enzymes involved in metabolism. We discovered an unexpected link between intracellular FAD levels and HCV replication. This study shows remarkable divergence in host dependency factors between DENV and HCV and illuminates novel host targets for antiviral therapy.
CRISPR is revolutionizing the utility of genetic screens because the ability to completely knockout genes substantially increases the robustness of the phenotypes5,6. We compared the CRISPR approach in the hepatocyte cell line Huh7.5.1 with an alternative method to generate knockout alleles on a genome-wide scale: insertional mutagenesis in human haploid cells (HAP1)8,9 (Fig. 1a). Both methods generate libraries of cells with knockout mutations in all non-essential genes. To comprehensively identify cellular genes with critical roles in the Flaviviridae life cycles, we first infected pools of mutagenized cells with DENV serotype 2 (DENV2). The two types of genetic screening methods showed a strong concordance in the genes enriched in the DENV2 resistant population. Many could be functionally classified into three distinct categories, each important for proper expression of ER-targeted glycoproteins (Fig. 1b–c, Supplementary Data Tables 1–2). The Translocon Associated Protein (TRAP) complex (SSR1, SSR2, SSR3) plays an elusive role in stimulating co-translational translocation mediated by several, but not all signal sequences10 (Fig. 1b–c, blue). Genes involved in protein quality control and ER-Associated protein Degradation (ERAD) pathway also scored high (Fig. 1b–c, green). Strikingly, in both the haploid and CRISPR screens, the most significantly enriched genes were subunits of the OST complex, an enzyme essential for N-linked glycosylation (Fig. 1b–c, red). This dependence on ER cellular genes is likely related to the expression of the DENV genome, which encodes an ER-targeted viral polyprotein containing signal sequences and viral glycoproteins. Given the similarities in DENV and HCV polyprotein expression we expected these genes to also be represented in the HCV CRISPR screen. Surprisingly, there was no overlap between the DENV and HCV core set of enriched genes, suggesting that these members of the Flaviviridae evolved divergent host factor dependencies (Fig. 1c–e, Extended Data Fig. 1a–b, Supplementary Data Tables 3–4). Indeed, cross comparison of the most significant hits with both viruses suggested specific dependencies, although minor quantitative effects cannot be excluded (Extended Data Fig. 1c). The robustness of the CRISPR approach was further underscored by the consistent identification of the core dependency factors in three independent replicate screens performed for each virus (Extended Data Fig. 2). We validated the novel DENV host factors in isogenic knockout cells using plaque-forming assay and observed a dramatic reduction in particle formation (Extended Data Fig. 3, 4a). Importantly, complementation of knockout cells restored DENV infection (Extended Data Fig. 4b–c). The relevance of the identified host factors was further confirmed in Raji DC-SIGN, a B cell line commonly used to study DENV (Extended Data Fig. 4d).
Figure 1. Haploid and CRISPR genetic screens identify essential host factors of DENV and HCV infections.

(a) Schematic for genome-wide screening approach. (b) Haploid genetic screen for DENV host factors. The y-axis represents significance of enrichment of gene-trap insertions in genes in DENV resistant population compared to unselected HAP1 cells. Each circle represents a specific gene and size corresponds to number of independent gene-trap insertions. All genes with p-value <0.05 were colored and grouped by function. The screen was performed once. (c) CRISPR genetic screen for DENV and (d) for HCV host factors in Huh7.5.1 cells. Significance of enrichment was calculated by RIGER analysis. The screens were performed in three replicates and the mean of the RIGER score is represented on the y-axis. The 30 most enriched genes were colored and grouped by function. (e) Comparison of the 30 most enriched genes from the DENV and HCV CRISPR screens and their position based on the mean RIGER score.
Struck by the distinct host factor requirements of DENV2 and HCV, we sought to evaluate selected DENV2 dependency factors against other mosquito-borne flaviviruses that are closely related to DENV (Fig. 2a). Using QPCR in isogenic knockout cells we found that West Nile virus (WNV), but not Yellow Fever virus (YFV) and Zika virus (ZIKV), was as sensitive as DENV2 to disruption of the tested ERAD genes, which is in line with previous reports implicating ERAD in WNV infection11,12. A functional TRAP complex is important for DENV2, YFV and ZIKV RNA replication whereas WNV RNA abundance is only slightly reduced. Individual subunits of the OST complex displayed strikingly different phenotypes for the four related flaviviruses. Whereas knockout of STT3A and STT3B both completely abolished DENV2 replication, only STT3A knockout affected YFV, WNV and ZIKV replication. When probing HCV replication in STT3A- and STT3B-knockout Huh7 cells using luciferase virus, we did not observe a substantial decrease (Extended Data Fig. 4e).
Figure 2. ER protein complexes play a crucial role in the replication phase of DENV and are also important for YFV, WNV and ZIKV infection.

(a) QPCR of DENV16681, YFV17D, WNVKUNJIN and ZIKVUganda RNA in knockout HAP1 cells. (b) QPCR of prototypic strains of DENV serotypes 1–4 RNA in knockout Huh7 cells. (c) Confocal Microscopy of STT3B-KO Huh7 cells immunostained for DENV Envelope protein immediately or 30 minutes after DENV infection. (d) Luminescence of DENV replicon RNA expressing luciferase in knockout Huh7 cells. The DENV NS5-GDD mutant served as replication-deficient control. Data depict average with s.e.m. (QPCR) or s.d. (RLU) for triplicate infections.
Intrigued by the differential sensitivity to the catalytic OST subunits we focused our mechanistic studies on the OST complex, which has not been linked to viral replication before. The highly conserved catalytic subunit of the OST complex, STT3, is duplicated into two paralogues STT3A and STT3B in mammalian cells and each isoform is present in distinct protein complexes13 (Extended Data Fig. 4f). The STT3A complex is important for the co-translational N-linked glycosylation of the majority of the glycoproteins whereas the STT3B complex is important for the co-translational or post-translational glycosylation of acceptor sites that have been skipped by the STT3A complex14. Despite their partially redundant functions in N-linked glycosylation, we found that all DENV serotypes required the presence of both catalytic subunits as well as MAGT1, the highest scoring gene in the CRISPR screen (Fig. 2b).
To pinpoint which step in the viral life cycle requires the OST complex, we first focused on viral entry. We did not observe major differences in viral particle entry in OST-deficient cells (Fig. 2c). Next, we used a replicon assay that bypasses viral entry by electroporation of dengue RNA. Translation of the viral genome, apparent at time points up to 10 hours, was equally efficient in OST-knockout cells compared to wild type cells (Fig. 2d). In stark contrast, viral RNA replication (apparent at time points after 10 hours) was completely abolished. This mirrored the expression pattern observed with a replication-deficient dengue mutant in the viral polymerase (NS5GDD). Thus, we show that the OST complex plays a critical role in viral RNA replication, after entry and translation of the viral genome.
The majority of glycoproteins can be efficiently modified by both OST isoforms and only few are preferentially modified by either STT3A or STT3B14. Concordant with this, knockout of either STT3A or STT3B did not lead to loss of cellular viability whereas double knockout was lethal (Fig. 3a). We demonstrated that OST catalytic activity is required for cellular function using STT3A and STT3B mutants containing mutations in the residues that coordinate the binding of the divalent cation required for catalysis15 (Fig. 3a, Extended Data Fig. 5). The functional redundancy between isoforms in global N-linked glycosylation is in contrast with the extreme dependence on each individual isoforms of the OST complex for DENV replication. To investigate whether the effect of the OST complex is mediated by the necessity to properly glycosylate viral proteins, we focused on NS1, an enigmatic dengue glycoprotein with essential roles in RNA replication16 and pathogenesis17. NS1 was fully glycosylated in STT3A and STT3B knockout cells in contrast to the hypo-glycosylation of control cellular proteins known to be preferentially glycosylated by STT3A (pSAP) or STT3B (SHBG) (Fig. 3b). This led us to speculate that the OST complex itself rather than its catalytic activity is required for dengue replication. To explore this hypothesis, we used the catalytic mutants of STT3A and STT3B (Fig. 3a,c, Extended Data Fig. 5). Surprisingly, the catalytically dead mutants were able to fully restore DENV replication in STT3A and in STT3B knockout cells (Fig. 3d). We thus concluded that DENV RNA replication has hijacked a function of the human OST complex that is independent from its canonical role in N-linked glycosylation. The dispensability of the catalytic function of the OST complex further suggests a more structural role of OST in viral replication. The OST complex forms a stoichiometric complex at the ER membrane where DENV establishes a functional replication complex to initiate RNA replication. Our electron microscopy studies using APEX2 confirmed the localization of STT3B in the ER membrane, in close proximity to the membranous replication vesicles in DENV2 infected cells (Extended Data Fig. 6a–c).
Figure 3. DENV RNA replication requires a non-canonical function of OST, and DENV non-structural proteins interact with OST.

(a) Viability of STT3A and STT3B double knockout cells complemented with wild-type (wt) or catalytic (cat) mutant cDNA. (b) Glycosylation of DENV protein NS1, SHBG, and pSAP in STT3A and STT3B knockout Huh7 cells. Different glycoforms are indicated by arrowheads. Tun. = Tunicamycin (c) Glycosylation state of pSAP and SHBG in STT3A- and STT3B-KO cells complemented with catalytic mutants. (d) DENV infection of knockout Huh7 cells complemented with WT or catalytic mutants of STT3A and STT3B. Data depict average with s.d. for triplicate infections. (e) Co-immunoprecipitations of STT3A-FLAG and STT3B-FLAG from DENV infected cell lysates. LE = long exposure.
