Abstract
Presence of excess unaltered, wild-type (WT) DNA providing no information of biological or clinical value often masks rare alterations containing diagnostic or therapeutic clues in cancer, prenatal diagnosis, infectious diseases or organ transplantation. With the surge of high-throughput technologies there is a growing demand for removing unaltered DNA over large pools-of-sequences. Here we present nuclease-assisted minor-allele enrichment with probe-overlap (NaME-PrO), a single-step approach with broad genome coverage that can remove WT-DNA from numerous sequences simultaneously, prior to genomic analysis. NaME-PrO employs a double-strand-DNA-specific nuclease and overlapping oligonucleotide-probes interrogating WT-DNA targets and guiding nuclease digestion to these sites. Mutation-containing DNA creates probe-DNA mismatches that inhibit digestion, thus subsequent DNA-amplification magnifies DNA-alterations at all selected targets. We demonstrate several-hundred-fold mutation enrichment in diverse human samples on multiple clinically relevant targets including tumor samples and circulating DNA in 50-plex reactions. Enrichment enables routine mutation detection at 0.01% abundance while by adjusting conditions it is possible to sequence mutations down to 0.00003% abundance, or to scan tumor-suppressor genes for rare mutations. NaME-PrO introduces a simple and highly parallel process to remove un-informative DNA sequences and unmask clinically and biologically useful alterations.
INTRODUCTION
Traces of altered DNA in clinical samples provide vital clues regarding disease states and preferred treatments. Detecting such information-rich, altered DNA sequences, which often exist within a large excess of normal wild-type (WT) DNA, pose a persistent technical challenge in several fields of biology, biotechnology and medicine. These include cancer, prenatal diagnosis, infectious diseases, organ transplantation and forensics (1–5). Removing the vast majority of normal DNA that does not provide meaningful scientific or clinical information would be a major boon to revealing and capturing the wealth of information that altered DNA offers. For example in liquid biopsy of cancer using circulating-free DNA (cfDNA) (6–10), traces of somatic mutations serve as biomarkers for early detection (10) or tumor response to treatment (11,12), yet the high excess of circulating WT DNA often dims the opportunities for diagnosis and treatment. While several methods for reducing WT DNA before, during or after polymerase chain reaction (PCR) amplification have been reported (13–18), these usually apply to single or limited numbers of PCR amplicons or are only applicable to a minority of DNA targets recognized by sequence-specific enzymes (19–23). Additionally, PCR amplification itself may contribute to false positive calls in view of polymerase mis-incorporations that are indistinguishable from DNA alterations (24). With the recent surge of high-throughput technologies (25–27) there has been an even greater demand for removing unaltered DNA over large pools of sequences, e.g. prior to performing massively parallel sequencing (MPS). Under standard operating conditions MPS cannot discriminate reliably mutations lower than about 2% abundance (24,28). While the accuracy of sequencing calls can be improved with molecular barcoding and multiple sequence readout (24,29–33) these approaches uniformly require an excessive number of reads for each target, the vast majority of which are non-informative, thus wasting time and reagents while also limiting throughput.
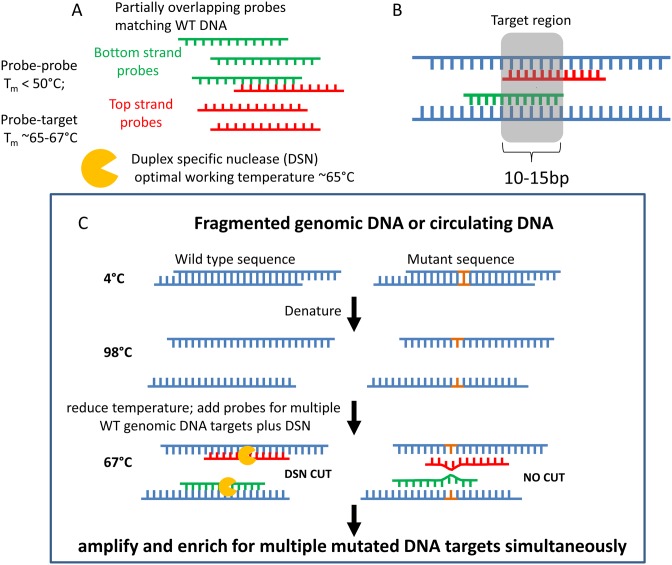
Here we present a new technology, Nuclease-Assisted Minor-Allele enrichment using Overlapping Probes (NaME-PrO) for removing excess unaltered DNA from an almost unlimited number of DNA targets simultaneously, thereby uniformly improving detection limits for all endpoint detection technologies. NaME-PrO is a single step approach that operates at the level of genomic DNA prior to performing DNA amplification hence it is not impacted by polymerase errors. The technique (Figure 1A–C) employs a thermostable duplex-specific nuclease (DSN) (34) that digests dsDNA with high preference over single stranded DNA and requires fully matched template i.e. mismatches near its binding site inhibit enzymatic action (34). DSN has no observed sequence specificity, thereby double stranded regions are digested irrespective of sequence. NaME-PrO harnesses these DSN properties to enable preferential digestion of WT DNA at any desired target. For each DNA target interrogated for mutations, a distinct pair of oligonucleotide probes overlapping the target region is designed. The probes are complementary to the WT DNA and bind respectively the top and bottom DNA template strands with an overlap ‘target’ region of about 10–15 bp (Figure 1A–C). Excess amount of probes are mixed with fragmented genomic DNA or circulating DNA and denatured for 2 min at 98°C. The temperature is then reduced to 67°C, while single-copy DNA sequences remain single stranded due to low concentration and slow re-association kinetics. The probes form preferential binding to target DNA strands since they are designed to have melting temperatures (Tm) of ∼65°C when bound to WT target DNA while probe-probe interactions melt below ∼50°C, by design. Consequently, probes hybridize stably to opposite WT DNA strands, while they generate opposing mismatches when contacting DNA strands with mutations in the probe overlap region, resulting in unstable hybrids. When DSN is added in the reaction, duplexes between probe and WT DNA strands are digested preferentially due to DSN's strong preference for fully-matched double strand DNA over single stranded or mismatch-containing DNA, thus mutant DNA strands remain substantially intact. Since probes overlap the target sequence on opposite strands, both WT DNA strands undergo preferential digestion when a mutation is present in the probe-overlap region. If at least one of two mutated DNA strands survives DSN digestion, subsequent DNA amplification amplifies exponentially these sites, thereby resulting in mutation enrichment of all targets simultaneously. The method creates the potential for massively parallel mutation enrichment prior to sequencing and engenders a new paradigm whereby rare mutations do not require deep sequencing for their detection.
