SUMMARY
The molecular circuits by which antigens activate quiescent T cells remain poorly understood. We combined temporal profiling of the whole proteome and phosphoproteome via multiplexed isobaric labeling proteomics technology, computational pipelines for integrating multi-omics datasets, and functional perturbation to systemically reconstruct regulatory networks underlying T cell activation. T cell receptors activated the T cell proteome and phosphoproteome with discrete kinetics, marked by early dynamics of phosphorylation and delayed ribosome biogenesis and mitochondrial activation. Systems biology analyses identified multiple functional modules, active kinases, transcription factors and connectivity between them, and mitochondrial pathways including mitoribosomes and complex IV. Genetic perturbation revealed physiological roles for mitochondrial enzyme COX10-mediated oxidative phosphorylation in T cell quiescence exit. Our multi-layer proteomics profiling, integrative network analysis and functional studies define landscapes of the T cell proteome and phosphoproteome and reveal signaling and bioenergetics pathways that mediate lymphocyte exit from quiescence.
Graphical abstract

INTRODUCTION
Naïve T cells exist in a quiescent state characterized by small cell size and exit from the active cell cycle (G0) (Hamilton and Jameson, 2012). Upon antigen stimulation, engagement of T cell receptors (TCRs) triggers a signaling cascade culminating in the induction of interleukin 2 (IL-2) and cell surface receptors, initiation of cell growth and proliferation, and ultimately differentiation into effector cells. Despite our knowledge of early TCR signaling events and subsequent clonal expansion and differentiation of activated T cells, the transitional state linking these processes, i.e. the process for naïve T cells to exit from quiescence, remains poorly understood.
Recent studies highlight the importance of metabolic reprogramming in T cell responses (Buck et al., 2015; MacIver et al., 2013). While naïve T cells mainly rely on mitochondria-dependent oxidative phosphorylation (OXPHOS) as the energy source, activated T cells markedly increase bioenergetic and biosynthetic activities, especially aerobic glycolysis (the Warburg effect). Despite the glycolytic nature of T cell activation, activated T cells also upregulate OXPHOS, and inhibition of mitochondrial function impairs T cell proliferation (Chang et al., 2013; Sena et al., 2013). How mitochondrial functions intersect with immune signals and molecular regulators remain elusive. Moreover, it is often difficult to understand whether an observed metabolic shift or ‘switch’ is the cause or consequence of a change in the cellular phenotype. From this perspective, genetic dissection of metabolic enzymes could provide the critical functional insight.
Transcriptional profiling and network analysis are instrumental to our understanding of molecular pathways and signaling networks in immunity (Amit et al., 2011). However, transcript levels are insufficient to predict protein levels in many scenarios especially during the dynamic transitional state when there is a temporal delay between transcription and translation (Liu et al., 2016). Moreover, posttranslational modifications, such as phosphorylation, are crucial regulators of protein functions and signaling. With the recent advancement in mass spectrometry-based analytical technologies (Mann et al., 2013), deep proteomic profiling with extensive coverage (the number of proteins identified) and throughput (the number of samples analyzed) provides an exciting opportunity to comprehensively characterize proteome dynamics during T cell activation.
Here we present the global analysis of whole proteome and phosphoproteome of T cell activation by the combination of the tandem-mass-tag (TMT) method and two dimensional liquid chromatography-tandem mass spectrometry (LC/LC-MS/MS), and computational pipelines for multi-tier integrative analyses of signaling networks. Our results revealed dynamic reprogramming of proteome and phosphoproteome in TCR-stimulated cells, and identified multiple functional modules, the connectivity between kinases and transcription factors (TFs), and in particular, activation of mitochondrial pathways including mitoribosomes and complex IV (cytochrome c oxidase). To further dissect bioenergetics pathways, we generated T cells lacking COX10, a critical regulator of complex IV, and established mechanisms of OXPHOS activation and the indispensable role of OXPHOS in T cell quiescence exit. These results establish dynamic signaling networks and selective bioenergetics pathways underlying T cell exit from quiescence.
RESULTS
Multiplexed quantitative analysis of whole proteome and phosphoproteome during T cell activation
To identify protein expression and phosphorylation events during T cell activation, we used multiplexed TMT and LC/LC-MS/MS approaches to quantify the proteome and phosphoproteome of naïve T cells from wild-type (WT) mice and those stimulated with anti-CD3 and anti-CD28 (α-CD3-CD28) for 2 h, 8 h and 16 h with biological replicates. As depicted in Figure 1A, samples were lysed, digested, labeled with different TMT tags, then pooled and analyzed by LC/LC-MS/MS (Wang et al., 2015). Five percent of the pool was used for whole proteome analysis, and the remaining 95% was subjected to phosphoproteome profiling. In total, we quantified 8,431 proteins and 13,755 phosphopeptides (Data S1A and S1B, < 1% false discovery rate (FDR)). Expression of specific proteins derived from proteomic profiling (Figure S1A) was in agreement with immunoblot results previously described (Yang et al., 2013). To further validate our data, we performed immunoblot analyses of TXNIP and PDCD4 expression and PDHA-1 and CAD phosphorylation. The results of immunoblot analysis (Figure 1B) were highly consistent with proteomic data (Figure 1C), which further validated the reliability of our profiling approach.
Figure 1. Temporal profiling of whole proteome and phosphoproteome during T cell activation.
(A) Experimental scheme.
(B) Immunoblot analysis of selected proteins and phosphorylation events.
(C) MS quantification of the selected proteins and phosphorylation events is highly consistent with immunoblot analysis in (B).
(D) Representative null comparisons of whole proteome and phosphoproteome (0h.a/0h.b and 16h.a/16h.b) display markedly distinct patterns from true comparisons (16h.a/0h.a and 16h.b/0h.b replicates). The color scheme indicates different numbers of proteins or phosphopeptides. Correlation coefficient (r) for each comparison is indicated.
(E) Principal component analysis of all identified proteins and phosphopeptides.
(F) Cluster analysis of differentially expressed proteins and phosphopeptides.
To differentiate experimental variances from biological alterations, we compared the replicate data of 0 h and 16 h from both whole proteome and phosphoproteome. The null comparisons of the biological replicates showed random distribution (Figure 1D, left panels), with the phosphoproteome data being more variable than the whole proteome data (Figure S1B), probably due to the transient nature of protein phosphorylation. In contrast, data comparisons of the two time points showed significant differences between naïve and activated T cells but consistency between replicates (Figures 1D, right panels). Principal component analysis (PCA) of all quantified proteins and phosphopeptides (Figure 1E) and clustering analysis of dysregulated proteins and phosphopeptides (Figure 1F) further verified the reproducibility of these results.
Recent studies suggest that translational control plays a more dominant role than transcription for cellular abundance of proteins (Schwanhausser et al., 2011), although opposing evidence also exists (Li and Biggin, 2015). We directly compared differential protein expression induced by 8 h of TCR stimulation with messenger (m)RNA expression at the same condition (Yang et al., 2013). Correlation analysis of quantified proteome and transcriptome induced by TCR stimulation (8 h vs. 0 h) showed a weak correlation (Figure S1C, r = 0.49). When applying a cutoff of 1.5-fold change, only a small fraction of differentially expressed (DE) RNAs and proteins were overlapped (n = 232). These results indicate that unique regulation of protein abundance is associated with TCR stimulation.