We next interrogated the physical interaction of the viral proteins with the OST complex by immunoprecipitation (IP) of STT3A-FLAG and STTB-FLAG in the context of viral infection. Western blot analysis following IP showed that the nonstructural proteins NS2B and NS3, components of the DENV replication complex, associate specifically with STT3A and STT3B (Fig. 3e, Extended Data Fig. 6d). Next, eluates of the immunoprecipitations (Extended Data Fig. 6e–f) were subjected to tryptic proteolysis and the resulting peptides were analyzed by mass spectrometry. Identification of tryptic peptides from NS2A, NS3 and NS4A in the eluates pointed to their association with the OST complexes formed by STT3A and STT3B (Extended Data Fig. 6g, Supplementary Data Table 5). Taken together, this suggests a structural role for the OST complex in dengue RNA replication through interactions with nonstructural proteins that form the RNA replication complex. Our data indicates that the OST complex fulfills specialized roles in host pathogen interactions and is more multifaceted than previously recognized. In support of this emerging notion, two recent studies uncovered unexpected roles of the OST complex in immunity. The OST complex was found to be crucial for innate immune responses triggered by LPS18, and in a separate study OST dysregulation was identified as cause for autoimmune disorders triggered by certain TREX1 mutations19.
The HCV-resistant cell population was highly enriched in guides targeting the known HCV receptors20 CD81, OCLN and CLDN1 confirming their non-redundant role in entry for HCV (Fig. 1d, blue), and highlighting the validity of the screen results. The completeness of the screen was further underscored by the identification of microRNA-12221 and DGCR8 (Fig. 1d, green), which is part of the microRNA processing machinery, as key factors for HCV replication. Several dependency factors of HCV were validated in Huh7.5.1 cells, where knockout significantly reduced viral RNA and particle formation (Fig. 4a–b, Extended Data Fig. 7, 8a). After CLDN1, the second most significantly enriched gene was ELAVL1 (also named HuR), an RNA binding protein involved in mRNA stabilization22. In isogenic ELAVL1 knockout cells, HCV RNA replication was nearly abolished, while we did not observe a decrease with other RNA viruses (Extended Data Fig. 8b–c) including the alphavirus Sindbis, which contains strong ELAVL1 binding sites23. We used an HCV replicon assay to show that ELAVL1 plays a critical role in HCV RNA replication, which is in line with a recent report24 (Extended Data Fig. 8d–e). Enzymatically active HCV dependency factors (Fig. 1d, red) are by far the most important category of host factors for identifying antiviral drug targets. A case in point is cyclophilin A (PPIA) that has been actively pursued until phase III clinical trials25. We discovered enzymes with novel putative roles in HCV replication and explored these potentially druggable factors further. We focused on RFK and FLAD1, enzymes involved in the two-step conversion of riboflavin (vitamin B2) to flavin adenine dinucleotide (FAD) (Fig. 4c). RFK and FLAD1 knockout cells were resistant to HCV replication but not DENV (Extended Data Fig. 9a). As predicted from their sequential role in FAD biogenesis, exogenous flavin mononucleotide (FMN) or FAD rescued HCV replication in RFK knockout cells, whereas FAD but not FMN rescued viral replication in FLAD1 knockout cells (Fig. 4d, Extended Data Fig. 9b). This demonstrates that HCV replication is dependent solely on sufficient FAD levels. Modulation of intracellular FAD levels can be achieved by treatment of the cells with lumiflavin, a cellular uptake inhibitor of riboflavin26. Treatment of cells with lumiflavin greatly reduced HCV replication, while other RNA viruses were less sensitive to lumiflavin treatment (Fig. 4e, Extended Data Fig. 9b–f). We further pinpointed RNA replication as the step of the life cycle that requires FAD using a replicon system (Fig. 4f). This highlights that knockout screens can identify specific host targets for antiviral drug discovery. Taken together, we used comparative genome-scale knockout screens to identify human genes with critical roles in the replication of Flaviviridae. Despite previous extensive interrogation of human host factors for these viruses through genomic and proteomic approaches11,27–30, we discovered dramatic dependencies on several host processes that had not been linked to flaviviral replication before (Fig. 4g, Extended Data Fig. 10).
Figure 4. FAD biosynthesis is required for HCV replication and can serve as antiviral target.

(a) QPCR of HCV RNA in Huh7.5.1 cell lines. (b) HCV particle formation measured by focus-forming units (FFU) assay. ND indicates that no foci were detected (threshold of detection is 50 FFU/ml). (c) Biosynthesis pathway of FAD. Lumiflavin (LF) competitively inhibits uptake of riboflavin. (d) QPCR of HCV RNA in untreated, FMN- or FAD-treated RFK- and FLAD1-KO Huh7.5.1 cells. (e) QPCR of DENV or HCV RNA in lumiflavin-treated Huh7.5.1 cells. For each concentration the significance of the effect on HCV versus DENV was determined. (f) HCV replicon assay in untreated and lumiflavin-treated Huh7.5.1 cells using WT sgJFH1 replicon. (g) Model of identified DENV and HCV host factors. Data depict average with s.e.m. (QPCR) or s.d. (FFU, RLU) for triplicate infections. The p-values were determined using an unpaired, parametric, two-sided student t-test, with a Welch post-correction, where * – P <0.05, ** – P <0.01, *** – P <0.001. ns = non-significant
Methods
Haploid Genetic Screen
The haploid genetic screen was performed as previously described9. In short, 100 million gene-trap mutagenized HAP1 cells were seeded and infected with DENV2 16681 (MOI=5). Eight hours after infection media was aspirated and replaced with IMDM containing 2% FBS. Clear cytopathic effects were observed after three days of infection leading to the death of the majority of cells. Clusters of cells resistant to DENV2 infection became apparent during further culture and 9 days after infection cells were harvested as a pool (yield ~30 million cells) and genomic DNA was isolated using a QIAamp DNA column. Gene-trap insertion sites were determined by linear amplification of the genomic DNA (gDNA) flanking regions of the gene-trap DNA insertion sites and sequenced on a Genome Analyzer II. Reads were aligned to the human genome using Bowtie and enrichment of independent insertions was calculated as previously described9.
CRISPR Genetic Screens
Huh7.5.1 cells were stably transduced with lentiCas9-Blast and subsequently selected using blasticidin. Next, a total of 300 million Huh7.5.1 cells that constitutively express Cas9 were transduced with the lentiGuide-Puro from the GeCKO v2 library31 at a MOI of 0.3. Cells were selected with puromycin and pooled together. The CRISPR genetic screens were started 10 days post transduction. 60 million mutagenized cells for each library (A and B) were infected with DENV2 16681 (replicate 1 and 2) or DENV2 strain 429557 (replicate 3) using a MOI of 1, or with HCV JFH1 at a MOI of 0.3. Cytopathic effect was visible 2 days and 5 days post infection for DENV and HCV, respectively. Huh7.5.1 cells grow slower than HAP1 cells and clusters of resistant cells took longer to develop. The selected cells were harvested 16 days after infection. As uninfected reference we chose the unselected starting population because in these strong positive selection screens the selection pressure of the viral infections renders potential small growth differences of the mutagenized cells inconsequential. For both selected and uninfected control cells gDNA was isolated using a QIAamp DNA columns, and the inserted guideRNA sequences were amplified from the gDNA by flanking primers and prepared for next-generation sequencing. Resulting amplicons were sequenced on a MiSeq or NextSeq platform (Illumina) and the enrichment of each guideRNA was calculated by comparing the relative abundance in the selected and unselected population. RIGER analysis was performed on guide RNAs (with at least 10 reads) ranked by enrichment using the weighted sum statistical method32. Each CRISPR screen was performed in 3 replicates and the mean of the 3 RIGER scores was calculated.
Cell culture
HAP1 cells were derived from the near-haploid chronic myeloid leukemia cell line KBM7 as described earlier9. HAP1 cells and knock out derivatives were cultured in IMDM supplemented with 10% fetal bovine serum, penicillin-streptomycin and L-glutamine. STT3A/STT3B double knockout HAP1 cells were cultured in IMDM supplemented with 10% fetal bovine serum, penicillin-streptomycin, L-glutamine and 25ng/ml doxycycline. Huh7, Huh7.5.1 (both gifts from Dr. Frank Chisari) and HEK293FT (Thermo Scientific) cells and knock out derivatives were grown in DMEM supplemented with 10% fetal bovine serum, penicillin-streptomycin, non-essential amino acids and L-glutamine. HEK293FT cells were used to generate lentivirus vectors for cellular transductions. Raji-DC-SIGN cells (a generous gift from Dr. Eva Harris) were cultured in RPMI-1640 supplemented with 10% fetal bovine serum, penicillin-streptomycin and L-glutamine. The cell lines have not been authenticated. Parental cell lines have been tested negative for mycoplasma.
Viral serotypes and strains
DENV2 infectious clone 16681 was a generous gift from Dr. Karla Kirkegaard, Stanford University. DENV2 from infectious clone 16681 was adapted to HAP1 cells through serial passaging. Viral whole genome sequence analysis revealed three coding mutations compared to the original clone 16681: Q399H in Envelope, L180F in NS2A and S238F in NS4B. DENV1 Hawaii 1944 (#NR82), DENV2 strain 429557 (#NR-12216), DENV2 New Guinea C 1944 (#NR-84), DENV3 Philippines/H871856 (#NR-80) and DENV4 H241 Philippines 1956 (#NR-86) were ordered from BEI resources (NIH, NIAID). Yellow Fever Virus was generated by culturing the Yellow Fever Vaccine YF-VAX 17D-204 vaccine. West Nile Virus (Kunjin strain CH 16532) was a generous gift from Dr. John F. Anderson, The Connecticut Agricultural Experiment Station. Zika virus (strain MR766) was kindly provided by Dr. Scott Weaver and Dr. Robert Tesh, University of Texas Medical Branch (UTMB), Galveston, Texas. Hepatitis C Virus JFH1 and HCV-Luc pFL-J6/JFH-5′C19Rluc2AUbi vector was a generous gift from Dr. Charles Rice, the Rockefeller University. Sindbis virus strain Ar-339 (TC adapted) Egypt 1952 (ATCC VR-1585) and Human Rhinovirus 14 (ATCC VR-284) were ordered from the American Type Culture Collection. Poliovirus type 1 strain Mahoney was a generous gift from Dr. Hidde Ploegh, Whitehead Institute. Venezuelan equine encephalitis virus TC-83 (pVEEV/GFP) was a generous gift from Dr. Ilya Frolov, University of Alabama at Birmingham.