Figure 1.
Components and workflow for nuclease-assisted minor-allele enrichment using probe-overlap (NaME-PrO). (A) Components of the assay: partially overlapping oligonucleotide probes included in excess as compared to target DNA molar ratio; and duplex-specific nuclease (DSN). (B) The probes are designed to bind the WT sequence of top and bottom target DNA strands. The overlap region of the probes defines the DNA region targeted for mutation enrichment. (C) NaME-PrO workflow; following fragmented genomic DNA denaturation, the excess probes bind their targets. Addition of DSN generated preferential digestion at double stranded regions defined by the probes, but not when there are mutation-caused mismatches. Multiple sites can be targeted simultaneously in this manner. Subsequent amplification enhances DNA target strands that escape DSN digestion, thereby enriching for mutated DNA.
MATERIALS AND METHODS
Genomic DNA and circulating DNA samples
Human male genomic DNA (Promega Corporation, Madison, WI, USA) was used as WT DNA control and was mixed with DNA from multiple mutated cell lines or standard reference DNA with multiple mutations (Horizon Discovery HD728) (Supplementary Table S1), to create DNA mixtures with gradually decreasing mutation abundances. Frozen tissue was obtained from clinical lung and colon tumor specimens (Supplementary Table S1) provided by the Massachusetts General Hospital Tumor Bank and used following approval from the Internal Review Board of the Dana Farber Cancer Institute. Genomic DNA was isolated using the DNAeasy™ Blood & Tissue Kit (Qiagen), following manufacturer's instructions.
Blood samples were obtained from colon cancer patients and healthy volunteers (Supplementary Table S1) after informed consent and Dana Farber-Cancer Institute Institutional Review Board approval. Blood samples were centrifuged at 1600 g for 20 min at room temperature within 3 h from collection. Plasma was carefully removed and re-centrifuged at 1600 g for 15 min at 4°C. Plasma was carefully removed and stored at −80°C. On thawing a third and final centrifugation was performed at 16 000 g for 5 min at room temperature and plasma was carefully removed into a separate tube away from any residual debris before extraction of circulating DNA. Cell-free circulating DNA (cfDNA) was isolated from plasma using the QIAamp Circulating Nucleic Acid Kit (Qiagen), and the concentration of cfDNA was measured on a Qubit 3.0 fluorometer (Thermo Fisher Scientific) using a dsDNA HS assay (Q32854).
NaME-PrO oligonucleotide probe design
Probes comprised oligonucleotides 20–25 bp long such that they have a Tm of an average 65°C and range 63–67°C when fully matched to WT DNA. Probes can optionally contain a polymerase block on their 3′ end to prevent polymerase extension in subsequent amplification reactions. Probe to probe interactions including those from top and bottom strand overlap were designed to have Tm < 50°C. Integrated DNA Technologies OligoAnalyzer 3.1 was used to calculate probe Tm. A computer program was written in Python version 2.7 for automatic probe design when tiling of probe-overlap regions along the entire length of a DNA sequence is desirable. The program is initialized with a genomic sequence and a start and stop index indicating the region to be tiled. The algorithm then begins by iterating over the sequence, constructing a set of contiguous oligonucleotides which serve as the overlap regions for NaME-PrO probes and automatically adjusting their lengths to ensure Tm < 50°C for each one. Once this is complete, each of the oligonucleotides is extended toward its 5′-end to form a pair of overlapping, isothermal probes with a Tm ∼65 ± 2°C. The program outputs a spreadsheet containing a comprehensive list of sense probes, antisense probes and their overlapping regions and respective Tm. A list of overlapping probes used for singlet, multiplexed and tiling NaME-PrO reactions are listed in Supplementary Table S2. Single nucleotide polymorphisms (SNPs) may, in rare cases, reside close to the targeted mutations and within the overlap region of either the upper or lower strand probes. Such SNPs may prevent DSN from digesting WT samples effectively, thereby leading to reduced mutation enrichment. To circumvent this hurdle, additional probes that match each of SNP versions may be included in the assay, such that irrespective of what SNP is present the WT sample will always have a fully-matched probe version. In the present work, for simplicity, we avoided targeting somatic mutations that are very close to mapped SNP positions.
NaME-PrO assay
Genomic DNA was treated with dsDNA Shearase Plus (ZYMO Research, CA, USA) to generate random fragments in the size range 100–500 bp. A total of 10 ng fragmented genomic DNA and overlapping oligonucleotides (probes) were mixed in 1× DSN buffer to a final 10 µl volume. For the lowest 0.03 and 0.01% mutation abundance serial mutation dilution studies, 100 ng genomic DNA was used as starting material. The final probe concentrations ranged from 1 to 100 nM depending on whether the intent was high enrichment with little or no quantification (50–100 nM) or modest enrichment with better quantification (1–20 nM). It is noteworthy that excessive amounts of probe directed a given target can lead to elimination of mutated, as well as WT sequences, hence increasing probe concentrations beyond 50–100 nM under current assay conditions is not recommended. The sample was placed in a SmartCycler real-time PCR system (Cepheid) for denaturation at 98°C for 2 min. The temperature was then reduced to 67°C and 0.2 units of DSN (Evrogen) was added into the mixture followed by 20 min incubation at 67°C and 2 min at 95°C for DSN inactivation. Samples where DSN was omitted (No-DSN-controls) were run in parallel in all reactions. NaME-PrO products were used in amplification reactions without purification as described below.
PCR amplification
PCR reactions were performed on SmartCycler real-time PCR system (Cepheid) or CFX Connect™ real-time PCR (Bio-Rad Labratories) using Phusion High-Fidelity DNA polymerase (New England Biolabs) and LCGreen Plus+ as a fluorescence dye (BioFire Diagnositics). Primers (synthesized by Integrated DNA Technologies Inc.) are listed in Supplementary Table S3. For single-target PCR, 1 μl of DSN treated samples or No-DSN control samples were added into a 24-μl PCR master mix containing 1× Phusion HF buffer, 200 nM of each primer, 200 μM of each of the four dNTPs, 0.8× LCGreen and 0.5 unit of Phusion polymerase (Thermo Fisher Scientific). For PCR amplification, an initial denaturation step was performed for 2 min at 98°C, followed by 45 cycles of 10 s denaturation at 98°C, 20 s annealing at 58°C and 10 s elongation at 72°C. The final step included melting curve analysis (0.2°C step increments, 2 or 4 s hold before each acquisition) from 65 to 95°C. Multiplex PCR amplification for NaME-PrO products using Ion Ampliseq™ Cancer Hotspot Panel v2 (207 amplicons covering approximately 2800 COSMIC mutations from 50 oncogenes and tumor suppressor genes) was performed according to the Ion AmpliSeq library preparation user guide (Thermo Fisher Scientific).