TCR stimulation reprograms the proteome and engages multiple functional modules
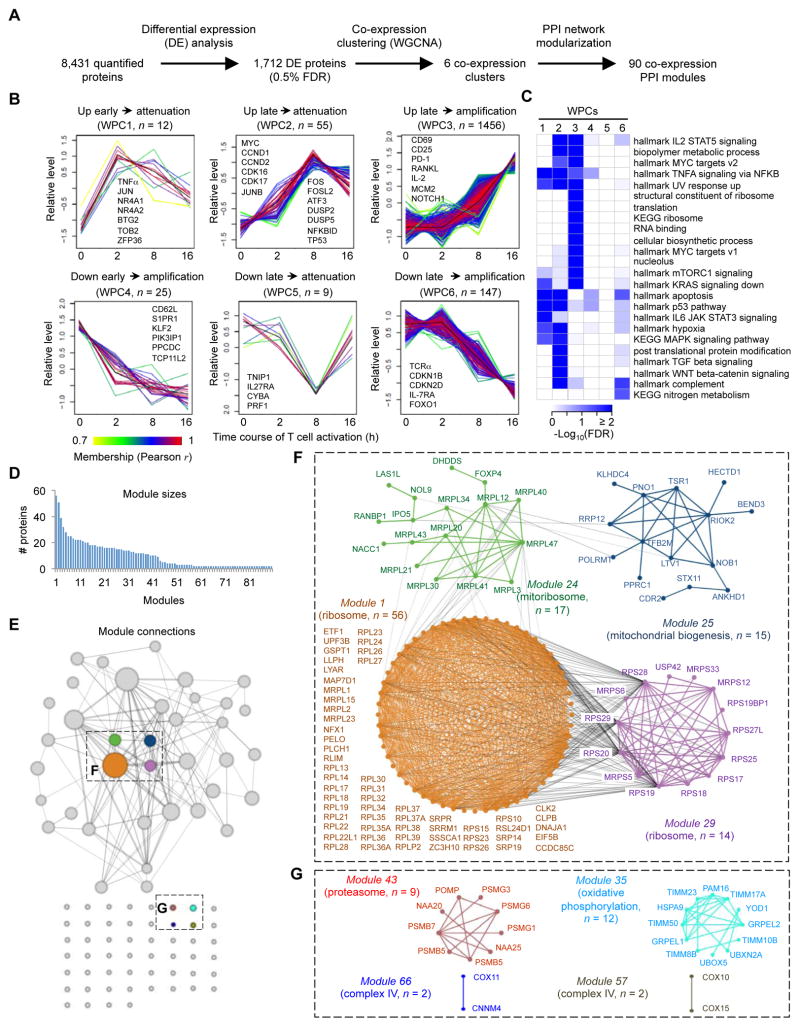
To model dynamics of proteome changes upon T cell activation, we developed a three-step computational pipeline by integrating co-expression clustering analysis and the protein-protein interaction (PPI) network (Figure 2A). We first identified proteins significantly changed between any two time points using one-way ANOVA with stringent criteria, resulting in 1,712 DE proteins during T cell activation (0.5% FDR) (Figure 1F and Data S1A).
Figure 2. Temporal expression profiling of whole proteome reveals co-expression clusters and functional modules during T cell activation.
(A) Overview of computational analysis for whole proteome.
(B) Six co-expression clusters of whole proteome (WPCs). Each line indicates the relative abundance of each protein and is color-coded by the cluster membership. Selective proteins in each WPC are shown.
(C) Functional annotations of WPCs by Gene Ontology (GO), KEGG and Hallmark databases (FDR < 0.05).
(D) Distribution of module size, with modules identified by superimposition of proteins in each WPC onto the protein-protein interaction (PPI) network. The numbers of proteins from each module are shown.
(E) Diagram of the individual modules and their interactions. Circles (nodes) represent the 90 modules, with the circle size proportional to module size. Edges connect modules that share PPIs. Boxed modules are further expanded in panels (F) and (G).
(F) Four interconnected modules of cytoplasmic and mitochondrial ribosomes derived from WPC3. The names of proteins and the representative functional term for each module are shown.
(G) Modules of proteasome, OXPHOS and mitochondrial complex IV pathways derived from WPC3.
In the second-step computational pipeline, we applied weighted gene correlation network analysis (WGCNA) for these DE proteins to identify clusters of highly correlated proteins, which often share similar biological functions or mechanisms of regulation (Jansen et al., 2002). We were able to group the majority of the DE proteins (1,704) into six co-expression clusters (named WPCs, whole proteome clusters), with WPC1-3 displaying upregulated and WPC4-6 downregulated patterns (Figure 2B). WPC1 showed an early and pronounced upregulation at 2 h and then gradual donwnregulation (termed “attenuation”). WPC2 had a small change at 2 h but a marked late upregulation at 8 h followed by attenuation at 16 h. WPC3 was largely unaltered at 2 h, upregulated at 8 h, and further increased (termed “amplification”) at 16 h. Associated with the kinetics of rapid induction, WPC1 contained the cytokine TNFα and TFs that represent immediate early genes (IEGs), including JUN and nuclear receptor NR4A subgroup NR4A1 and NR4A2. The anti-proliferative BTG/TOB protein family members BTG2 and TOB2 and cytokine mRNA-destabilizing factor ZFP36 (Tristetraprolin) also fall into WPC1, which probably suggests a control mechanism to prevent premature cell cycle progression and cytokine expression. WPC2 included crucial regulators of cell metabolism and cell cycle progression, including MYC, CCND1, CCND2, CDK16 and CDK17, as well as AP-1 and ATF TFs including JUNB, FOS, FOSL2 and ATF3. Negative regulators of TCR signaling were also identified in WPC2, including MAPK phosphatases DUSP2 and DUSP5, NF-κB inhibitor NFKBID (I BNS), and cell cycle inhibitor TP53. Functional annotation of these proteins revealed the involvement of cytokine (IL-2 and TNFα) signaling (Figure 2C), consistent with the transition from TCR signaling in WPC1 to cytokine signaling in WPC2. This notion was further supported by hallmark analysis that reveals activation of distinct NF-κB targets in WPC1 and WPC2 (Figure S2A and Data S2A).
WPC3 contained the majority (85%) of DE proteins and included classical T cell activation markers CD69, IL2RA (CD25), PDCD1 (PD-1), TNFSF11 (RANKL) and IL-2 (Figure 2B). Pathway analysis of these proteins showed significant enrichment in protein translation (ribosome), cell metabolism, cell cycle, and protein degradation (Figure 2C and Data S2A). Hallmarks analysis showed MYC targets and mTORC1 signaling as the two most predominant pathways, followed by cell cycle pathways, unfolded protein response, metabolic pathways, cytokine signaling (IL-2 and TNFα signaling), and DNA repair (Figure 2C and Data S2A). Thus, TCR activation for 16 h resulted in profound reprogramming of the translational and metabolic machineries, associated with activation of MYC and mTORC1 pathways (Figure S2A).