QPCR Infectivity Assays
Cells were plated in 96 well plates and infected with an MOI of 0.1 of virus. Cells were harvested as outlined in Ambion Power SYBR Green Cells-to-Ct kit (Ambion 4402954). Cells were harvested 8 hours post infection with Polio virus, 24 hours post infection with Sindbis virus, Venezuelan equine encephalitis virus and Human Rhinovirus 14, 2 days post infection with DENV2 16681 and Zika virus, Yellow Fever Virus, and West Nile Virus Kunjin strain, 3 days post infection with HCV JFH1 and 5 days post infection with DENV2 New Guinea. For comparison of DENV serotypes, cells were infected with DENV1 Hawaii 1944, DENV2 New Guinea C 1944, DENV3 Philippines/H871856 and DENV4 H241 Philippines 1956 for 2 days. All samples were normalized to 18S expression. Two independent experiments were performed with triplicate infections and one representative is shown.
The following QPCR primers were used:
DENV2-Forward: GCCCTTCTGTTCACACCAT
DENV2-Reverse: GGCTCTGCCAATCAGTTCAT
Universal-DENV-F GGTTAGAGGAGACCCCTCCC
Universal-DENV-R GGTCTCCTCTAACCTCTAGTCC
Yellow Fever-Forward: GAAATGCCTGCCCTTTATGA
Yellow Fever-Reverse: GCACATGGCAACAGAAGCTA
Kunjin-Forward: GCTTTGCCACCTCTCTTCAC
Kunjin-Reverse: CGGTTGATGGTTTCCACTCT
ZIKV-Forward: ACCATACGGCCAACAAAGAG
ZIKV-Reverse: TCCACAGCCAGGAAGAGACT
HCV-Forward: TCTCTCAGTCCTTCCTCGGA
HCV-Reverse: AAGCCGGCTAGAGTCTTGTT
SINV-Forward: CGCGGTCACGTAAGGATAAT
SINV-Reverse: TTTGGCATTCTTCAGCACAG
Polio-Forward: CAACCTCCCACTGGTGACTT
Polio-Reverse: ATTTCCCCTGCTCAACCTTT
18S-Forward: AGAAACGGCTACCACATCCA
18S-Reverse: CACCAGACTTGCCCTCCA
VEEV-Forward: CAGGACGATCTCATTCTCAC
VEEV-Reverse: TCATTCACCTTGTACCGAACG
HRV-14-Forward: aagcaatttggtggtccaag
HRV-14-Reverse: acactggggtttgaagcact
Crystal violet staining
For virus infections WT and KO cell lines were plated out in either 24 well or 96well plates. Cells were infected with DENV2 16681, HCV, SINV or Polio using a MOI of 1. Huh7-STT3A-KO-STT3B-KO-pLenti-TRE3G-CMV-STT3B cells were cultured in presence or absence of 25ng/ml doxycycline. Cells were incubated for 48 to 120hrs then fixed using 4% formaldehyde in PBS. Cell viability at time of fixation was determined by crystal violet staining.
Plaque-forming units assay
Plaque assays were performed on BHK-21 cells. Briefly, BHK-21 monolayers were grown to 80% confluency in 24-well plates and incubated for 1 h at 37°C in 5% CO2 with serially diluted virus supernatants from WT and mutant HAP1 cells infected with DENV, MOI 0.1 for 48 hours. The wells were then overlaid with Dulbecco’s modified Eagle’s medium, 0.8% Aquacide II (EMD Millipore), and 10% FBS; incubated for 7 days; and fixed with 10% formaldehyde. The cells were then stained overnight with crystal violet. Next day the wells were extensively washed with water then dried and the resulting plaques were counted and plaque-forming units (PFU) per ml were calculated. Two independent experiments were performed with triplicate infections and one representative is shown.
Focus-forming units assay
WT and KO Huh7.5.1 cells were plated in 24 well plates and infected with HCV with a MOI of 0.1. 3 days post infection supernatant was collected and added to WT Huh7.5.1 cells in a 10-fold dilution series. After 3 days cells were fixed, stained with mouse-anti-HCV-core (Abcam ab2740) and anti-mouse-IgG-Alexa-488 (Life technologies) and fluorescent colonies were counted. Two independent experiments were performed with triplicate infections and one representative is shown.
Luciferase reporter virus assays
Cells were plated out in 96 well plates in triplicates and infected with dengue luciferase reporter virus at an MOI of 0.01. Cells were incubated with dengue luciferase reporter virus at 37C 5%CO2 and cell lysates were harvested. Luciferase expression was measured using Renilla Luciferase Assay system (Promega E2820). Cells were lysed using Renilla lysis buffer and luciferase activity measured by addition of substrate and immediate luciferase readings were taken using Glomax 20/20 luminometer using a 10sec integration time. For the cross-comparison of the effects of host factors on DENV and HCV Huh7.5.1 KO cell lines were infected with dengue luciferase reporter virus at an MOI of 0.01 or with HCV luciferase virus at an MOI of 0.2. For the validation of HCV host factors 4 different KO cell lines (created using lentiCRISPRv2) were infected with HCV luciferase virus at an MOI of 0.2. Two independent experiments were performed with triplicate infections and one representative is shown, with the exception of the experiment shown in Extended Data Fig. 4e which was performed once with triplicate infections.
Infection of Raji DC-SIGN cells
Raji DC-SIGN host factor KO cell lines were created by transduction of lentiCRISPRv2 and subsequent puromycin selection. Resulting cell lines were infected with dengue luciferase virus at an MOI of 0.05 and harvested 3 days post infection. Three independent experiments were performed with triplicate infections and one representative is shown.
Internalization assay and Confocal Microscopy
10,000 Huh7 were seeded on Lab-TekII Chamber slides (Thermo Fisher Scientific). Next day, cells were incubated on ice for 15min, infected with DENV (MOI=60) and incubated on ice for 1h. Cells were washed 3 times with ice cold PBS and subsequently incubated at 37°C for 0 or 30min. At each time point, 10μg/ml wheat germ agglutinin-Alexa-594 (Life technologies, W11262) was added for 10 min at room temperature prior to 3 washes with PBS and fixation with 4% paraformaldehyde. Dengue virus was stained with a rabbit-anti-Dengue-envelope antibody (Genetex 127277) in block/perm buffer (1% saponin, 1% Triton-X-100, 5% FBS) for 1h followed by incubation with goat anti-rabbit-IgG-Alexa-488 (Life technologies, A-11008) and DAPI (Insitus, F203) for 30min. After 3 washes with PBS cells were visualized using confocal microscopy.
Replicon assays
Dengue Replicon Plasmid was linearized using XbaI restriction enzyme. Replicon RNA was generated using the MEGAscript T7 High Yield Transcription Kit (Ambion AM1334) with the reaction containing 5mM m7G(5′)ppp(5′)G RNA Cap Structure Analog (NEB S1405S). Resulting RNA was purified by sodium acetate ethanol precipitation. HCV sgJFH1 replicon33 RNA was prepared as described for DENV with the exception of adding the Cap Structure Analog. Cells were washed twice with PBS and re-suspended in electroporation buffer (Teknova E0399). 3μg of purified replicon RNA was mixed with cells and cells were electroporated using Bio-Rad Gene Pulser Xcell electroporator using square wave protocol. Electroporated cells were resuspended in cell culture medium without antibiotics and plated into 24well plates. Luciferase expression was measured using Renilla Luciferase Assay system (Promega E2820). Cells were lysed using Renilla lysis buffer and luciferase activity measured by addition of substrate and luciferase readings were taken immediately using Glomax 20/20 luminometer using a 10sec integration time. For lumiflavin treatment cells were electroporated with 2μg of viral RNA and 1μg of firefly mRNA (Trilink) to normalize for effects on cell proliferation. For the lumiflavin treatment two independent experiments with three electroporations each were performed. One representative is shown. For the replicon assay in ELAVL1-KO cells three independent experiments with a single electroporation were performed. The average of the experiments is shown. For the DENV-Replicon assay three independent experiments were performed. One representative experiment was shown.
Immunoblot Analysis
Cell pellets were lysed using laemmli SDS sample buffer containing 5% beta-mercaptoethanol and boiled for 10min. Lysates were separated by SDS-PAGE on pre-cast Bio-Rad 4–15% poly-acrylamide gels in bio-rad miniprotean gel system. Proteins were transferred onto PVDF membranes using bio-rad trans-blot protein transfer system. PVDF membranes were blocked with PBS buffer containing 0.1% tween-20 and 5% non-fat milk. Blocked membranes were incubated with primary antibody diluted in blocking buffer and incubated overnight at 4 degrees Celsius rotating. Primary antibodies were detected using HRP-conjugated secondary anti-mouse and anti-rabbit antibodies Genetex by incubating membranes with 1:5000 dilution for 1 hr at room temperature. Antibody bound proteins were detected by incubating with pierce west pico and extended duration peroxide solutions and visualized on film. WT cells were treated with 10μg/ml Tunicamycin and treated for 3–4hrs at 37°C 5% CO2. To visualize proteins by immunoblotting the following antibodies were used anti-SHBG (Genetex GTX63795) at a dilution of 1:2500. Anti-P-Sap (Genetex GTX101064) at a dilution of 1:2500. Anti-HA C29F4 (Cell Signaling 3724P) at a dilution of 1:2500. Anti-Mouse M2-FLAG (Sigma F1804-200UG) at a dilution of 1:2500. Anti-DYKDDDK (FLAG) (Cell signaling 2368s) at a dilution of 1:2500. Anti-RPN1 (generous gift from Martin Ivessa) at a dilution of 1:2000. Anti-NS1 (Genetex GTX124280) at a dilution of 1:2500. Anti-P84 (Genetex GTX70220) at a dilution of 1:3500. Anti-DENV-ENV (Genetex GTX127277) at a dilution of 1:2500. Anti-prM (Genetex GTX128092) at a dilution of 1:2500. Anti-NS2B (Genetex GTX124246) at a dilution of 1:2500. Anti-NS3 (Genetex GTX124252) at a dilution of 1:2500. Anti-RFK (Sigma SAB1409492) at a 1:500 dilution. Anti-FLAD1 (Santa Cruz Bio sc-376819) at a 1:250 dilution. Anti-STT3B (Sigma HPA036646) at a dilution of 1:1000. Anti-MAGT1 (Proteintech Group 17430-1-AP) at a dilution of 1:1000, Anti-RPS25 (Abcam 102940) at a dilution of 1:1000. Anti-HUR (Santa Cruz sc-5261) at a dilution of 1:200. Anti-SRRD (Sigma HPA002945) at a dilution of 1:500.