Droplet digital PCR for mutation abundance validation
Droplet digital PCR (ddPCR) reactions were carried out to verify mutation abundance before and after NaME-PrO, as well as in the No-DSN control samples. Sequences of primers and probes (Integrated DNA Technologies Inc.) are listed in Supplementary Tables S3 and 4. Amplifications were performed in a 20 μl volume containing 1× ddPCR Supermix for probes (Bio-Rad), 900 nM forward and reverse primers, 250 nM 6-carboxyfluorescein (FAM) and 6 - carboxy - 2′,4,4′,5′,7,7′ - hexachlorofluorescein (HEX) probes (Integrated DNA Technologies) and 10 ng genomic DNA or real-time PCR products (using 1–1 000 000 final dilution for PCR products). Droplets were then generated using the DG8™ droplet generator cartridges (Bio-Rad) which contained 20 μl aqueous phase with 70 μl of droplet generation (DG) oil (Bio-Rad). Samples were transferred to a 96-well reaction plate and then sealed using the PX1 PCR plate sealer (Bio-Rad) for 10 s at 180°C prior to thermal cycling. The thermal cycling program was performed on an Eppendorf Mastercycler ep Gradient S (Eppendorf) with an initial denaturation step at 95°C for 10 min, followed by 40 cycles of 30 s denaturation at 94°C, 60 s annealing at 58°C and with a final step holding at 98°C for 10 min. Then the plate was transferred to QX100 droplet reader (Bio-Rad) for endpoint reading. Calculation of absolute number of positive events for a given channel (FAM or HEX), the ratio and the fractional abundance of mutation for each sample were performed by the Quantasoft Software (Bio-Rad). The determination of number of target copies per droplet (number of copies of target molecule) was adjusted by the software to fit a Poisson distribution model with 95% confidence level.
Fifty-plex PCR preamplification for examining the NaME-PrO enrichment limits
Fifty-plex PCR primers (Supplementary Table S5) were designed for commonly mutated exonic regions identified in lung and esophageal cancers according to the COSMIC database, as previously reported (17). Multiplex pre-amplification from 20 ng genomic DNA was performed in a total volume of 25 microlitters using a mixture of 100 primers (50 paired sets) at a final concentration of 0.3 mM for each primer with 0.3 mM dNTPs, 3 mM MgCl2, 1× Kapa HiFi buffer and 0.5U of Kapa HiFi HotStart DNA polymerase (Kapa Biosystems) reported to have an average error rate of 2.8 × 10−7 mis-incorporations/bp. Multiplex PCR cycling was performed according to the manufacturer's recommendations (Kapa Biosystems) for a total of 20 PCR cycles using 63°C as optimal annealing temperature. Amplicon lengths ranged from 120 to 190 bp in size depending on the amplicon (Supplementary Table S5). Following multiplex cycling, 1 ml of exonuclease I (New England Biolabs) was added to each reaction and incubated at 37°C for 30 min and 80°C for 15 min to remove unincorporated primers.
COLD-PCR
COLD-PCR for KRAS exon 2 DNA fragments was conducted either directly from genomic DNA or following NaME-PrO treatment of genomic DNA from lung tumor sample TL119, as previously described (35,36). COLD-PCR products were analyzed via Sanger sequencing.
SNP genotyping assay
TaqMan® SNP genotyping assays were purchased from Thermo Fisher Scientific for a number of SNP loci, including rs1050171. Genotyping reactions for cfDNA samples were conducted following manufacturer instructions on CFX Connect™ real-time PCR. Thermo cycling comprised initial denaturation for 10 min at 95°C, followed by 40 cycles of 15 s denaturation at 95°C, 1 min annealing and extension at 60°C. Cell-free circulating DNA (cfDNA) from healthy volunteer blood samples were tested via SNP genotyping assays to identify samples that differ at the tested SNP positions. Samples with different genotypes of rs1050171 were mixed to obtain a gradually increasing 1 to 5% SNP-abundance. The probe and primer sets used in TaqMan® SNP genotyping assays were also used in ddPCR format to quantify the genotype abundances of cfDNA mixtures formed by mixing cfDNA samples carrying different genotypes. SNP abundance was quantified via ddPCR before and after NaME-PrO reactions.
NaME-PrO applied to error-prone PCR (EP-PCR) products
Multiplexed-PCR amplicons containing random mutations were created using error-prone PCR (EP-PCR) following a published protocol (37). Briefly, PCR reactions were performed on SmartCycler real-time PCR system (Cepheid). A total of 100 ng of Human genomic DNA (Promega Corporation) were added into 99 μl EP-PCR master mix containing 10 mM Tris⋅HCl (pH 8.3), 7 mM MgCl2, 1 mM dCTP, 1 mM dTTP, 0.2 mM dATP, 0.2 mM dGTP, 55-plex reaction primers (final 0.3 μM each, sequences listed in Supplementary Table S5), 0.5 mM MnCl2 and 5 unit of GoTaq Flexi DNA polymerase (Promega Corporation). To achieve EP-PCR products with different mutation abundances, 10, 20 and 30 PCR cycles (94°C for 1 min, 60°C for 1 min, 72°C for 3 min) were performed respectively. The products were treated with 30 units of Exonuclease I (New England Biolabs Inc) to remove the excess primers. Mutations in EP-PCR products were tested via high resolution melting analysis (HRM) before and after application of NaME-PrO.
High resolution melting (HRM) analysis
Ten-microliter of PCR product were transferred to a 96-well plate, and 20 μl of mineral oil was added to each well. HRM was performed on a 96-well LightScanner® system (Idaho Technology). The software sensitivity level was set as 1.2 for computing DNA variant groups. All experiments were replicated at least three independent times for assessing the reproducibility of the results.
Sanger sequencing
The PCR products were digested by Exonuclease I and Shrimp Alkaline Phosphatase (New England Biolabs) and processed for Sanger sequencing at Eton Bioscience Inc. To enable sequencing of short PCR amplicons, a 30-T tail was added to the 5′-end of the forward primer.