Among the three downregulated clusters, WPC4 showed rapid downregulation at 2 h and amplification at 8 h and 16 h. This cluster contained important immune regulators known to be dampened upon T cell activation, including trafficking molecules CD62L and S1PR1, and TF KLF2 that controls their expression (Hamilton and Jameson, 2012). WPC5 was downregulated at 8 h but was subsequently reversed, and included TNIP1 (ABIN-1), an inhibitory molecule for NF-κB signaling, and IL-27 receptor IL27RA (WSX-1) that has both pro- and anti-inflammatory activities. WPC6 was the largest downregulated cluster that initiated downregulation at 8 h and was further amplified at 16 h. WPC6 contained TCRα itself, cell cycle inhibitors CDKN1B (p27) and CDKN2D (p19), cytokine receptor IL7RA (IL-7Rα), and TF FOXO1 that mediates IL7RA expression. Therefore, TCR stimulation leads to dynamic downregulation of immune regulators required for naïve T cell homeostasis as well as negative regulators of TCR signaling and cell cycle regulation.
As the third step of our computational pipeline, we superimposed proteins within each WPC onto the PPI network to identify functional modules. We identified 90 modules within which the protein members were highly inter-connected (Data S2B), with the number of proteins within each module ranging from 2 to 56 (Figure 2D). We were able to associate the majority of these modules with known gene annotation terms or signaling pathways (Data S2B). For example, the largest module (#1) was mainly comprised of ribosome proteins (Figures 2E and 2F, orange; Figures S2B and S2C, orange) in WPC3. Additionally, WPC3 was enriched with modules in cell metabolism including steroid biosynthesis and nucleotide and glucose metabolism (#8, #20 and #47), cell cycle (#13 and #27), autophagy (#21), and translation initiation (#5) (Data S2B). Of note, 80 out of these 90 modules were derived from WPC3 (Data S2B), indicating the concordant regulation of these functional modules during T cell activation.
Analogous to the interaction between proteins that are functional units at a molecular level, modules are the functional units at a systems level and can interact with each other to form a module-level network (Figures 2E and S2B; see also grey lines in Figure 2F). For example, the modules enriched in ribosome proteins (Data S2B, modules 1 and 29) and the module of mitochondrial ribosome proteins (Data S2B, module 24) were inter-connected (Figures 2E and 2F), suggesting coordinated upregulation of both cytoplasmic and mitochondrial ribosomes. Our unbiased approach also allowed us to identify hitherto unknown functional modules during T cell activation. For example, the proteasome was unexpectedly activated (Figure 2G and Data S2B, module 43). Cells must possess mechanisms to coordinate protein synthesis with protein turnover to maintain amino acid and protein homeostasis, as an imbalance between these processes is detrimental to cellular homeostasis (Zhang et al., 2014).
Our integrative network analysis further identified six mitochondrial pathways in TCR-stimulated cells (Data S2B), including: i) mitoribosomes (module 24; Figure 2F); ii) mitochondrial biogenesis (module 25; Figure 2F); iii) cytochrome c oxidase (complex IV in OXPHOS) (modules 57 and 66; Figure 2G); iv) oxidative phosphorylation (module 35; Figure 2G); v) mitochondrial one-carbon folate metabolism (Ron-Harel et al., 2016) (modules 23 and 73; Figure S2D); and vi) apoptotic mitochondrial changes (module 31; Figure S2D). Consistent with these analyses, imaging studies showed that activated T cells increased expression of mitoribosome protein MRPL20 (Figure S2E), and proteins SHMT2 and MTHFD2 involved in one-carbon folate metabolism (Figure S2F). Taken together, extensive reprogramming of the proteome and concordant regulation of multiple and interconnected functional modules are key features of naïve T cell activation.
Phosphoproteome profiling identifies kinase pathways and kinase interaction network
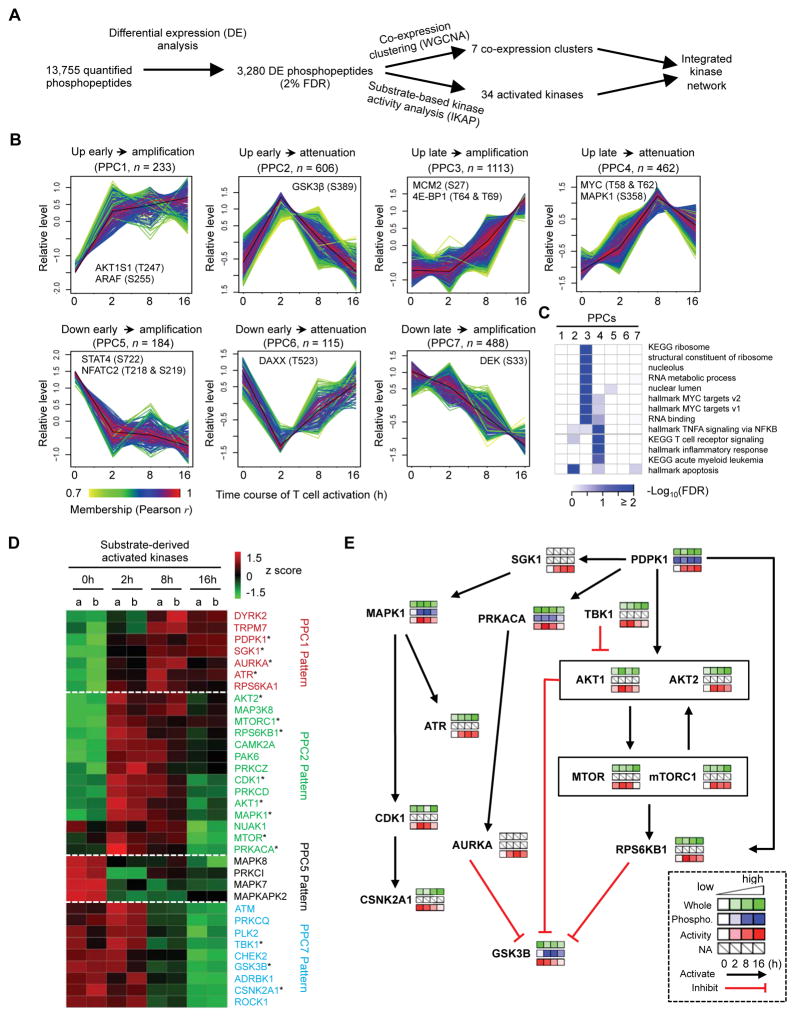
In our phosphoproteome profiling, we quantified 13,755 unique phosphopeptides from WT cells, of which 3,280 phosphopeptides were differentially expressed (Figure 3A and Data S1B). WGCNA co-expression clustering of phosphorylation sites identified seven phosphoproteome clusters (PPCs; Figure 3B), and functional enrichment was performed for each cluster (Figure 3C and Data S3A). In contrast to the limited number of upregulated proteins in the whole proteome before 8 h (WPC1 from Figure 2B), hundreds of phosphorylation events were induced immediately after TCR stimulation (see PPC1 and PPC2 from Figure 3B). These clusters of early upregulated phosphorylation were enriched for proteins localized in the cytoplasm and cytoskeletal proteins (Data S3A). The largest cluster of phosphorylated peptides was PPC3, which was induced at 8 h and further amplified at 16 h and thus resembled the pattern of whole proteome WPC3, and was enriched for MYC targets, translation and nucleus localizing proteins (Figure 3C). The similar temporal pattern observed for whole proteome and phosphoproteome for these pathways suggested coordinated regulation between translation and phosphorylation. For example, MCM2, a protein important for DNA replication, showed increases in protein abundance (Figure 2B: WPC3) and S27 phosphorylation that positively regulates MCM2 function (Hornbeck et al., 2015) (Figure 3B: PPC3). Also, MYC protein expression and T58/S62 phosphorylation were of WPC2 and PPC4, respectively, which shared the same pattern of delayed upregulation at 8 h followed by attenuation.