Lentiviral or retroviral complementations
Lentiviral or retroviral transduction was used to create stable cell lines expressing a selected gene of interest. Respective genes of interest (see Construction of lenti- or retroviral constructs section) were cloned into the pLenti-CMV-Puro-DEST vector (w118-1) (a gift from Eric Campeau), or the PMX-IRES-BLAST-DEST (see Construction of lenti- or retroviral constructs). Lentivirus or retrovirus produced in HEK293FT cells were used to transduce respective cell lines overnight. Cells stably expressing the gene of interest were selected by treatment with 1–4 μg/ml puromycin or 10–50μg/ml Blasticidin over 2 days (InvivoGen) along with untransduced cells as negative control.
Genome Engineering
CRISPR gRNA sequences were designed using the Zhang lab CRISPR design tool (see Extended Data Fig. 3 for CRISPR target sites). Corresponding oligos or geneblocks containing U6 promoter sequence and U6 termination sequence were ordered from IDT. Oligos were cloned into the Zhang lab generated Cas9 expressing pX458 guide RNA plasmid (Addgene) as previously described using Gibson assembly reaction New England Biolabs. Geneblocks were cloned into pCR-Blunt II-TOPO vector (Life Technologies). TOPO-cloned geneblocks were co-transfected into respective cells with a mCherry-expressing construct and hCas9-expressing vector (Addgene 41815 hCas9 Church pcDNA3.3-Topo) guideRNA encoded in the pX458 plasmids were transfected alone using lipofectamine 2000 (Life technologies) according to manufactures guidelines. Transfected cells were single cell sorted based on GFP or mCherry expression into 96well plates using BD influx cell sorter. Clonal cell lines were allowed to expand and genomic DNA was isolated for sequenced based genotyping of targeted allele. For this, a 500–700 base-pair (bp) region that encompassed the gRNA-targeted site was amplified and the PCR product was Sanger sequenced. In haploid cells (HAP1), only one mutated allele was present in the sequenced PCR product and cellular subclones containing a frame shift mutations or large indels were selected. In aneuploid Huh7.5.1 cells, we regularly observed that the PCR product contained more than 1 trace, suggesting non-identical mutations in multiple alleles. In this case, the PCR product was cloned into a plasmid vector and colonies were sequenced to separate allele specific mutations. Subclones were chosen where all alleles were mutated. It should be noted that in aneuploid Huh7.5.1 cells, we sometimes observed cellular subclones where all mutant alleles contained the same mutation (e.g. CD81 and ELAVL1). It has been reported that CRISPR/Cas9 technology can generate homozygous biallelic mutations more frequently than expected in diploid cells or cancer cells34,35, perhaps because both alleles were independently repaired in an identical manner or because one allele served as a template for homology-directed repair of the other allele. To create KO cell lines using lentiCRISPRv2 (Addgene) the following guideRNAs below were cloned into the vector, Huh7.5.1 cells were lentivirally transduced and selected with Puromycin. The following guideRNA sequences were used:
ANKRD49: AGAAAGGAGTCTCCGCACTG
ANKRD49 guide2: ATGAACCGTTACGTCAAACC
ANKRD49 guide3: GCCCAAAGAAGCAATCTGCT
ANKRD49 guide4: AGAAAGGAGTCTCCGCACTG
CD81: GCGCCCAACACCTTCTATGT
CLDN1: CGATGGCGCCGATCCATCCC
ELAVL1: TTGGGCGGATCATCAACTCG
ELAVL1 guide2: TGTGAACTACGTGACCGCGA
ELAVL1 guide3: GGGCCTCCGAACCGTCGCGC
ELAVL1 guide4: AGAGCGATCAACACGCTGAA
EMC1: AGGCCGAATCATGCGTTCCT
EMC2: GATTGCCATTCGAAAAGCCC
EMC3: GTGCCACCTTCTCCTATGAC
EMC4: TGCTTGTCCAAGTAACCGAC
FKBPL: GTCAAGAAGATCGTAATCCG
FKBPL guide2: GAAGAGCCCGTCCATAGCAT
FKBPL guide3: ACAGAGCTAACTATGGGCGT
FKBPL guide4: GTTTCGGTAGGAGGGTCTCG
FLAD1: ACAGACCATTGAGACCTCCC
FLAD1 guide2: CATGCGCATCAACCCACTGC
FLAD1 guide3: TACAGGAGTAGGGGTCAGTC
FLAD1 guide4: TGTGTCCCTGGGGGTTGAAG
MAGT1: GAGCGAACATGGCAGCGCGT
MIR-122: GAGTTTCCTTAGCAGAGCTG
MMGT1: CAGGCACTTACGCTGCGCAG
Non-targeting: GCCCAGACGCCCTAGAATAG
OCLN: ACGTAGAGTCCAGTAGCTGC
OSTC: TCAGTCATAGAACCGACACT
PPIA: GTACCCTTACCACTCAGTCT
RFK: TATCATGCATACCTTCAAAG
RFK guide2: GGTCAAGTGGTGCGGGGCTT
RFK guide3: CTATGGGGAAATCCTCAATG
RFK guide4: CCAACCATAGTAAATACCAG
RPN2: TCGCTACCACGTGCCAGTTG
SRRD: GACTGTTCTCAGTGAGAACG
SRRD guide2: GATAGATACCTTTGCAATGT
SRRD guide3: ATTGAAGTCCTTAACACCCT
SRRD guide4: AACAACTGAAGGCCCCTGTG
SSR2: CAATAGCAGGGGGATGCCGA
SSR3: GACCCTAGTAAGCACATATT
STT3A: GTACTCACGGATCAAACTCA
STT3B : TACAGCAAAAGAGTCTACAT
ZEB1: TGAAGACAAACTGCATATTG
TALENs targeting AUP1 in HAP1 cells were generated as indicated in Extended Data Fig. 3. Cells were co-transfected with Left and Right TALEN containing constructs and an mCherry expressing construct using lipofectamine 2000 (Life technologies) according to manufactures guidelines. Transfected cells were single cell sorted based on mCherry expression into 96well plates using BD influx cell sorter. Subclones were allowed to expand and genomic DNA was isolated for sequenced based genotyping of AUP1 allele. HAP1 cells containing gene-trap insertions in STT3A, STT3B, RPN1, SSR2, SSR3, ASCC2 and RPS25 were isolated by picking resistant colonies from the DENV2 haploid genetic screen. Picked colonies were screened for gene-trap insertions utilizing PCR with primers directed to the genetrap and the flanking region of the gene of interest.
Co-immunoprecipitation
WT HAP1 cells were transduced with STT3A-FLAG, STT3B-FLAG or RPS25-FLAG lentiviral vectors (see “Construction of lenti- or retroviral constructs” section). HAP1 cells expressing FLAG tagged proteins were trypsinized and washed once with PBS. Cells were lysed with TNM buffer (25mM Tris-HCL, 15mM NaCl, 5mM MgCl2) containing 1% digitonin, 1mM PMSF, and Halt™ Protease and Phosphatase Inhibitor Cocktail (Life Technologies 78440) for 1hr on ice gently vortexing every 15min. Cell lysates were clarified by centrifugation at 15000×g for 10min. Clarified lysates were incubated at 4C overnight with Anti-FLAG® M2 Magnetic Beads (Sigma M8823-5ml). Incubated beads were washed 3 times with TNM buffer containing 0.1% Digitonin, 1mM PMSF, 1X halt protease and phosphatase inhibitor. Cells were washed once with TNM buffer and competitively eluted with TNM buffer containing 150ng/ul 3X FLAG® Peptide (Sigma F4799) for 30min on ice. For immunoblotting elutions were denatured by boiling in 5x Sample buffer and analyzed by SDS-PAGE using antibodies against DENV non-structural proteins. Elutions were also prepared for Mass spec analysis. Cells expressing RPS25-FLAG, a host protein with a likely different mechanistic action, as well as untransduced cells, were used as a negative control in these experiments.
SYPRO Ruby Staining
After electrophoresis gel was fixed for 30min in fixative buffer (50% Methanol, 7% Acetic acid) and incubated with SYPRO Ruby Stain (Fischer #S-12000) overnight. Gels were then washed once with wash buffer (10% Methanol, 7% Acetic acid) and twice with distilled water. Gel was imaged on Molecular Dynamics Storm scanner.
Construction of STT3A/STT3B double knockout cell line with STT3B conditionally expressed using a Tet-On system
HAP1 cells stably transduced with the transactivator pLenti-CMVrtTA3G-Blast (R980-M38-658) Addgene# 31797 with the endogenous STT3B gene disrupted by CRISPR/Cas9 were lentivirally transduced with pLenti-CMVTRE3G-Puro-STT3B-FLAG which drives STT3B under a doxycycline inducible promoter. Transduced cells were then transfected with pX458 plasmid encoding a guideRNA targeted to the STT3A gene. Transfected cells were then subcloned based on GFP expression of PX458 plasmid into 96well plates containing IMDM+10% fetal bovine serum+Penn/Strep, L-glut, and 25ng/ml doxycycline. Subclones were allowed to grow for 2 weeks, then replica plated and in one replicate the doxycycline medium was washed away and replaced with regular growth medium without doxycycline and incubated for 5 days. Cells that were dependent on doxycycline for growth were genotyped to verify both endogenous STT3A and STT3B had double frame shifting CRISPR/Cas9 editing events. STT3A/STT3B endogenous double knock out cells were then lentivirally transduced with wild-type or mutant STT3A or STT3B under the CMV promoter. The lentivirally transduced cell lines were then plated in 96 well plates and the doxycycline was washed away and incubated for 5 days. Cells were then fixed and stained with crystal violet to assess cell viability.