Illumina MiSeq sequencing of PCR products after NaME-PrO assay
Following NaME-PrO-PCR, mutation enriched products were processed using the standard barcoding and library preparation protocol for Illumina MiSeq sequencing. Library preparation was performed at the Center for Cancer Computational Biology at the Dana-Farber Cancer Institute. Libraries with ligated Illumina adapters were assessed for DNA quality and quantity on Agilent bio-analyzer, and then pooled together into a single tube prior to MiSeq sequencing. Data analysis was conducted on MiSeq Reporter software and the alignment sequencing data were loaded into Integrative Genome Viewer 2.3 (IGV, Broad Institute) using human genome hg19 as reference. Reads for each non-reference bases at the nucleotide of interest as well as the adjacent nucleotides were recorded and plotted in a noise plot to distinguish the true mutation versus background noise.
RESULTS
Single target NaME-PrO: proof of principle
We first applied NaME-PrO to genomic DNA with KRAS exon 2 (p.G12V, c.34G > T) mutations. WT DNA was mixed with SW480 cell line DNA, resulting in DNA with ∼0.5% abundant KRAS mutation. Following random fragmentation of genomic DNA, NaME-PrO was applied along with control experiments. Samples were PCR amplified and ddPCR was employed to quantify the KRAS mutation abundance before and after NaME-PrO. The probe sequences for NaME-PrO, primers and hydrolysis probes for PCR and ddPCR are listed in Supplementary Tables S3 and S4. When DSN and both overlapping probes are present, a 167-fold enrichment mutation enrichment is achieved (Figure 2) converting an original 0.5% mutation abundance to 83.7% abundance. In the presence of only one probe, or when a probe overlapping the mutation is combined with an opposite strand probe that does not overlap, the mutation enrichment is lower than when both probes overlap the mutation location. When NaME-PrO is applied on DNA with KRAS mutations using probes specific to a different location (TP53 exon 8) the KRAS mutation abundance is not affected, indicating the specificity of the process for the sequence targeted by the overlapping probes.
Figure 2.

NaME-PrO applied to a single target. Genomic DNA with ∼0.5% mutation in KRAS exon 2 plus a mutation in TP53 exon 8 was tested for mutation abundance before and after KRAS-directed NaME-PrO using droplet digital PCR (ddPCR). Presence of two KRAS-specific overlapping probes plus DSN nuclease generate the highest KRAS mutation enrichment (∼167-fold, resulting to a clonal mutation). Omission of any system component, or using only one probe, two non-overlapping probes (‘probe plus cutter’) or probes directed to the wrong target (TP53) result to lower mutation enrichment or no enrichment at all. Experiments were repeated five independent times.
Multiplexed NaME-PrO
To apply NaME-PrO in a multiplexed fashion, we first designed a duplex assay for KRAS exon 2 and TP53 exon 8 mutations. Supplementary Figure S1 shows that both KRAS and TP53 mutations are enriched, for two distinct sets of mutations encompassed under a single set of probes for each gene. Next we used genomic DNA containing multiple clinically relevant mutations at ∼1% abundance (HDx DNA), in order to examine a 9-plex NaME-PrO assay, using 50Mm concentration for all probes. All mutations targeted by NaME-PrO probes are significantly enriched to clonal mutation status (>50% abundance, Figure 3A). When downstream analysis is performed using (38) HRM or Sanger sequencing, detection sensitivity is enhanced via NaME-PrO for both these detection methods (Supplementary Figure S2). To understand the dependence of mutation enrichment on probe concentration we performed ddPCR evaluations versus probe concentration and initial mutation abundance. Probes for two targets, IDH1 and BRAF were followed within a 9-plexed reaction. For probe concentration of 50 nM or higher, mutation enrichment is high but not quantitatively related to original mutation abundance Supplementary Figure S3. In contrast, probe concentrations in the region 3–10 nM for the two targets provide modest enrichment bearing proportionality to the original mutation abundance. Accordingly, individual probe concentrations within multiplexed NaME-PrO reactions can be modulated to yield the desired endpoint. We then designed probes for another 41 target sequences to generate a 50-plex probe pool for simultaneous enrichment to these targets. Figure 3B depicts mutation abundance for the nine targets examined. The 50-plex NaME-PrO resulted to similar levels of enrichment as the 9-plex reaction, converting mutations from 1% initial mutation abundance to clonal status. These data reveal the specificity of the process and the absence of significant illegitimate interactions with probes targeting different DNA targets (Figure 3B). Following 50-plex NaME-PrO on HDx DNA containing 1% original mutation abundance, we also utilized a commercially available panel, Ion Ampliseq Cancer Hotspot Panel v2, to conduct 207-plex PCR. This resulted to 50–100-fold mutation enrichment in the targets followed (Supplementary Figure S4). In summary, we demonstrated that NaME-PrO enables multi-target mutation enrichment by ∼50-fold to over ∼200-fold depending on conditions and can be combined with targeted cancer panel applications.
Figure 3.
Multiplexed NaME-PrO directed to 9–50 clinically relevant DNA targets. (A) 9-plex-NaME-PrO applied to genomic DNA containing mutations at ∼1% abundance at nine clinically relevant sites. Mutation enrichment to 80–95% mutation abundance is observed for all targets via droplet-digital-PCR (ddPCR). (B) 50-plex-NaME-PrO applied to genomic DNA containing mutations at ∼1% abundance at the same nine clinically relevant sites as in (A). Mutation enrichment to 80–95% mutation abundance is observed for all targets. (C) Serial dilution study to examine NaME-PrO mutation enrichment at low mutation abundances. Genomic DNA contained serial mutant dilutions with mutation abundance of ∼1, 0.3, 0.1, 0.03 and 0.01% in all nine DNA targets tested. For the lowest three dilutions the amount of starting DNA was increased to 100 ng to ensure adequate number of mutated sequences in the sample. (D) Plots of mutation abundance after NaME-PrO against mutation abundance in starting genomic DNA as quantified via ddPCR. Experiments were repeated three independent times.
NaME-PrO applied to serial mutation dilutions
To evaluate further the enrichment potential of NaME-PrO, we applied the technology to mutated DNA diluted serially into WT DNA. Fragmented genomic DNA containing multiple mutations at ∼5, 1, 0.3, 0.1, 0.03 and 0.01% mutation abundances was obtained by serial dilution of HDx DNA into WT DNA, then screened via multiplexed NaME-PrO and assessed via ddPCR (Figure 3C). The data indicate that the targeted mutations are enriched and distinguished from WT DNA down to ∼0.01% abundance. The original mutation abundance is logarithmically correlated (R2 > 0.9) to the NaME-PrO-derived mutation abundance with differences depending on mutation type and position, as well as on the individual probe concentrations used for the targeted site (Figure 3D and Supplementary Table S6).