Figure 3. T cell phosphoproteome profiling reveals co-expression clusters, multiple active kinases, and dynamically regulated kinase signaling networks.
(A) Overview of computational analysis for phosphoproteome.
(B) Seven co-expression phosphoproteome clusters (PPCs). Each line indicates the relative abundance of each protein and was color-coded by the cluster membership.
(C) Functional annotations of PPCs by Gene Ontology (GO), KEGG and Hallmark databases (FDR < 0.05).
(D) Kinase activity inference based on substrate phosphorylation levels. The asterisk indicates kinases that are shown in Figure 3E.
(E) Kinase-to-kinase network upon TCR stimulation.
Additionally, we observed 3 downregulated phosphoproteome clusters, PPC5-PPC7. Within PPC5, S722 phosphorylation of STAT4, the critical driver of Th1 differentiation, was reduced after T cell activation. S219 phosphorylation of NFATC2, a key TF in T cells, showed a rapid decrease upon T cell activation. Since S219 is a phosphorylation-dependent inhibition site (Hornbeck et al., 2015), NFATC2 is likely activated by reduced S219 phosphorylation. PPC7 was the largest downregulated cluster and showed a pattern of delayed and sustained downregulation, resembling the whole proteome counterpart WPC6. In summary, analysis of whole proteome and phosphoproteome indicates early dynamic phosphorylation events and late amplification of both protein phosphorylation and expression, which collectively drive T cell activation.
We next used a machine learning algorithm, IKAP (Mischnik et al., 2016), to infer kinase activity based on the substrate phosphorylation levels from phosphoproteome after normalization of phosphopeptide abundance by protein abundance (Wu et al., 2011). This analysis extracted the activities for 66 kinases, of which 34 were differential regulated and hierarchically clustered into four major clusters: patterns similar to PPC1, PPC2, PPC5 and PPC7 (Figure 3D and Data S3B). Most of the upregulated kinases had direct or indirect relationship to AKT and mTOR, master regulators of cell growth and metabolism (Chi, 2012), including PDPK1 (PDK1), SGK1, AURKA, AKT2, mTORC1, RPS6KB1 (S6K1), AKT1, and mTOR (see AKT1, mTORC1 and S6K substrate phosphorylation in Figures S3A–C). Additionally, other growth factor signaling kinases, including ERK/MAPK signaling (MAPK1 and the respective upstream and downstream kinases MAP3K8 (TPL-2) and RPS6KB1 (RSK1)), CDK1 and CAMPK2A (CaM kinase), were also upregulated upon T cell activation.
We also identified downregulation of a number of kinase activities during T cell activation. The kinase activity for GSK3β, which inhibits T cell proliferation, was downregulated after 8 h of TCR stimulation (Figure 3D; see GSK3β substrate phosphorylation in Figure S3D). Additionally, S389 phosphorylation of GSK3β, an inhibitory phosphorylation site (Thornton et al., 2016), was upregulated at 2 h and 8 h (Figure 3B and Data S1B). Kinases that negatively regulate AKT-mTOR were also downregulated, including TBK1 and PLK2, supporting AKT-mTOR signaling as a central pathway for T cell activation. Attenuated activity was also shown for kinases associated with DNA-damage response including GSK3β, ATM, CHEK2 (CHK2), and PLK2 (Figures 3D and S3E), indicating that DNA damage response might be tuned down upon T cell activation. Finally, activities of stress-activated MAPKs, including MAPK8 (JNK1), MAPKAPK2 (MK2, downstream of p38 MAPK; Figure S3F) and MAPK7 (ERK5) were reduced upon TCR stimulation (Figure 3D). Immunoblot experiments validated altered activities of ATR and TBK1 (Figure S3G). Thus, T cell activation involves downregulation of kinase activities of GSK3β, DNA-damage response and stress-activated MAPK signaling, as well as negative regulators of AKT-mTOR.
Finally, utilizing the temporal data and IKAP-inferred kinase activities, we reconstructed a kinase-to-kinase signaling network with each kinase representing a node with edges representing phosphorylation-dependent activation or inhibition between two kinases (Hornbeck et al., 2015). Additionally, temporal patterns of the kinase’s protein abundance, phosphorylation status, and kinase activity were shown in Figure 3E. This analysis revealed multiple crosstalk events but centrally positioned AKT-mTOR and GSK3β in the signaling network. For example, since the protein abundance of GSK3β appears to be stably expressed, AKT and S6K1, both negative regulators of GSK3β, likely drive the inactivation of GSK3β (Figure 3E). Altogether, our phosphoproteome profiling, kinase activity inference and kinase interaction network revealed dynamic rewiring of T cell phosphoproteome upon TCR stimulation and the central roles of AKT-mTOR and GSK3β as the respective positive and negative regulators of T cell activation.