Treatment of HCV infected cells with lumiflavin, FMN and FAD
WT Huh7.5.1 cells were treated with lumiflavin (Santa Cruz Bio sc-224045) ranging from 10–100μM and infected with HCV or DENV2 at a MOI of 0.1. For WNV, YFV, PV, SINV, VEEV and HRV-14 cells were treated with 50μM lumiflavin and infected at a MOI of 0.1. For rescue of HCV replication in lumiflavin-treated cells 100μM FMN and 10mM FAD were used. RFK- and FLAD1-KO Huh7.5.1 were cultured in absence or presence of 500μM FMN (TCI America R0023) or FAD (Sigma F8384) and subsequently infected with HCV at a MOI of 0.1. After 3 days of infection levels of infection were determined using immunofluorescence, Western blot and QPCR. Anti-HCV core 1b (Abcam ab2740) was used at 1:500 for IF and 1:1000 for WB. Anti-DENV2 NS5 (GeneTex GTX103350) was used at a 1:2500 dilution for WB. For the lumiflavin treatment two independent experiments with triplicate infections were performed and one representative is shown. For the FMN/FAD complementation two independent experiments were performed and the average is shown.
MTT assay
To test effects of lumiflavin on cell viability MTT assay was performed according to the manufacturer’s instructions (Sigma Cell Proliferation Kit I (MTT) 11465007001). Three independent experiments in triplicates each were performed and one representative is shown.
Transmission Electron Microscopy
Cells stably expressing STT3B-APEX2 were plated in 6well plates and infected with an MOI 5 of DENV2. 28hrs after infection, cells were washed with PBS and fixed with 2%glutaraldehyde in 100mM sodium cacodylate, 2mM CaCl2 pH 7.4 buffer. Cells were fixed at 4C for 60min then washed three times with PBS. Fixed cells were quenched with 100mM sodium cacodylate, 2mM CaCl2 pH7.4 and 20mM glycine. Quenched cells were washed twice with PBS and stained with using the KPL DAB reagent set (KPL#54-10-00) for 8hrs. After incubation with DAB, cells were rinsed twice with PBS and scraped off well using a cell scraper and pelleted. Pelleted cells were re-suspended in 10% Gelatin in 0.1M Sodium Cacodylate buffer pH 7.4 at 37°C and allowed to equilibrate 5 min. Cells were pelleted again, excess gelatin removed, then chilled in cold blocks and covered with cold 1% Osmium tetroxide (EMS Cat# 19100) for 2 hr rotating in a cold room. They were then washed 3X with cold ultrafiltered water, then en bloc stained overnight in 1% Uranyl Acetate at 4°C while rotating. Samples were then dehydrated in a series of ethanol washes for 20 minutes each @ 4°C beginning at 30%, 50%, 70%, 95% where the samples were then allowed to rise to RT, changed to 100% ethanol 2X, then Propylene Oxide (PO) for 15 min. Samples were then infiltrated with EMbed-812 resin (EMS Cat#14120) mixed 1:2, 1:1, and 2:1 with PO for 2 hrs each with leaving samples in 2:1 resin to PO overnight rotating at RT in the hood. The samples were then placed into EMbed-812 for 2 to 4 hours then placed into molds w/labels and fresh resin, orientated and placed into 65°C oven overnight. Sections were taken approx. 80nm, picked up on formvar/Carbon coated 100 mesh Cu grids, stained for 30seconds in 3.5% Uranyl Acetate in 50% Acetone followed by staining in 0.2% Lead Citrate for 3 minutes. Observed in the JEOL JEM-1400 120kV and photos were taken using a Gatan Orius 4k × 4k digital camera.
Mass spectrometry
Liquid chromatography-tandem mass spectrometry: Elutions from co-immunoprecipitations were trypsin digested and purified using Sep Pak C18 purification column. Peptides were analyzed using an LTQ Velos Orbitrap mass spectrometer (Thermo Fisher Scientific, San Jose, CA) coupled to an Agilent 1100 high performance liquid chromatography pump (Agilent Technologies, Santa Clara, CA) and a MicroAS autosampler (Thermo Fisher Scientific, San Jose, CA). Peptide mixtures were introduced into the mass spectrometer via a fused silica microcapillary column (100 μm inner diameter) ending in an in-house pulled needle tip (internal diameter ≈ 5 μm). Columns were packed to a length of 17 cm with a C18 reversed-phase resin (Magic C18AQ; Michrom Bioresources, Auburn, CA). Peptides were loaded onto the column and then eluted into the nanospray ionization source of the mass spectrometer via a two-step gradient of 7–25% buffer B (2.5% water and 0.1% formic acid in acetonitrile (v/v)) in buffer A (2.5% acetonitrile and 0.1% formic acid in water (v/v)) over 60 min followed by a second phase of 25–45% buffer B over 20 min. Eluting peptides were measured by the LTQ Velos Orbitrap operating in a data-dependent mode in which 10 ion-trap MS/MS spectra were acquired per data-dependent cycle from a high-resolution (R = 60,000) precursor spectrum (mass range = 360–1600 m/z).
Mass spectrometry data processing: Raw data files produced by the mass spectrometer were converted to the mzXML format using in house software, MS Convert. MS and MS/MS data were extracted from mzXML files with in-house software. MS/MS spectra were analyzed using Sequest algorithm searching a composite target-decoy protein sequence database. The target sequences consisted of human proteins downloaded from the Uniprot database (11-17-2014) and protein sequence corresponding to the Dengue virus 2 16681 polyprotein. Decoy sequences were created by reversing the orientation of all target sequences. Parameters used for all searches included the requirement of trypsin peptide cleavage, two missed cleavages allowed, peptide mass tolerance of 20 ppm, variable oxidation of methionine residues (+15.99491 Da), and static carbamylation modification of cysteine residues (+57.02146). Decoy peptide identifications guided the creation of filtering criteria delivering preliminary sets of peptide-spectrum matches (PSMs) with estimated false discovery rate <1%. Spectral counts for each condition were combined at a protein level and normalized by protein length to infer protein abundances in each case.
DENV Reporter virus and DENV Replicon design and generation
Construction of pDENV-Luc replicon: The design of the DENV replicon was based on DVRep described previously36: The viral 5′UTR was followed by a duplication of the first 102 nucleotides of the C coding region, which contain cis-acting elements required for replication (CAE). The CAE was fused to the renilla luciferase coding region followed by the DENV open reading frame (ORF) starting at the signal peptide preceding NS1. Between the luciferase and the DENV structural proteins a foot and mouth disease virus (FMVD) 2A sequence was introduced to provide cotranslational cleavage and release of luciferase. The construct was based on pD2/IC-30P, which contains a full-length infectious clone encoding dengue virus serotype 2 strain 1668137. We also included the amino acid mutation Q399H in the Envelope protein. We gene-synthesized a fragment containing the T7 polymerase promoter sequence followed by the first 102 nucleotides of the C coding region in frame with Renilla luciferase and FMDV 2A followed by the DENV open reading frame (ORF) starting at the signal peptide preceding NS1 until an internal HpaI site. This fragment was released by SacI (preceding the T7 promoter) and HpaI and cloned in pD2/IC-30P in a three point ligation with KpnI/SacI and KpnI/HpaI fragments.
Construction of pDENV-Luc infectious clone: The design of the DENV reporter was based on mDV-R described previously38: The viral 5′UTR was followed by a duplication of the first 104 nucleotides of the C coding region, which contain cis-acting elements required for replication (CAE). The CAE was fused to the renilla luciferase coding region followed by the complete DENV open reading frame (ORF). Between the luciferase and the DENV structural proteins a foot and mouth disease virus (FMVD) 2A sequence was introduced to provide cotranslational cleavage and release of luciferase. The construct was based on pD2/IC-30P, which contains a full-length infectious clone encoding dengue virus serotype 2 strain 1668137 in which an Envelope Q399H mutation was introduced that enhanced viral infection in mammalian cells using primers 5′-GGAAGTTCTATCGGCCACATGTTTGAGACAAC-3′ 5′-GTTGTCTCAAACATGTGGCCGATAGAACTTCC-3′ via the QuikChange Site-Directed Mutagenesis kit (Agilent Technologies, Santa Clara, CA). We gene-synthesized a fragment containing the T7 polymerase promoter sequence followed by the first 104 nucleotides of the C coding region in frame with Renilla luciferase and FMDV 2A. This fragment was PCR amplified, introducing a SacI site at the 5′end and a NheI site (present in the FMDV 2A sequence) at the 3′end using primers: 5′-CGAAATTCGAGCTCACGCG-3′ and 5′-TCCTGCTAGCTTGAGCAAATCAAAGTTC-3′. To create and in frame fusion of FMDV 2A with the DENV-ORF a second DNA fragment was amplified using pD2/IC-30P as template with primers: 5′-TCAAGCTAGCAGGAGACGTTGAGTCCAACCCCGGGCCCATGAATAACCAACGGAAAAAGGCG-3′ and 5′-GGAAGAGCATGCAGTCGGAAATG-3′ introducing 5′ NheI and 3′ SphI restriction sites. The two fragments were cut with the respective restriction enzymes and ligated into pD2/IC-30P cut with SacI and SphI to create pDENV-Luc. DENV-Luc virus was produced by cutting with XbaI to linearize plasmid and in vitro transcription performed of pDENV-Luc and transfection into BHK cells using lipofectamine 2000.