To determine the technical limits of NaME-PrO mutation enrichment we also conducted an extended dilution series, down to one mutant molecule in 3 × 106 WT alleles for IDH1 mutations. Since it is difficult to perform PCR directly on genomic DNA containing 3 × 106 human genomes (18 μg), we performed a few cycles of high fidelity 50-plex PCR pre-amplification separately for DNA with 5% mutation abundance and for WT DNA. The pre-amplified mutated PCR products were then serially diluted in WT pre-amplification products to form the desired final mutant ratios. Special precautions to exclude airborne contamination were adopted. To account for potential interference by polymerase-introduced errors we amplified WT DNA in parallel, in five independent experiments. NaME-PrO was then applied to these samples under conditions for maximum enrichment, followed by HRM and Sanger sequencing to detect IDH1 mutations (c. 394C > T, p.R132C, Supplementary Figure S5). Both HRM and Sanger sequencing were able to identify IDH1 mutations down to 0.00003%, (∼1 mutant allele in 3 million WT alleles), while WT-only samples did not indicate mutations. In summary, under highest-enrichment conditions NaME-PrO can result to almost complete isolation of mutated samples from large excess WT DNA.
NaME-PrO applied to tumor samples and plasma-circulating DNA
To generate cfDNA with mutations at specific sites we extracted cfDNA from blood of healthy individuals and then ‘spiked’ fragmented genomic DNA containing mutations, to generate 1% mutation abundances at 10 known positions. Then a 10-plex NaME-PrO assay was applied to enrich mutations, followed by PCR amplification and ddPCR detection. Similar levels of mutation enrichment using cfDNA, as with fragmented genomic DNA were observed (Figure 4A).
Figure 4.
Multiplexed NaME-PrO applied to plasma-circulating DNA (cfDNA) and genomic DNA from colon/lung tumors obtained under consent and IRB approval. (A) Multiplexed-NaME-PrO applied to cfDNA from normal volunteers spiked with fragmented genomic DNA to generate ∼1% mutation abundance at 10 clinically relevant sites. ddPCR was used to evaluate mutation abundance. Mutation enrichment is evident at all sites tested. (B) Multiplexed NaME-PrO applied to cfDNA obtained from nine colon cancer patients, two healthy volunteers and standard reference DNA, HMC. Two cfDNA samples were deemed as KRAS G12V mutation-positive via ddPCR while the rest exhibited signals below the ddPCR detection threshold and deemed to be negative. NaME-PrO-treated samples enriched highly the two KRAS positive samples but not the negative samples, thus boosting the signal to noise ratio. (C) Multiplexed NaME-PrO applied to clinical tumor genomic DNA samples previously shown to harbor low-level mutations at multiple TP53 exons. Sanger sequencing following NaME-PrO enables detection of the enriched low-level mutations. (D) Multiplexed NaME-PrO followed by ddPCR, applied to a cfDNA from two normal volunteers differing at SNP rs1050171. cfDNA was mixed to generate SNP proportions increasing gradually from 1 to 5%, to simulate differences observed in stable versus acute solid organ transplantation rejection(4). NaME-PrO conditions were adjusted in favor of preserving quantification rather than maximizing enrichment, using 10 nM probe concentration. Application of NaME-PrO enriches DNA alterations and enlarges the clinically significant differences between 1 and 5% ‘donor’ cfDNA. Experiments were repeated three to five independent times.
Next we examined cfDNA extracted from plasma of Stage III colon cancer patients. We conducted 10-plex NaME-PrO for cfDNA from nine patients and two healthy volunteers, and applied ddPCR to identify KRAS hotspot mutations both with and without application of NaME-PrO (Figure 4B). Using ddPCR alone, we found that two patients had very low levels KRAS G12V mutations while the remaining samples and negative controls yielded ddPCR signals below the ddPCR detection limit (∼0.05%). Accounting for background ‘limit-of-blank’ signals, ddPCR derived mutations have an abundance of about 0.05%. Samples treated with NaME-PrO identified the same positive and negative cfDNA samples with ddPCR and enriched mutations in the two mutant samples to about 4 and 6%, respectively (Figure 4B). T he NaME-PrO-projected original mutation abundances using Supplementary Table S6 are 0.05 and 0.07%, respectively, in agreement with ddPCR quantification. The mutations become clearly more evident using NaME-PrO-ddPCR instead of ddPCR alone.
Next, NaME-PrO was applied to genomic DNA from colon and lung tumor samples previously shown to harbor clonal or low-level mutations in KRAS and TP53 (28). One of these samples TL119 harbors low abundance GG > TT KRAS mutations (39) which was not detectable on Sanger sequencing but was enriched and became clearly visible following NaME-PrO-PCR-sequencing (Supplementary Figure S6). When PCR was replaced by COLD-PCR, which provides additional mutation enrichment (36,40,41), the resulting mutation abundance in TL119 was ∼90% indicating synergism between NaME-PrO and COLD-PCR. Two additional samples, CT20 and TL121 previously shown to harbor two low-level mutations in distinct exons of TP53 (39) were screened following NaME-PrO containing probes targeting TP53 exons 7–9, Figure 4C. Enrichment was obtained on both mutation sites for each sample.
Circulating DNA may serve as an early biomarker for solid organ transplant rejection (4). An increase of donor circulating DNA as a fraction of total recipient cfDNA in the narrow range from 1 to 5% is an indicator of impending acute rejection (4). To simulate a transplant rejection situation we identified SNPs that differ in cfDNA from two healthy donors, using ddPCR; we then mixed stochiometrically the two cfDNA samples such that SNPs appear in fractions of 1–5% of the total representing. These samples were then examined either directly via ddPCR, or by applying NaME-PrO-ddPCR to derive the fraction of donor DNA. NaME-PrO conditions were adjusted in favor or preserving quantification rather than maximizing enrichment. Figure 4D indicates that, following NaME-PrO, the quantitative difference corresponding to stable disease vs. acute rejection is broadened, thus potentially facilitating detection of clinically significant cfDNA increase in transplantation patients.
NaME-PrO application with targeted re-sequencing
Combination of NaME-PrO with MPS is suitable since both technologies address multiple targets simultaneously. 50-plex NaME-PrO was applied to fragmented genomic DNA or plasma-circulating DNA containing mutations at 0.5% prevalence, followed by targeted PCR amplification and library preparation for MiSeq sequencing, following our previously described protocol (28). Amplicon regions produced by targeted PCR amplification were paired-end sequenced on the Illumina Miseq and aligned to the reference genome (GRh37/hg19). Frequency calls were generated for each nucleotide aligned within the amplicon locations in order to develop ‘variant-and-noise plots’ to display both the frequencies of mutation calls and the background signals. Figure 5 and Supplementary Figure S7 show representative variant-and-noise plots for genomic DNA and circulating DNA containing KRAS, BRAF, MEK1 and IDH1 mutations with or without NaME-PrO in the workflow. Consistent with previous observations (28), in the absence of NaME-PrO, targeted re-sequencing cannot distinguish 0.5% mutations from noise levels. Following NaME-PrO, the mutation abundance increases to levels that are clearly distinguished from noise.