Reconstruction of signaling and transcriptional networks by integrative multi-level omics profiling
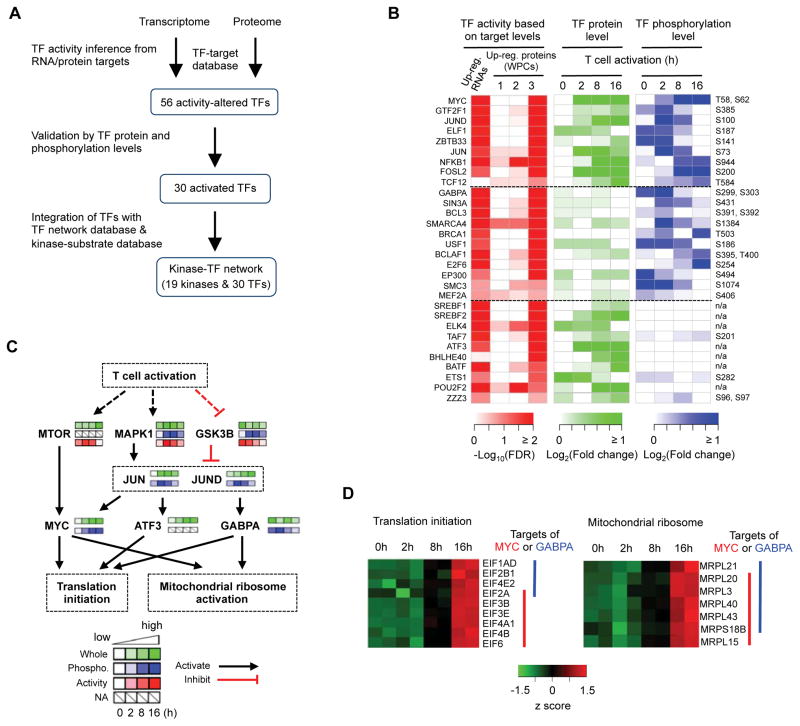
We hypothesize that integrative analysis of proteome and phosphoproteome via systems biology approaches could provide new insights into functional interaction linking kinases to TFs and cellular activities. As a proof of principle, we reconstructed the connectivity between TFs and kinases by integrating proteome and phosphoproteome profiling data with known TF-target and kinase-substrate databases (Gerstein et al., 2012; Hornbeck et al., 2015) (Figure 4A). First, we predicated TF activity in TCR-stimulated cells based on mRNA or protein expression of target genes, assuming that the expression dynamics of co-expressed transcripts and proteins (i.e. each WPC) is controlled by specific TFs (Figure 4B and Data S4). These candidate TFs were further analyzed for their protein abundance and phosphorylation levels. Second, we linked these TFs to reconstruct T cell-specific TF networks based on known TF-TF interactions (Neph et al., 2012). Finally, we deduced the upstream kinase(s) for each TF by screening the functional phosphosites of TFs using a known kinase-substrate database (Hornbeck et al., 2015). Altogether, we reconstructed signaling networks that connect 19 kinases and 30 TFs that are activated during T cell activation (Figures 4B and 4C; Data S4).
Figure 4. Integrated network analysis reveals key transcription factors and kinases and the connectivity between them in T cell activation.
(A) Overview of the integrative analysis based on transcriptome, whole proteome, phosphoproteome, TF-target database, and kinase-substrate database.
(B) Activated TFs during T cell activation.
(C) Prediction of a putative signaling cascade comprised of key kinases and TFs in the control of protein translation machinery.
(D) Mitochondrial ribosome and translation initiation proteins are activated by MYC and GABPA. The protein levels of the TF downstream targets are indicated.
These unbiased integrative analyses revealed that the activities of TFs are temporally and differentially regulated at the levels of either expression, or phosphorylation, or both. The first group contained TFs with dynamic regulation of both protein expression and phosphorylation. This group included MYC, NF-κB, FOSL2, TCF12 and JUND (Figures 4B, S4A and S4B). Second, GABPA, SMARCA4 (BRG1), BCLAF1, BRCA1 and SIN3A constituted a group of TFs that showed altered regulation of phosphorylation but no substantial changes in protein abundance (Figure 4B). Some of these TFs have also been implicated in T cell biology, including GABPA with important roles in T cell development (Xue et al., 2004), and our results provided a potential mechanism for the regulation of these TFs (Figure 4B). Third, SREBF1, SREBF2, POU2F2, ATF3 and ETS1 were altered in protein expression but without detectable changes of phosphorylation (Figures 4B and S4C). SREBF1 and SREBF2 are TCR-inducible factors mediating lipid synthesis (Kidani et al., 2013), while expression of TCF12, POU2F2, and FOSL2 was verified by immunoblots (Figure S4D). Collectively, these results highlight diverse mechanisms of controlling TF activities for quiescence exit of antigen-stimulated T cells.
We next reconstructed TF-kinase signaling networks by considering two scenarios of TF-TF and kinase-TF interaction: (i) an upstream TF binds to the DNA elements of another TF for transcriptional activation; and (ii) an upstream kinase modifies a TF to alter its activity (Gerstein et al., 2012; Hornbeck et al., 2015; Neph et al., 2012) (Figure 4C and Data S4). In one representative signaling network, TCRs activated mTOR and MAPK1, which in turn phosphorylate and activate TFs (MYC S62, JUN S73 and JUND S100). Certain TFs (e.g. JUN and JUND) induce expression of other TFs (e.g. ATF3, GABPA and MYC), which in turn drive the expression of target genes involved in translation (Figure 4C). Specifically, our model predicted that MYC and GABPA mediate the expression of ribosomes, mitochondrial ribosomes and translation initiation factors in TCR-stimulated cells (Figure 4D). Collectively, our unbiased analysis identifies specific TFs and kinases and their interplays in mediating transcriptional, translational and post-translational controls of T cell activation.
Proteomics analysis of Rptor-deficient cells identifies mTORC1-dependent mitochondrial pathways
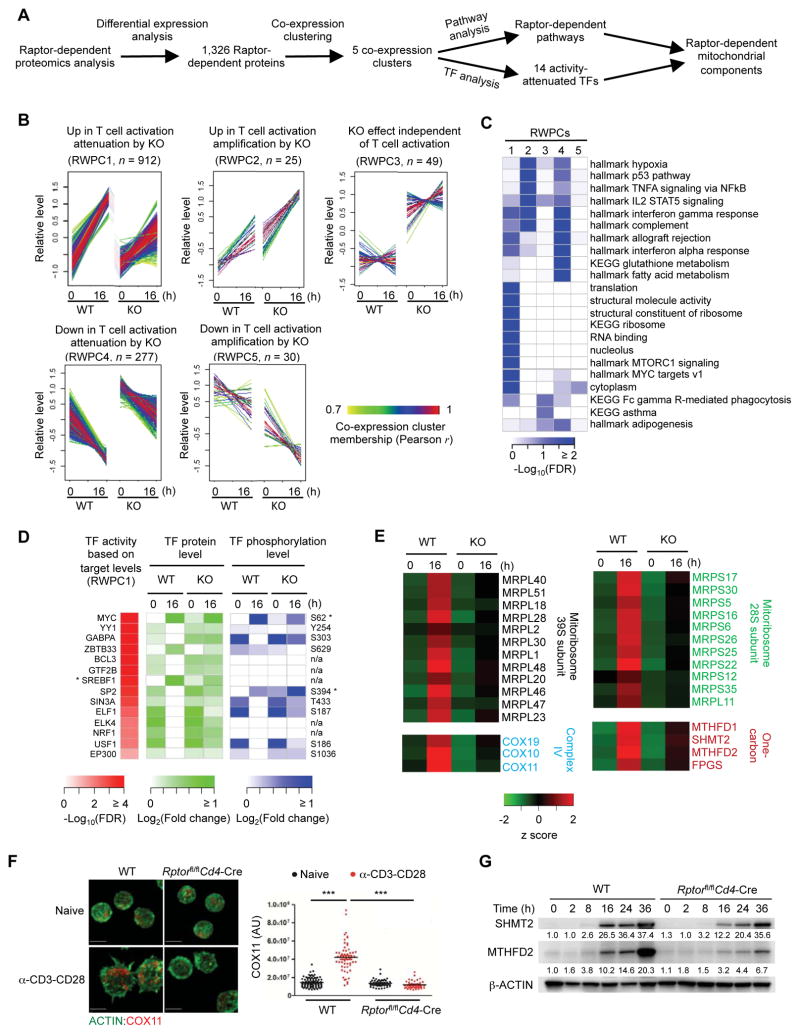
Raptor-mTORC1 is a central regulator of T cell quiescence exit (Yang et al., 2013). In our initial profiling, we included T cells from Rptorfl/flCd4-Cre mice that were activated by α-CD3-CD28 for 2 h and 16 h (Figure S5A; experiment 1), and found that Raptor deficiency led to profound proteomic changes only at 16 h (data not shown). We therefore performed an independent experiment by profiling whole proteome and phosphoproteome of naïve WT and Rptor-deficient T cells, as well as those stimulated with α-CD3-CD28 for 16 h, with biological replicates included (Figure S5A; experiment 2). In total, we quantified 9,178 proteins and 10,159 phosphopeptides (Data S5A and S5B). PCA and clustering analyses showed tight clustering of biological replicates (Figures S5B and S5C), and comparison of the two independent profiling experiments revealed that the results were highly reproducible (Figure S5D).