Construction of lenti- or retroviral constructs
PMX-IRES-BLAST-DEST was made by cutting pMXs-IRES-Blasticidin Retroviral Vector (cell biolabs CATALOG NUMBER: RTV-016) with SnaBI and the Gateway destination cassette (reading frame A) was blunt cloned in to this vector according to manufacturer’s protocol (Gateway® Vector Conversion System; Invitrogen catalogue number 11828-029)
To generate a lentiviral construct expressing STT3A-FLAG, Dharmacon cDNA BC020965 was used as template to generate a PCR product using primers 5′-CACCATGACTAAGTTTGGATTTTTGCG-3′ and 5′-TTACTTATCGTCGTCATCCTTGTAATCTGTCCTTGACAAGCCTCGATT-3′
Amplified PCR product was then topo cloned into Gateway compatible entry vector pENTR™/D-TOPO® Cloning Kit (Life Technologies K2400-20) and gateway reaction (Life Technologies) used to insert into pLenti-CMV-Puro-Dest (w118-1).
To generate a lentiviral construct expressing STT3B-FLAG, Dharmacon cDNA BC052433 was used as template to generate PCR product using primers forward primer 5′-CACCATGTCTTGGTGGGATTATGGC-3′ and reverse primer 5′-TTACTTATCGTCGTCATCCTTGTAATCAACAGTCTTCTTAGAGGTCTTCTT-3′. It should be noted that we used Mus musculus STT3B because we were unable to clone human STT3B.
Amplified PCR product was then topo cloned into Gateway compatible entry vector pENTR™/D-TOPO® Cloning Kit (Life Technologies K2400-20) and gateway reaction was used to insert into pLenti-CMV-Puro-Dest(w118-1).
To generate an SHBG expressing construct SHBG was ordered as two Geneblocks and used to generate PCR product using primers 5′-TGTGGTGGAATTCTGCAGATACCTGTGGTGGAATTCTGCAGATACC-3′ with 5′-ATCCAGCACAGTGGCGG-3′. PCR product was Gibson cloned Gibson assembly reaction kit (New England Biolabs, UK) into pLenti-CMV-Puro-Dest(w118-1) that was EcoRV digested.
To generate a FLAG3x-RPS25 expression construct, entry vector PENTR-FLAG3x-RPS25 was generated as described39 using the forward primer CACCATGGACTACAAAGACCATGACGG. PENTR-FLAG3x-RPS25 was then used in a Gateway reaction (Life Technologies) to introduce FLAG3x-RPS25 into PMX-IRES-BLAST-DEST retroviral expression construct.
To generate a construct expressing STT3B fused with APEX2 and FLAG tagged PCR products were generated using pLenti-STT3B expression construct described above and APEX2 addgene plasmid 4938640,41 as templates to generate PCR products using primers 5′-GACTCTAGTCCAGTGTGGTG-3′ with 5′-AACAGTCTTCTTAGAGGTCTTC-3′ and 5′-GAAGACCTCTAAGAAGACTGTTATGGACTACAAGGATGACGA-3′ with 5′-CGGCCGCCACTGTGCTGGATTTAGGCATCAGCAAACCCAAG-3′. PCR products were Gibson assembled Gibson assembly reaction kit (New England Biolabs, UK) into pLenti-CMV-Puro-Dest(w118-1) that was EcoRV digested.
To generate a STT3B-Doxycycline-lenti construct, Dharmacon cDNA BC052433 was used as template to generate PCR product using primers 5′-CACCATGTCTTGGTGGGATTATGGC-3′ and 5′-TTACTTATCGTCGTCATCCTTGTAATCAACAGTCTTCTTAGAGGTCTTCTT-3′
Amplified PCR product was then topo cloned into Gateway compatible entry vector pENTR™/D-TOPO® Cloning Kit (Life Technologies K2400-20) and gateway reaction was used to insert into the doxycycline inducible lentiviral vector pLenti CMVTRE3G Puro DEST (w811-1) Addgene#27565.
To generate catalytic site mutants (Extended Data Fig. 5 and refs15,42) STT3A and STT3B expressing constructs DNA fragments were generated using pLenti-STT3B-FLAG described above as template using pLenti-EcoRV primers and mutant primers to generate two PCR products (see primers below). Both PCR products for each mutation were Gibson cloned (Gibson assembly reaction kit (New England Biolabs, UK) into pLenti-CMV-Puro-Dest(w118-1) that was EcoRV digested.
Primers Forward and Reverse
pLenti EcoRV-F GACTCTAGTCCAGTGTGGTG
pLenti EcoRV-R ATCCAGAGGTTGATTGTCGAG
STT3A Mutations
E63A-F CAGGTTCCTGGCTGAGGCCGGGTTTTATAAATTCCATAACTGG
E63A-R CCGGCCTCAGCCAGGAACCTG
D167A-F CTGTGGCTGGCTCCTATGCCAATGAAGGGATTGCCATCTTTTG
D167A-R CATTGGCATAGGAGCCAGCCACAG
E351Q-F CCATCATTGCTTCTGTGTCTCAGCATCAGCCCACAACCTG
E351Q-R ATGCTGAGACACAGAAGCAATGATGG
STT3B Mutations
D100A-F ATCATCCACGAGTTCGCCCCGTGGTTTAACTATAG
D100A-R CTATAGTTAAACCACGGGGCGAACTCGTGGATGAT
D218A-F CAGTGGCGGGATCCTTTGCCAATGAAGGCATTGCCATT
D218A-R AATGGCAATGCCTTCATTGGCAAAGGATCCCGCCACTG
E402Q-F CAATTATTGCATCAGTGTCTCAGCATCAGCCTACGACATGG
E402Q-R CCATGTCGTAGGCTGATGCTGAGACACTGATGCAATAATTG
To generate ELAVL1 fused with FLAG cDNA was prepared from total RNA of Huh7.5.1 using Biorad RT Superscript and ELAVL1 was PCR amplified with 5′-TGTGGTGGAATTCTGCAGATACCATGTCTAATGGTTATGAAGACCA-3′ and 5′-CGGCCGCCACTGTGCTGGATTTACTTATCGTCGTCATCCTTGTAATCTTTGTGGGACTTG-3′. Next, using Gibson Assembly the PCR product was cloned into pLenti-CMV-Puro-Dest(w118-1) that was digested with EcoRV.
To generate RPN1 fused to 2Strep cDNA BC010839 was PCR amplified with 5′-CACCATGGAGGCGCCAGCCGC-3′ and 5′-CTACAGGGCATCCAGGATG-3′. The amplified fragments were cloned into P-ENTR-D-Topo (Invitrogen) then a Gateway LR reaction was performed to shuttle cDNA into Plenti-CMV-puro expression vector.
To generate SSR2 fused to 2Strep cDNA NM_003145.3 was PCR amplified with 5′-CACCATGAGGCTGCTGTCATTTGTG-3′ and 5′-TCAGTTCTTCTTCGTTTTGGGAG-3′. The amplified fragments were cloned into P-ENTR-D-Topo (Invitrogen) then a Gateway LR reaction was performed to shuttle cDNA into Plenti-CMV-puro expression vector.
To generate SSR3 fused to 2Strep cDNA NM_003145.3 was PCR amplified with 5′-CACCATGGCTCCTAAAGGCAGCTC-3′ and 5′-CTATTTGGAGCCAGTAGACAG-3′. The amplified fragments were cloned into P-ENTR-D-Topo (Invitrogen) then a Gateway LR reaction was performed to shuttle cDNA into Plenti-CMV-puro expression vector.
To generate ASCC2 fused to 2Strep BC025368 was PCR amplified with 5′-CACCATGCCAGCTCTGCCCCTGG-3′ and 5′-TCAGGATGGGATCATGCCTTTGCT-3′. The amplified fragments were cloned into P-ENTR-D-Topo (Invitrogen) then a Gateway LR reaction was performed to shuttle cDNA into Plenti-CMV-puro expression vector.
To generate RPS25 fused to 2Strep NM_001028 was PCR amplified with 5′-CACCATGGACTACAAAGACCATGACG-3′ and 5′-TTAATTAACCTCGAGTTTAAACGCG-3′. The amplified fragments were cloned into P-ENTR-D-Topo (Invitrogen) then a Gateway LR reaction was performed to shuttle cDNA into Plenti-CMV-puro expression vector.
To generate UBE2J1 expression contruct, cDNA kindly provided by Ron Kopito (Stanford University) was PCR amplified with 5′-TGTGGTGGAATTCTGCAGATACCATGGAGACCCGCTACAACCTG-3′ and 5′-CGGCCGCCACTGTGCTGGATTTATAACTCAAAGTCAAATATGTATTC-3′. The amplified fragments were cloned into Plenti-CMV-puro expression vector by a Gibson Assembly reaction.
To generate SEL1L expression contruct, cDNA kindly provided by Ron Kopito (Stanford University) was PCR amplified with 5′-TGTGGTGGAATTCTGCAGATACCATGCGGGTCCGGATAGGGCTG-3′ and 5′-CGGCCGCCACTGTGCTGGATTTAAAGTCTACTTACCAAAACCATG-3′. The amplified fragments were cloned into Plenti-CMV-puro expression vector by a Gibson Assembly reaction.
To generate AUP1, glycerol stocks containing AUP1 cDNA in pENTR entry vector were ordered from Darmacon (#OHS5894-99868092) a Gateway LR reaction was performed to shuttle cDNA into Plenti-CMV-puro expression vector.
Comparison of knockout screens to siRNA screens
To compare the top 30 host factor genes of the knockout screens to results from previous siRNA screens, we used the following data: (1) Sessions et al. (Supplementary Table 2)27. To rank the list by strength of phenotype, we used the p-value. If multiple siRNA sequences per gene were present, we used the one with the stronger effect. (2) Krishnan et al. (Supplementary Table 1)11. To rank the identified DENV host factor, column AM was filtered for “Required by both WNV and dengue”. The remaining genes were sorted by “Pooled siRNA Fold reduction of DENV” (column AN). (3) Tai et al. (Table S2)29. To sort by phenotype, we chose the validated genes scoring with at least 2 siRNA and ranked by p-value. (4) Li et al. (from Dataset S1)28. In order to rank the genes, we took the mean of the Average Normalized Percent Infected Cells Part One or Two of the four siRNAs. The top10 genes based on phenotype as explained above are shown in Ext. Data Fig. 10.