Figure 5.
Multiplexed NaME-PrO combined with targeted re-sequencing. Multiplexed-NaME-PrO applied to cfDNA from normal volunteers spiked with fragmented genomic DNA to generate ∼0.5% mutation abundance at multiple clinically relevant sites. Following NaME-PrO, samples were PCR-amplified and processed for library preparation. Variant to noise plots derived from Illumina MiSeq sequencing data are depicted. (A and B) KRAS G12V, G12R, G13D mutations in fragmented genomic DNA with and without application of NaME-PrO, respectively. (C and D) IDH1 R132C mutation in cfDNA with and without NaME-PrO, respectively. The original mutation abundance is 0.5% in both cases, while the read depth is about 1000–2000× depending on sequence. Inserts in B and D represent magnified versions of the same graphs to depict clearly the effect of sequencing noise. Application of NaME-PrO increases the signal-to-noise ratio and reveals clearly the rare mutation. Experiments were repeated two independent times.
NaME-PrO assay ‘tiling’ for enrichment at multiple mutation positions
To examine the ability of NaME-PrO to enrich mutations at multiple positions on target sequences, as opposed to targeting ‘hotspot’ mutations, we applied a single set of overlapping probes for enriching three distinct KRAS mutations falling within the probe overlap region. Mutations at all positions were enriched (Supplementary Figure S8), indicating that mismatches of diverse types and positions within the probe overlap region inhibit DSN digestion effectively.
In order to further examine the ability of NaME-PrO to enrich diverse mutations, we also generated random mutagenesis DNA templates by EP-PCR, followed by HRM of the resulting amplicons. The mis-incorporation-error abundance of an EP-PCR amplicon depends on the number of PCR duplications (42). As shown in Supplementary Figure S9, in the absence of NaME-PrO, amplicons required 30 cycles of EP-PCR to become distinguished from WT DNA via HRM, for both KRAS and IDH1. When NaME-PrO was applied, 10-cycle EP-PCR products were clearly distinguished from WT, consistent with NaME-PrO enrichment of EP-PCR generated random mutations and enhancement of HRM sensitivity for detecting polymerase-induced base changes.
Finally, we adapted NaME-PrO to encompass mutation enrichment along all sequence positions in TP53 exons 6 to exon 9 by ‘tiling’ probe overlap regions along exons of interest and performing multiple NaME-PrO assays in parallel. The strategy is depicted in Supplementary Figure S10. Each exon is divided into 7–10 successive target regions and overlapping probes corresponding to contiguous 10–15 bp regions are designed to ‘tile’ the entire interrogated DNA fragment. Probes are then mixed to form 7–10 groups such that every group contains a single pair of overlapping probes for each interrogated DNA fragment. The interrogated DNA samples are then split in 7–10 parallel reactions and NaME-PrO is applied separately to them, enriching mutations on a single portion from every exon simultaneously. By pooling results from all NaME-PrO reactions, mutations from all positions on the sequences are enriched. Since genomic DNA is fragmented to an average size of ∼150 bp, a limited number of tiled NaME-PrO reactions ensure all sequences on an average size fragment are addressed by a pair of overlapping probes. To include longer exons or higher numbers of exons, only the number of probes within each group needs to increase, while the number of distinct NaME-PrO reactions need not change. Supplementary Figure S10A depicts simultaneous enrichment of mutations along TP53 exons 6–9 in a human DNA sample containing single mutations in all four exons. Supplementary Figure S10B depicts enrichment of multiple random mutations generated by EP-PCR along TP53 exon 8. By tiling NaME-PrO probes in seven groups, HRM-based identification of 0.1–1% mutation abundance in all TP53 exon 8 segments is demonstrated.
DISCUSSION
We introduced and experimentally validated NaME-PrO, a highly parallel approach for eliminating excess WT DNA that does not provide incremental biological or clinical value, in order to boost analysis of clinically significant rare DNA variants. Previous enzymatic approaches for removing WT DNA (19–23) relied on enzymes that digest only at specific sequences, limiting their broad application. The non-selective, DNAse-like activity of DSN in combination with ∼10–15 bp target regions defined by overlapping probes engender NaME-PrO with the potential for massively parallel degradation of WT DNA. Indeed, the main condition required is the hybridization of appropriately designed probes to the chosen targets in the starting DNA material (Figure 1C). This step is already established in existing sample preparation workflows for thousands of targets including the whole exome (43). The potential implication for MPS is that a limited read depth may be adequate to identify low-level events reliably, and that deep sequencing may not be necessary, leading to substantial speed and cost benefits. For example, using NaME-PrO 0.5% abundance mutations are enhanced by orders of magnitude, such that they would be detected with just 10–30 reads (Figure 5 and Supplementary Figure S7). By adjusting probe concentration and potentially including modified bases in probe design it may eventually be possible to enable near complete separation of large genomic fractions out of mixed clinical samples, as shown in Supplementary Figure S5 for IDH1, thereby resulting to highly simplified and cost-effective sequencing. To preserve quantification of original mutation abundances on the other hand, lower probe concentrations combined with modest, quantitative enrichment can be adopted.
Since NaME-PrO is applied at the genomic DNA level, prior to other treatments, it combines with downstream sample preparation, amplification or endpoint detection methods, with almost no change in the existing workflows. For example, boosting the sensitivity of sequencing, HRM, ddPCR, COLD-PCR (Figures 2–5) did not require a change in the established processes. Of note, the endpoint mutation readout method still determines the final outcome, e.g. a 0.1% mutation enriched via NaME-PrO to a level of 7–10% mutation abundance may still be missed via Sanger sequencing, while it would be detectable via next-generation sequencing. While techniques like real time PCR (44,45), ddPCR (46) or BEAM-ing (47) also allow high mutation detection sensitivity, these are limited to examining a single target per reaction, as opposed to NaME-PrO that can address a broad target footprint. In summary, NaME-PrO provides a simple and powerful process to remove WT-DNA in large numbers of sequences that raises long-standing limitations in identification and tracking of rare, clinically useful DNA sequences.