We defined Raptor-dependent events based on proteins or phosphopeptides that were consistently and significantly changed in both experiments. This analysis revealed 1,326 proteins (Figure 5A and Data S5A) and 547 phosphopeptides as Raptor-dependent, including known mTORC1 substrates RPS6, CAD and 4EBP1 (EIF4EBP1) (Data S5B). We further performed co-expression clustering analysis by applying temporal points from experiment 2, and identified five Raptor-dependent whole proteome clusters (RWPCs; Figure 5B) and eight Raptor-dependent phosphoproteome clusters (RPPCs; Figure S5E), and enriched pathways associated with each cluster (Figures 5C and S5F). Raptor deficiency strongly attenuated TCR-mediated protein expression and phosphorylation, as indicated by two of the largest RWPCs (RWPC1 and 4; Figure 5B) and RPPCs (RPPC1 and 5; Figure S5E). In particular, ribosomes and transcriptional processes were significantly attenuated at both protein and phosphorylation levels (Figure 5C and Data S5C, Figure S5F and Data S5D). In contrary, Raptor deficiency upregulated immune response-related pathways, including complement system (RWPC2 and 4), immune system process (RWPC4), and fatty acid oxidation (RWPC4) (Figure 5C and Data S5C), suggesting a negative role of mTORC1 in these immune and metabolic pathways.
Figure 5. Integrative analysis of Rptor-deficient proteom0e identifies mTORC1-dependent mitochondrial pathways.
(A) Overview of Raptor-dependent proteomics analysis.
(B) Five Raptor-dependent whole proteome co-expression clusters (RWPCs) defined by WGCNA.
(C) Functional annotations of RWPCs by Gene Ontology (GO), KEGG and Hallmark databases.
(D) TFs attenuated by Raptor deficiency. For TFs with an asterisk, a larger scale (e.g. maximum fold change of 10 instead of 2 folds) is used to reflect the large change of protein expression or phosphorylation abundance.
(E) Raptor-dependent mitochondrial pathways. The related protein levels are shown.
(F) Immunofluorescence images of ACTIN (green) and COX11 in naïve and activated WT and Rptor-deficient CD4+ T cells (scale bars, 5 μm). Right, statistical analysis of mean fluorescence intensity (MFI) of COX11 in WT and Rptor-deficient CD4+ T cells.
(G) Immunoblot of SHMT2 and MTHFD2 expression in WT and Rptor-deficient CD4+ T cells stimulated with α-CD3-CD28 for the indicated time points. Numbers below the lanes of SHMT2 and MTHFD2 indicate band intensity relative to that of the loading control-β-ACTIN.
Data are representative of two (F–G) independent experiments. P values are determined by one-way ANOVA with Tukey post-tests (F). ***P < 0.001.
To explore the potential mechanism of mTORC1-dependent transcriptional regulation, we focused on RWPC1 and performed TF analysis. We found that Raptor deficiency significantly attenuated 14 TFs (Figure 5D), 10 of which were activated in WT T cells after TCR stimulation (Figure 4B), highlighting the key contribution of mTORC1 to TCR-induced transcriptional responses. MYC and GABPA within this list have been shown to activate transcription of genes encoding components of the translational machinery (Figure 4D), suggesting that mTORC1-mediated transcriptional responses likely act in synergy with translational effects to boost protein synthesis during T cell activation. Moreover, we observed a crucial role of mTORC1 in the regulation of mitochondrial function. First, of the 14 TFs attenuated in Rptor-deficient cells (Figure 5D), at least four TFs, including MYC (Li et al., 2005), YY1 (Cunningham et al., 2007), GABPA (Yang et al., 2014) and SREBF1 (Gilardi et al., 2014), have been implicated in mitochondrial functions. Second, focusing on mitochondrial proteins in the MitoCarta database (Calvo et al., 2016), we identified 129 mitochondrial proteins as Raptor-dependent. Third, several TCR-activated mitochondrial pathways and components, including mitoribosomes (Figure 2F), complex IV (Figure 2G), and one-carbon metabolism (Figure S2D), were attenuated in Rptor-deficient cells (Figure 5E). We validated the inhibitory effect of Raptor deficiency on the induction of COX11 (complex IV), SHMT2 and MTHFD2 in TCR-stimulated cells (Figures 5F and 5G). Moreover, mTORC1 inhibitor rapamycin suppressed the expression of SHMT2 and MTHFD2 in activated T cells (Figure S5G). Taken together, these results demonstrate crucial roles of mTORC1 in coordinating transcriptional, translational and post-translational events and contributing to activation of mitochondrial function upon TCR stimulation.
Differential roles of mitoribosomes, COX10 and HK2 in T cell quiescence exit
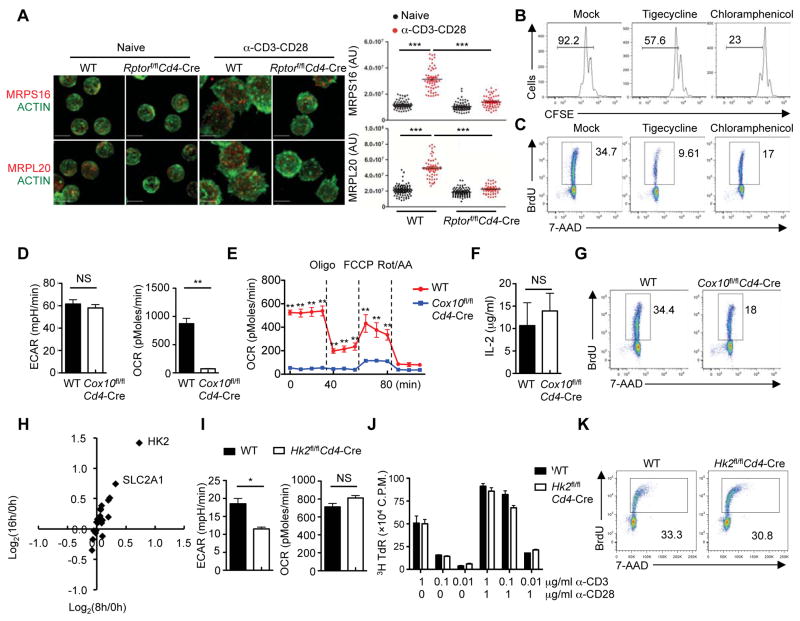
The specific mitochondrial pathways and upstream signals involved in T cell activation remain poorly understood. We found that TCR-stimulated cells increased mitochondrial biogenesis, but this was substantially dampened in the absence of Raptor (Figure S6A). Moreover, expression of mitochondrial ribosomal proteins MRPS16 and MRPL20 was reduced in TCR-stimulated Rptor-deficient (Figure 6A) and rapamycin-treated cells (Figure S6B), suggesting impaired mitoribosome biogenesis. In T cells treated with tigecycline, an inhibitor that targets mitochondrial ribosome (Skrtic et al., 2011), cell division was diminished (Figure 6B). Also, tigecycline reduced bromodeoxyuridine (BrdU) incorporation at 24 h after TCR stimulation (Figure 6C), indicative of defective S phase entry. Similar phenotypes were observed after treatment with chloramphenicol, another inhibitor of mitochondrial translation (Figures 6B and 6C), whereas these inhibitors did not affect activation marker CD25 (Figure S6C) or cell survival (Figure S6D). Thus, mitochondrial translation contributes to quiescence exit of TCR-stimulated cells.