Extended Data
Extended Data Figure 1. Divergence of DENV and HCV host factors.

(a) Gene ontology (GO) analysis for DENV and HCV CRISPR screens on the ranked gene lists. Curated (by redundancy) enriched GO terms are shown. A complete list of all enriched GO terms can be found in Supplementary Data Table 4. (b) Distribution of the subcellular location of the 30 most enriched host factors for DENV and HCV. (c) Cross-comparison of the effects of DENV or HCV host factor knockout in Huh7.5.1 cells on the replication of DENV or HCV using reporter viruses expressing luciferase. Data depict average with s.d. for triplicate infections.
Extended Data Figure 2. Reproducibility of CRISPR screens.

(a) Ranked lists of the 30 most enriched DENV and HCV host factors and their rankings in the individual replicate screens. The color code reflects in what percentile the gene scored in the replicate. (b) Gene enrichment based on RIGER score for the individual replicate screens. Red dots highlight where the 30 most significant host factors ranked in the individual replicates.
Extended Data Figure 3. Genotyping of cell lines for DENV host factors.

(a) The site of gene trap insertion in HAP1 cell lines was determined using a PCR based method. Bases depicted in red are the flanking sequences upstream of the genetrap insertion. Bases depicted in green are downstream gene-trap flanking sequences. (b) TALENs were used to edit the genomic region of AUP1 in HAP1 cells. Bases depicted in red are the left TALEN binding site, bases depicted in blue are the right TALEN binding site. Bases depicted in green are the TALEN target site. Arrow indicates site of 260bp insertion. (c) CRISPR Cas9 nuclease was targeted to bases depicted in red in HAP1 cells. Editing events are depicted at the gRNA target sites below the WT sequence. (d) CRISPR Cas9 nuclease was targeted to bases depicted in red in Huh7 cells. Editing events are depicted at the gRNA target sites below the WT sequence. (e) Immunoblots of wild type and KO cell lines.
Extended Data Figure 4. Validation of DENV host factor genes.

(a) Plaque-forming units (PFU) assay of DENV infection. ND indicates no plaques were detected (threshold of detection of the assay is 6 PFU/ml). (b) DENV luciferase levels in HAP1 isogenic knockout cells complemented using lentiviral stable expression of corresponding genes. (c) Crystal violet of complemented Huh7 knockout cells infected with DENV. (d) DENV luciferase levels in Raji DC-SIGN cells with KO in DENV host factors (lentiCRISPRv2). Empty denotes an empty vector control (expressing Cas9 but no guideRNA) and NT a cell line expressing a non-targeting guideRNA. (e) Time course of DENV and HCV expressing Renilla luciferase in Huh7 knockout cells. (f) Schematic diagram of the STT3A and STT3B isoforms. Gene names in red indicate OST subunits identified in the DENV screens. Data depict average with s.d. for triplicate infections.
Extended Data Figure 5. Catalytic site mutations introduced in mammalian STT3A and STT3B.

(a) Catalytic site amino acids highlighted in red as identified in the bacterial STT3 (C. lari pglb). Strong conservation allows their identification in other species. Alignments of STT3 isoforms across different species highlight the conserved catalytic sites that were mutated. The table specifies the amino acid position and the specific triple mutations that were made to abolish catalytic activity. (b) Huh7 STT3A and STT3B KO cells expressing FLAG tagged STT3A and STT3B WT and catalytic mutants.
Extended Data Figure 6. Physical interaction between the OST complex and the replication complex of DENV.

(a) APEX2, a protein tag for electron microscopy was fused to the C-terminus of STT3B enabling the imaging of subcellular protein localization by deposition of a polymer of 3,3′-diaminobenzidine (DAB). (b) Luminescence of Huh7 STT3B-KO cells complemented with STT3B-APEX2 and infected with DENV expressing Renilla luciferase. Data depict average with s.d. for triplicate infections. (c) STT3B localizes on ER membranes in the vicinity of DENV-induced vesicle packets as shown by transmission EM micrograph of DENV-infected or uninfected Huh7 cells expressing the STT3B-APEX2 construct. N represents the cell Nucleus and the arrowheads in samples transfected with STT3B-APEX2 represent APEX polymerized DAB staining in the lumen of the Endoplasmic Reticulum (ER) or around DENV-induced vesicle packets (VP). (d) Co-immunoprecipitations (IP) of STT3A-FLAG and STT3B-FLAG from DENV infected cell lysates. LE = long exposure. (e) Anti-FLAG Western blots of IP elutions of DENV infected cells stably expressing FLAG tagged STT3A, STT3B and RPS25. (f) SYPRO Ruby staining of elutions and inputs of IP of DENV infected cell lysates. (g) Co-IP elutions of DENV infected lysates were analyzed by mass spectrometry and DENV specific peptides aligned to DENV polyprotein.
Extended Data Figure 7. Analysis of HCV host factor knockout cell lines.
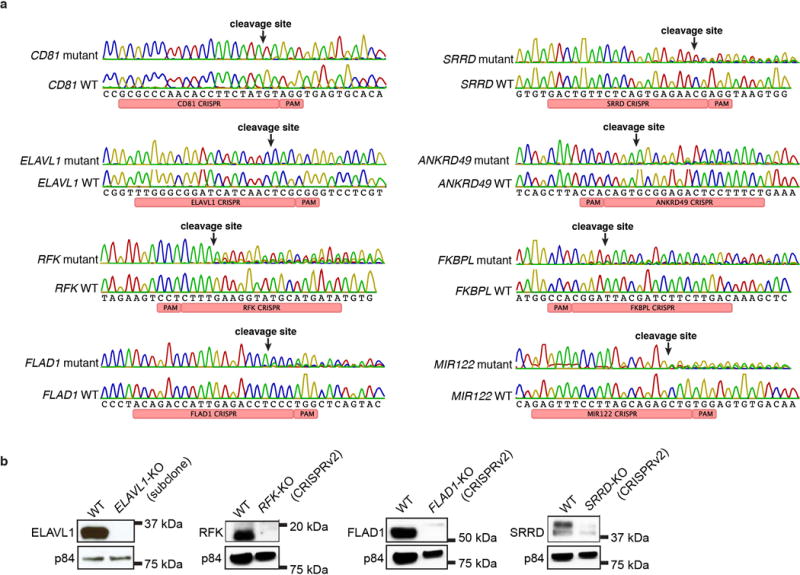
(a) Genotyping of CRISPR-induced KO Huh7.5.1 cells by Sanger sequencing showing the mutated locus and the WT reference. CRISPR/Cas9 induces mutations close to the PAM site resulting in frameshifts. CD81 and ELAVL1 KO cell lines are subclones whereas others are populations of cells mutagenized with lentiCRISPRv2. (b) Immunoblots of CRISPR-induced KO cells.
Extended Data Figure 8. ELAVL1 is a critical host factor for HCV replication.

(a) HCV luciferase infection in KO cell lines using 4 different guideRNAs per gene. NT = non-targeting guideRNA. (b) QPCR of viral RNA in WT or ELAVL1-KO Huh7.5.1 cells. (c) Crystal violet assay for different RNA virus infections. (d) HCV replicon assays using wild-type sgJFH1 (left) or GND sgJFH1 replicon. Note that one downward error bar for the right panel (6 hours, ELAVL1-KO) was not plotted because it would reach a negative value, which cannot be plotted on a logarithmic scale. (e) Transfection of ectopically expressed ELAVL1 restores HCV replication. Western blot of ELAVL1-FLAG transfected and untransfected Huh7.5.1 ELAVL1-KO cells. Data depict average with s.e.m. (QPCR) or s.d. (FFU, RLU) for triplicate infections, except e, which was a single infection.
Extended Data Figure 9. Lumiflavin inhibits the replication of HCV but not other RNA viruses.
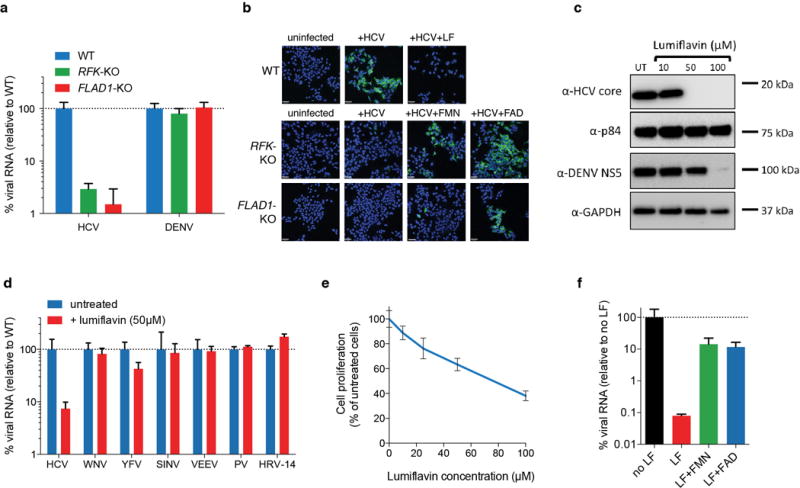
(a) QPCR of HCV or DENV RNA replication in WT, RFK-KO or FLAD1-KO Huh7.5.1. (b) Immunofluorescence of HCV infection in WT, RFK-KO and FLAD1-KO Huh7.5.1 cells under treatment with lumiflavin, FMN or FAD. HCV core protein (green). Blue=DAPI. (c) Western blot for HCV core and DENV NS5 in untreated (UT) and lumiflavin-treated Huh7.5.1 cells. p84 and GAPDH served as loading controls. (d) QPCR of RNA viruses in untreated or lumiflavin-treated Huh7.5.1 cells. (e) MTT cell proliferation assay for lumiflavin-treated Huh7.5.1 cells. (f) Restoration of HCV replication in lumiflavin-treated cells by exogenous addition of FMN or FAD. Data depict average with s.e.m. (QPCR) or s.d. (MTT) for triplicate infections/treatments.