Supplementary Material
Disclaimer: The contents of this manuscript do not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Cancer Institute [R21CA-175542, in part]; R.F. was supported by NCI/CANCURE to Northeastern University [R25 CA17465002]. Funding for open access charge: Departmental funds.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kobayashi S., Boggon T.J., Dayaram T., Janne P.A., Kocher O., Meyerson M., Johnson B.E., Eck M.J., Tenen D.G., Halmos B. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 2.Hoffmann C., Minkah N., Leipzig J., Wang G., Arens M.Q., Tebas P., Bushman F.D. DNA bar coding and pyrosequencing to identify rare HIV drug resistance mutations. Nucleic Acids Res. 2007;35:e91. doi: 10.1093/nar/gkm435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lo Y.M., Corbetta N., Chamberlain P.F., Rai V., Sargent I.L., Redman C.W., Wainscoat J.S. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350:485–487. doi: 10.1016/S0140-6736(97)02174-0. [DOI] [PubMed] [Google Scholar]
- 4.Snyder T.M., Khush K.K., Valantine H.A., Quake S.R. Universal noninvasive detection of solid organ transplant rejection. Proc. Natl. Acad. Sci. U.S.A. 2011;108:6229–6234. doi: 10.1073/pnas.1013924108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong S.M., Traverso G., Johnson C., Geng L., Favis R., Boynton K., Hibi K., Goodman S.N., D'Allessio M., Paty P., et al. Detecting colorectal cancer in stool with the use of multiple genetic targets. J. Natl. Cancer Inst. 2001;93:858–865. doi: 10.1093/jnci/93.11.858. [DOI] [PubMed] [Google Scholar]
- 6.Diehl F., Schmidt K., Choti M.A., Romans K., Goodman S., Li M., Thornton K., Agrawal N., Sokoll L., Szabo S.A., et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 2008;14:985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thierry A.R., Mouliere F., El Messaoudi S., Mollevi C., Lopez-Crapez E., Rolet F., Gillet B., Gongora C., Dechelotte P., Robert B., et al. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat. Med. 2014;20:430–435. doi: 10.1038/nm.3511. [DOI] [PubMed] [Google Scholar]
- 8.Newman A.M., Bratman S.V., To J., Wynne J.F., Eclov N.C., Modlin L.A., Liu C.L., Neal J.W., Wakelee H.A., Merritt R.E., et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 2014;20:548–554. doi: 10.1038/nm.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bettegowda C., Sausen M., Leary R.J., Kinde I., Wang Y., Agrawal N., Bartlett B.R., Wang H., Luber B., Alani R.M., et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diehl F., Li M., Dressman D., He Y., Shen D., Szabo S., Diaz L.A., Jr, Goodman S.N., David K.A., Juhl H., et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc. Natl. Acad. Sci. U.S.A. 2005;102:16368–16373. doi: 10.1073/pnas.0507904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kimura T., Holland W.S., Kawaguchi T., Williamson S.K., Chansky K., Crowley J.J., Doroshow J.H., Lenz H.J., Gandara D.R., Gumerlock P.H. Mutant DNA in plasma of lung cancer patients: potential for monitoring response to therapy. Ann. N.Y. Acad. Sci. 2004;1022:55–60. doi: 10.1196/annals.1318.010. [DOI] [PubMed] [Google Scholar]
- 12.Misale S., Yaeger R., Hobor S., Scala E., Janakiraman M., Liska D., Valtorta E., Schiavo R., Buscarino M., Siravegna G., et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486:532–536. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newton C.R., Graham A., Heptinstall L.E., Powell S.J., Summers C., Kalsheker N., Smith J.C., Markham A.F. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS) Nucleic Acids Res. 1989;17:2503–2516. doi: 10.1093/nar/17.7.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun X., Hung K., Wu L., Sidransky D., Guo B. Detection of tumor mutations in the presence of excess amounts of normal DNA. Nat. Biotechnol. 2002;20:186–189. doi: 10.1038/nbt0202-186. [DOI] [PubMed] [Google Scholar]
- 15.Li J., Wang L., Mamon H., Kulke M.H., Berbeco R., Makrigiorgos G.M. Replacing PCR with COLD-PCR enriches variant DNA sequences and redefines the sensitivity of genetic testing. Nat. Med. 2008;14:579–584. doi: 10.1038/nm1708. [DOI] [PubMed] [Google Scholar]
- 16.Wu L.R., Wang J.S., Fang J.Z., Evans E.R., Pinto A., Pekker I., Boykin R., Ngouenet C., Webster P.J., Beechem J., et al. Continuously tunable nucleic acid hybridization probes. Nat. Methods. 2015;12:1191–1196. doi: 10.1038/nmeth.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guha M., Castellanos-Rizaldos E., Liu P., Mamon H., Makrigiorgos G.M. Differential strand separation at critical temperature: a minimally disruptive enrichment method for low-abundance unknown DNA mutations. Nucleic Acids Res. 2013;41:e50. doi: 10.1093/nar/gks1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milbury C.A., Li J., Makrigiorgos G.M. PCR-based methods for the enrichment of minority alleles and mutations. Clin. Chem. 2009;55:632–640. doi: 10.1373/clinchem.2008.113035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parry J.M., Shamsher M., Skibinski D.O. Restriction site mutation analysis, a proposed methodology for the detection and study of DNA base changes following mutagen exposure. Mutagenesis. 1990;5:209–212. doi: 10.1093/mutage/5.3.209. [DOI] [PubMed] [Google Scholar]
- 20.Haliassos A., Chomel J.C., Grandjouan S., Kruh J., Kaplan J.C., Kitzis A. Detection of minority point mutations by modified PCR technique: a new approach for a sensitive diagnosis of tumor-progression markers. Nucleic Acids Res. 1989;17:8093–8099. doi: 10.1093/nar/17.20.8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ward R., Hawkins N., O'Grady R., Sheehan C., O'Connor T., Impey H., Roberts N., Fuery C., Todd A. Restriction endonuclease-mediated selective polymerase chain reaction: a novel assay for the detection of K-ras mutations in clinical samples. Am. J. Pathol. 1998;153:373–379. doi: 10.1016/S0002-9440(10)65581-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bielas J.H., Loeb L.A. Quantification of random genomic mutations. Nat. Methods. 2005;2:285–290. doi: 10.1038/nmeth751. [DOI] [PubMed] [Google Scholar]