Figure 6. mTORC1-dependent mitoribosome synthesis and complex IV, but not HK2-mediated glycolysis, contribute to T cell quiescence exit.
(A) Immunofluorescence images of ACTIN (green) and MRPS16 (red) or MRPL20 (red) in naïve and activated WT and Rptor-deficient CD4+ T cells (scale bars, 5 μm). Right, statistical analysis of mean fluorescence intensity (MFI) of MRPS16 and MRPL20 in WT and Rptor-deficient CD4+ T cells.
(B) Flow cytometry of CFSE-labeled CD4+ T cells stimulated with α-CD3-CD28 in the presence of tigecycline or chloramphenicol for 60 h.
(C) BrdU incorporation of activated CD4+ T cells in the presence of tigecycline or chloramphenicol (24 h).
(D) Extracellular acidification rate (ECAR) and oxygen consumption rate (OCR) of activated WT and Cox10-deficient CD4+ T cells (24 h).
(E) OCR of activated WT and Cox10-deficient CD4+ T cells (24 h) in response to the indicated mitochondrial inhibitors (Oligo, Oligomycin; FCCP, carbonyl cyanide p-trifluoromethoxyphenylhydrazone; and Rot/AA, Rotenone/Antimycin A).
(F) IL-2 secretion of activated WT and Cox10-deficient CD4+ T cells (24 h).
(G) BrdU incorporation of activated WT and Cox10-deficient CD4+ T cells (24 h).
(H) Expression of glycolytic enzymes and transporters in TCR-activated T cells detected by proteomics profiling. The x-axis shows the induction of protein expression at 8 h, and the y-axis shows the induction of protein expression at 16 h.
(I) ECAR and OCR of activated WT and Hk2-deficient CD4+ T cells (24 h).
(J) [3H]Thymidine incorporation of WT and Hk2-deficient CD4+ T cells stimulated with α-CD3 or α-CD3-CD28 for 64 h, and pulsed with [3H]thymidine for an additional 8 h.
(K) BrdU incorporation of activated WT and Hk2-deficient CD4+ T cells (24 h).
Data are representative of two (A–D, H–K) or three (E–G) independent experiments. Data are mean ± s.e.m. P values are determined by one-way ANOVA with Tukey post-tests (A), two-tailed Student’s t-test (D, I), or two-way ANOVA with Bonferroni post-tests (E). NS, not significant, *P < 0.05, **P < 0.005, and ***P < 0.001. Numbers in gates indicate percentage of cells.
See also Figure S6.
A key function of mitoribosomes is the translation of mitochondria-encoded electron transport components to support OXPHOS. To study additional pathways mediating OXPHOS activation, we explored mechanisms underlying enrichment of mitochondrial complex IV in activated T cells (Figure 2G). Cytochrome c oxidase 10 (COX10), an accessory factor for the expression and assembly of ETC complex IV, was upregulated upon T cell activation (Figures 2G and 5E). To explore the roles of COX10 in T cell immunity, we generated mice with Cox10 deleted in T cells (Cox10fl/flCd4-Cre). Activated Cox10-deficient T cells exhibited largely normal extracellular acidification rate (ECAR, an indicator of aerobic glycolysis), but abolished oxygen consumption rate (OCR, an indicator of OXPHOS) (Figure 6D). Moreover, Cox10-deficient T cells were unresponsive to treatment of mitochondrial inhibitors (Figure 6E). Immunofluorescence of TOM20 showed that Cox10-deficient T cells had largely normal quantity of mitochondria (Figure S6E). In contrast, Cox10 deficiency abolished the expression of MT-CO1, a key component of respiratory complex IV, but not proteins in other respiratory complexes (Figure S6F). Thus, COX10 is crucial for mitochondrial complex IV activity and OXPHOS.
We next assessed the role of COX10 in TCR-induced exit from quiescence. TCR-induced proliferation was considerably reduced in Cox10-deficient T cells (Figure S6G). For TCR-induced initial activation events at 24 h, Cox10-deficient T cells showed largely normal IL-2 expression (Figure 6F), induction of CD25 and CD98 expression (Figure S6H), and cell survival (Figure S6I), but BrdU incorporation was diminished in Cox10-deficient T cells (Figure 6G). When examined at 48 h after TCR stimulation, Cox10-deficient T cells showed greatly elevated cell death (Figure S6J), whereas remaining live cells did not proliferate effectively (Figure S6K). Thus, COX10 is required for cell cycle entry of naïve T cells after TCR stimulation and ensuing expansion and long-term survival. Altogether, these results point to the obligatory role of OXPHOS activation in TCR-induced quiescence exit, which requires remodeling of both mitoribosomes and complex IV activity (Figure S6L).
OXPHOS and glycolysis are two major bioenergetic pathways in mammalian cells (Buck et al., 2015; MacIver et al., 2013). Our proteomics profiling showed that hexokinase 2 (HK2), a key rate-limiting enzyme in glycolysis, was the most profoundly upregulated glycolytic enzyme after TCR stimulation (Figure 6H). We therefore generated Hk2fl/flCd4-Cre mice. Hk2-deficient cells had reduced ECAR activity (Figure 6I), but largely normal OCR activity (Figure 6I), proliferation (Figure 6J), and BrdU incorporation (Figure 6K) upon TCR stimulation. Also, expression of hexokinases HK1 and HK3 was comparable between WT and Hk2-deficient T cells (data not shown). Thus, T cell quiescence exit proceeds independently of HK2 or after substantial reduction of glycolysis.