Extended Data Figure 10. Comparison of knockout screen results to previous siRNA screens.

(a) Venn diagram comparing the hits from the CRISPR and haploid screens for DENV host factors to previous siRNA screens from Sessions et al.27 (from Supplementary Table 2) and Krishnan et al.11 (from Supplementary Table 1). The top ten validated host factors (by strength of phenotype in the validation screen) for each screen are shown next to circle. (b) Venn diagram comparing the hits from the CRISPR screen for HCV host factors to previous siRNA screens from Tai et al.29 (from Table S2) and Li et al.28 (from Dataset S1).
Supplementary Material
Acknowledgments
The authors thank Thijn Brummelkamp, Juliana Idoyaga and Shirit Einav for critically reading the manuscript and discussions; Xuhuai Ji (Stanford Functional Genomics Facility); The Stanford Transmission Electron Microscope facility; The Stanford Shared FACS Facility, and members of the Carette lab for discussions and support. Drs. Shirit Einav, Karla Kirkegaard, Frank Chisari, John F. Anderson, Charles Rice, Hidde Ploegh, Ron Kopito, Eva Harris, Martin Ivessa, Scott Weaver, Robert Tesh and Eric Campeau are acknowledged for generously providing materials. The work was funded in part by NIH DP2 AI104557 (JEC), NIH AI109662 (JEC), David and Lucile Packard Foundation (JEC), Stanford Graduate Fellowship (ASP), Boehringer Ingelheim Fonds (ASP) and NSF- GFRP (CDM).
Footnotes
Author contributions
CDM, ASP and JEC were responsible for overall design of the study. The majority of the experiments related to the DENV and HCV genetic screens were performed by CDM and ASP, respectively. KM designed and performed several validation experiments and JEC designed dengue constructs. YSO performed one of the DENV screens. PS provided expertise in design of HCV experiments and MAM and SMB performed several HCV experiments. Mass spectrometry experiments were performed and analyzed by CDM, GF, KS under the technical expertise of JEE. CDM, ASP, KM and JEC wrote the manuscript with input from all authors.
The authors declare no competing financial interest.
Accession codes
BioProjects
CRISPR genetic screens: PRJNA322191
Haploid genetic screens: PRJNA284536
References
- 1.Bhatt S, et al. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomas SJ, Rothman AL. Trials and Tribulations on the Path to Developing a Dengue Vaccine. American journal of preventive medicine. 2015;49:S334–344. doi: 10.1016/j.amepre.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 3.Lavanchy D. Evolving epidemiology of hepatitis C virus. Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2011;17:107–115. doi: 10.1111/j.1469-0691.2010.03432.x. [DOI] [PubMed] [Google Scholar]
- 4.Paul D, Bartenschlager R. Flaviviridae Replication Organelles: Oh, What a Tangled Web We Weave. Annu Rev Virol. 2015;2:289–310. doi: 10.1146/annurev-virology-100114-055007. [DOI] [PubMed] [Google Scholar]
- 5.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rasmussen SA, Jamieson DJ, Honein MA, Petersen LR. Zika Virus and Birth Defects – Reviewing the Evidence for Causality. The New England journal of medicine. 2016 doi: 10.1056/NEJMsr1604338. [DOI] [PubMed] [Google Scholar]
- 8.Carette JE, et al. Haploid genetic screens in human cells identify host factors used by pathogens. Science. 2009;326:1231–1235. doi: 10.1126/science.1178955. [DOI] [PubMed] [Google Scholar]
- 9.Carette JE, et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011;477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fons RD, Bogert BA, Hegde RS. Substrate-specific function of the translocon-associated protein complex during translocation across the ER membrane. The Journal of cell biology. 2003;160:529–539. doi: 10.1083/jcb.200210095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krishnan MN, et al. RNA interference screen for human genes associated with West Nile virus infection. Nature. 2008;455:242–245. doi: 10.1038/nature07207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma H, et al. A CRISPR-Based Screen Identifies Genes Essential for West-Nile-Virus-Induced Cell Death. Cell reports. 2015;12:673–683. doi: 10.1016/j.celrep.2015.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shrimal S, Cherepanova NA, Gilmore R. Cotranslational and posttranslocational N-glycosylation of proteins in the endoplasmic reticulum. Seminars in cell & developmental biology. 2014 doi: 10.1016/j.semcdb.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruiz-Canada C, Kelleher DJ, Gilmore R. Cotranslational and posttranslational N-glycosylation of polypeptides by distinct mammalian OST isoforms. Cell. 2009;136:272–283. doi: 10.1016/j.cell.2008.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lizak C, Gerber S, Numao S, Aebi M, Locher KP. X-ray structure of a bacterial oligosaccharyltransferase. Nature. 2011;474:350–355. doi: 10.1038/nature10151. [DOI] [PubMed] [Google Scholar]
- 16.Lindenbach BD, Rice CM. trans-Complementation of yellow fever virus NS1 reveals a role in early RNA replication. Journal of virology. 1997;71:9608–9617. doi: 10.1128/jvi.71.12.9608-9617.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beatty PR, et al. Dengue virus NS1 triggers endothelial permeability and vascular leak that is prevented by NS1 vaccination. Science translational medicine. 2015;7:304ra141. doi: 10.1126/scitranslmed.aaa3787. [DOI] [PubMed] [Google Scholar]
- 18.Parnas O, et al. A Genome-wide CRISPR Screen in Primary Immune Cells to Dissect Regulatory Networks. Cell. 2015;162:675–686. doi: 10.1016/j.cell.2015.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hasan M, et al. Cytosolic Nuclease TREX1 Regulates Oligosaccharyltransferase Activity Independent of Nuclease Activity to Suppress Immune Activation. Immunity. 2015;43:463–474. doi: 10.1016/j.immuni.2015.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scheel TK, Rice CM. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nature medicine. 2013;19:837–849. doi: 10.1038/nm.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 22.Brennan CM, Steitz JA. HuR and mRNA stability. Cellular and molecular life sciences : CMLS. 2001;58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sokoloski KJ, et al. Sindbis virus usurps the cellular HuR protein to stabilize its transcripts and promote productive infections in mammalian and mosquito cells. Cell host & microbe. 2010;8:196–207. doi: 10.1016/j.chom.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shwetha S, et al. HuR Displaces Polypyrimidine Tract Binding Protein To Facilitate La Binding to the 3′ Untranslated Region and Enhances Hepatitis C Virus Replication. Journal of virology. 2015;89:11356–11371. doi: 10.1128/JVI.01714-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin K, Gallay P. Curing a viral infection by targeting the host: the example of cyclophilin inhibitors. Antiviral research. 2013;99:68–77. doi: 10.1016/j.antiviral.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujimura M, et al. Functional characteristics of the human ortholog of riboflavin transporter 2 and riboflavin-responsive expression of its rat ortholog in the small intestine indicate its involvement in riboflavin absorption. The Journal of nutrition. 2010;140:1722–1727. doi: 10.3945/jn.110.128330. [DOI] [PubMed] [Google Scholar]
- 27.Sessions OM, et al. Discovery of insect and human dengue virus host factors. Nature. 2009;458:1047–1050. doi: 10.1038/nature07967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Q, et al. A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:16410–16415. doi: 10.1073/pnas.0907439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tai AW, et al. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell host & microbe. 2009;5:298–307. doi: 10.1016/j.chom.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramage HR, et al. A combined proteomics/genomics approach links hepatitis C virus infection with nonsense-mediated mRNA decay. Molecular cell. 2015;57:329–340. doi: 10.1016/j.molcel.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nature methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo B, et al. Highly parallel identification of essential genes in cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:20380–20385. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berger KL, et al. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:7577–7582. doi: 10.1073/pnas.0902693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Canver MC, et al. Characterization of genomic deletion efficiency mediated by clustered regularly interspaced palindromic repeats (CRISPR)/Cas9 nuclease system in mammalian cells. The Journal of biological chemistry. 2014;289:21312–21324. doi: 10.1074/jbc.M114.564625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horii T, et al. Validation of microinjection methods for generating knockout mice by CRISPR/Cas-mediated genome engineering. Scientific reports. 2014;4:4513. doi: 10.1038/srep04513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alvarez DE, De Lella Ezcurra AL, Fucito S, Gamarnik AV. Role of RNA structures present at the 3′UTR of dengue virus on translation, RNA synthesis, and viral replication. Virology. 2005;339:200–212. doi: 10.1016/j.virol.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 37.Kinney RM, et al. Construction of infectious cDNA clones for dengue 2 virus: strain 16681 and its attenuated vaccine derivative, strain PDK-53. Virology. 1997;230:300–308. doi: 10.1006/viro.1997.8500. [DOI] [PubMed] [Google Scholar]
- 38.Samsa MM, et al. Dengue virus capsid protein usurps lipid droplets for viral particle formation. PLoS pathogens. 2009;5:e1000632. doi: 10.1371/journal.ppat.1000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fuchs G, et al. Kinetic pathway of 40S ribosomal subunit recruitment to hepatitis C virus internal ribosome entry site. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:319–325. doi: 10.1073/pnas.1421328111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lam SS, et al. Directed evolution of APEX2 for electron microscopy and proximity labeling. Nature methods. 2015;12:51–54. doi: 10.1038/nmeth.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martell JD, et al. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nature biotechnology. 2012;30:1143–1148. doi: 10.1038/nbt.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jaffee MB, Imperiali B. Exploiting topological constraints to reveal buried sequence motifs in the membrane-bound N-linked oligosaccharyl transferases. Biochemistry. 2011;50:7557–7567. doi: 10.1021/bi201018d. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