- 23.Gu W., Crawford E.D., O'Donovan B.D., Wilson M.R., Chow E.D., Retallack H., DeRisi J.L. Depletion of Abundant Sequences by Hybridization (DASH): using Cas9 to remove unwanted high-abundance species in sequencing libraries and molecular counting applications. Genome Biol. 2016;17:41. doi: 10.1186/s13059-016-0904-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kinde I., Wu J., Papadopoulos N., Kinzler K.W., Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc. Natl. Acad. Sci. U.S.A. 2011;108:9530–9535. doi: 10.1073/pnas.1105422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mardis E.R. A decade's perspective on DNA sequencing technology. Nature. 2011;470:198–203. doi: 10.1038/nature09796. [DOI] [PubMed] [Google Scholar]
- 26.Shendure J., Lieberman Aiden E. The expanding scope of DNA sequencing. Nat. Biotechnol. 2012;30:1084–1094. doi: 10.1038/nbt.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Genomes Project C., Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Milbury C.A., Correll M., Quackenbush J., Rubio R., Makrigiorgos G.M. COLD-PCR enrichment of rare cancer mutations prior to targeted amplicon resequencing. Clin. Chem. 2012;58:580–589. doi: 10.1373/clinchem.2011.176198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Narayan A., Carriero N.J., Gettinger S.N., Kluytenaar J., Kozak K.R., Yock T.I., Muscato N.E., Ugarelli P., Decker R.H., Patel A.A. Ultrasensitive measurement of hotspot mutations in tumor DNA in blood using error-suppressed multiplexed deep sequencing. Cancer Res. 2012;72:3492–3498. doi: 10.1158/0008-5472.CAN-11-4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forshew T., Murtaza M., Parkinson C., Gale D., Tsui D.W., Kaper F., Dawson S.J., Piskorz A.M., Jimenez-Linan M., Bentley D., et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci. Transl. Med. 2012;4:136ra68. doi: 10.1126/scitranslmed.3003726. [DOI] [PubMed] [Google Scholar]
- 31.Lou D.I., Hussmann J.A., McBee R.M., Acevedo A., Andino R., Press W.H., Sawyer S.L. High-throughput DNA sequencing errors are reduced by orders of magnitude using circle sequencing. Proc. Natl. Acad. Sci. U.S.A. 2013;110:19872–19877. doi: 10.1073/pnas.1319590110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmitt M.W., Kennedy S.R., Salk J.J., Fox E.J., Hiatt J.B., Loeb L.A. Detection of ultra-rare mutations by next-generation sequencing. Proc. Natl. Acad. Sci. U.S.A. 2012;109:14508–14513. doi: 10.1073/pnas.1208715109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gregory M.T., Bertout J.A., Ericson N.G., Taylor S.D., Mukherjee R., Robins H.S., Drescher C.W., Bielas J.H. Targeted single molecule mutation detection with massively parallel sequencing. Nucleic Acids Res. 2016;44:e22. doi: 10.1093/nar/gkv915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shagin D.A., Rebrikov D.V., Kozhemyako V.B., Altshuler I.M., Shcheglov A.S., Zhulidov P.A., Bogdanova E.A., Staroverov D.B., Rasskazov V.A., Lukyanov S. A novel method for SNP detection using a new duplex-specific nuclease from crab hepatopancreas. Genome Res. 2002;12:1935–1942. doi: 10.1101/gr.547002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song C., Milbury C.A., Li J., Liu P., Zhao M., Makrigiorgos G.M. Rapid and sensitive detection of KRAS mutation after fast-COLD-PCR enrichment and high-resolution melting analysis. Diagn. Mol. Pathol. 2011;20:81–89. doi: 10.1097/PDM.0b013e3181fde92f. [DOI] [PubMed] [Google Scholar]
- 36.Murphy D.M., Bejar R., Stevenson K., Neuberg D., Shi Y., Cubrich C., Richardson K., Eastlake P., Garcia-Manero G., Kantarjian H., et al. NRAS mutations with low allele burden have independent prognostic significance for patients with lower risk myelodysplastic syndromes. Leukemia. 2013;27:2077–2081. doi: 10.1038/leu.2013.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilson D.S., Keefe A.D. Random mutagenesis by PCR. Curr. Potoc. Mol. 2001 doi: 10.1002/0471142727.mb0803s51. Chapter 8, Unit 8.3. [DOI] [PubMed] [Google Scholar]
- 38.Reed G.H., Wittwer C.T. Sensitivity and specificity of single-nucleotide polymorphism scanning by high-resolution melting analysis. Clin. Chem. 2004;50:1748–1754. doi: 10.1373/clinchem.2003.029751. [DOI] [PubMed] [Google Scholar]
- 39.Li J., Milbury C.A., Li C., Makrigiorgos G.M. Two-round coamplification at lower denaturation temperature-PCR (COLD-PCR)-based sanger sequencing identifies a novel spectrum of low-level mutations in lung adenocarcinoma. Hum. Mutat. 2009;30:1583–1590. doi: 10.1002/humu.21112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li J., Makrigiorgos G.M. COLD-PCR: a new platform for highly improved mutation detection in cancer and genetic testing. Biochem. Soc. Trans. 2009;37:427–432. doi: 10.1042/BST0370427. [DOI] [PubMed] [Google Scholar]
- 41.Galbiati S., Brisci A., Lalatta F., Seia M., Makrigiorgos G.M., Ferrari M., Cremonesi L. Full COLD-PCR protocol for noninvasive prenatal diagnosis of genetic diseases. Clin. Chem. 2011;57:136–138. doi: 10.1373/clinchem.2010.155671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCullum E.O., Williams B.A., Zhang J., Chaput J.C. Random mutagenesis by error-prone PCR. Methods Mol. Biol. 2010;634:103–109. doi: 10.1007/978-1-60761-652-8_7. [DOI] [PubMed] [Google Scholar]
- 43.Gnirke A., Melnikov A., Maguire J., Rogov P., LeProust E.M., Brockman W., Fennell T., Giannoukos G., Fisher S., Russ C., et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat. Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amicarelli G., Shehi E., Makrigiorgos G.M., Adlerstein D. FLAG assay as a novel method for real-time signal generation during PCR: application to detection and genotyping of KRAS codon 12 mutations. Nucleic Acids Res. 2007;35:e131. doi: 10.1093/nar/gkm809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li J., Wang F., Mamon H., Kulke M.H., Harris L., Maher E., Wang L., Makrigiorgos G.M. Antiprimer quenching-based real-time PCR and its application to the analysis of clinical cancer samples. Clin. Chem. 2006;52:624–633. doi: 10.1373/clinchem.2005.063321. [DOI] [PubMed] [Google Scholar]
- 46.Vogelstein B., Kinzler K.W. Digital PCR. Proc. Natl. Acad. Sci. U.S.A. 1999;96:9236–9241. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Diehl F., Li M., He Y., Kinzler K.W., Vogelstein B., Dressman D. BEAMing: single-molecule PCR on microparticles in water-in-oil emulsions. Nat. Methods. 2006;3:551–559. doi: 10.1038/nmeth898. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.