COX10 is crucial for Th1 cell differentiation and antibacterial immune responses
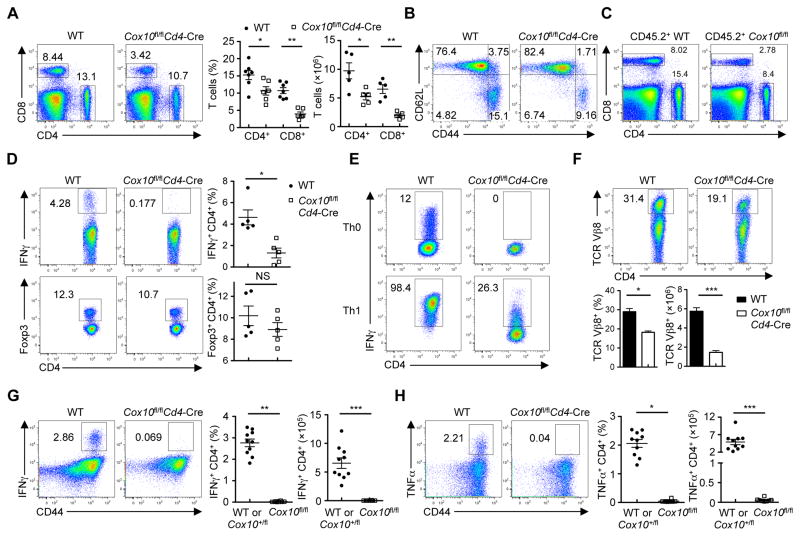
We next explored the physiological roles of COX10 in vivo. Cox10fl/flCd4-Cre mice had largely normal thymocyte development (Figure S7A) and diversity of TCR Vβ chain usage (Figures S7B and S7C), but they had significantly reduced numbers of splenic CD4+ and CD8+ T cells (Figure 7A), with a slightly reduced compartment of CD62LloCD44hi memory and effector T cells (Figure 7B). To test whether reduction of Cox10-deficient T cells was a cell-autonomous defect, we generated mixed bone marrow (BM) chimeras by reconstituting Rag1−/− mice with BM cells from CD45.1+ mice and those from WT or Cox10fl/flCd4-Cre CD45.2+ mice. The substantial reduction of Cox10-deficient donor T cells indicated that COX10 was intrinsically required for T cell homeostasis (Figure 7C).
Figure 7. COX10 deficiency impairs T cell homeostasis, Th1 cell differentiation and immune responses in vivo.
(A) Flow cytometry of splenic CD4+ and CD8+ T cells from WT and Cox10fl/flCd4-Cre mice. Right, proportions and numbers of CD4+ and CD8+ T cells (n = 5–7 mice each group).
(B) Expression of CD62L and CD44 on splenic CD4+ T cells from WT and Cox10fl/flCd4-Cre mice.
(C) Flow cytometry of splenic CD4+ and CD8+ T cells from WT:CD45.1+ and Cox10fl/flCd4-Cre:CD45.1+ mixed BM chimeras.
(D) Flow cytometry of the expression of IFNγ (upper) and Foxp3 (lower) in splenic CD4+ T cells from WT and Cox10fl/flCd4-Cre mice. Right, statistics of IFNγ-producing cells and Foxp3+ Treg cells.
(E) Flow cytometry of IFNγ production in WT or Cox10-deficient T cells cultured under Th0 and Th1 conditions for 3–4 days.
(F) Flow cytometry of splenic CD4+ TCR Vβ8+ T cells from WT and Cox10fl/flCd4-Cre mice treated with SEB (i.v. 100 μg/mouse) for 2 days. Bottom, proportion and number of CD4+ TCR Vβ8+ T cells.
(G, H) Flow cytometry of IFNγ-producing CD4+ (G) and TNFα-producing CD4+ (H) cells in the spleen from control (WT or Cox10+/flCd4-Cre) and Cox10fl/flCd4-Cre mice infected with L. monocytogenes-expressing OVA (LM-OVA) (WT, n = 4; Cox10+/fl, n = 6; Cox10fl/fl, n = 8 mice), after OVA peptide stimulation in vitro. Right, proportion and number of cytokine-producing T cells.
Data are representative of at least three (A) or two (B–H) independent experiments. Data are mean ± s.e.m. P values are determined by Mann-Whitney test (A, F–H, cell proportion) or two-tailed Student’s t-test (A, F–H, cell number). NS, not significant, *P < 0.05, **P < 0.005 and ***P < 0.0001. Numbers in quadrants or gates indicate percentage of cells.
See also Figure S7.
We next examined the role of COX10 in the generation of CD4 T cell subsets. Cox10-deficient T cells had diminished production of IFNγ (Figure 7D) upon ex vivo stimulation, whereas the proportion of IL-17 or IL-4-expressing cells was low and similar between WT and Cox10-deficient T cells (Figure S7D). Additionally, the proportion of Foxp3+ regulatory T (Treg) cells was comparable between these mice (Figure 7D). Moreover, in response to non-polarizing (Th0) or Th1-polarizing conditions, Cox10-deficient T cells showed a marked defect in differentiating into IFNγ-expressing Th1 cells (Figure 7E). However, Cox10-deficient T cells were able to develop into Foxp3+ induced Treg (iTreg) cells (Figure S7E), indicating a selective requirement of COX10 in T cell responses.
To investigated the physiological relevance of COX10 in T cell immunity, we challenged Cox10fl/flCd4-Cre mice with staphylococcal enterotoxin (SEB), a superantigen that expands CD4+ and CD8+ T cells bearing the TCR Vβ8 chain. Compared with WT controls, Cox10fl/flCd4-Cre mice had notable reduction of Vβ8+ CD4+ (Figure 7F) and CD8+ T cells (Figure S7F). Second, we infected mice with recombinant Listeria monocytogenes expressing ovalbumin (LM-OVA), and examined antigen-specific CD4+ T cells by restimulation with the listeriolysin O (LLO)189–201 peptide. Compared with controls, Cox10fl/flCd4-Cre mice showed significant fewer IFNγ and TNFα-producing CD4+ T cells (Figures 7G and 7H). Moreover, Cox10-deficient CD8+ T cells had substantially fewer tetramer-positive (H-2Kb-OVA+) cells than did control T cells (Figures S7G and S7H), and were defective to produce IFNγ and TNFα after OVA257–264 peptide stimulation (Figures S7I and S7J). Collectively, these results establish the crucial roles of COX10 in T cell responses in vivo.
DISCUSSION
A hallmark of adaptive immunity is antigen-triggered T cell activation, but the molecular circuits by which naïve T cells exit from quiescence remain poorly understood (Yang et al., 2013). We showed that the dynamics of T cell activation encompassed an early phase (2 h) with rapid induction of phosphorylation events but limited changes of protein abundance, and a late phase (8–16 h) that involved extensive reprogramming of both whole proteome and phosphoproteome. This late phase was marked by coordinated, massive upregulation of molecular machineries in protein translation and mitochondrial functions, and hallmarks of mTORC1 and MYC activation. Through systems biology approaches, we developed an analysis pipeline by integrating our multi-layer omics data with public databases, and identified signaling networks connecting key kinases and TFs. Finally, we applied genetic and pharmacological tools for experimental validation of hypotheses predicted from bioinformatics analyses, and revealed the crucial requirements of mitoribosomes, cytochrome c oxidase biogenesis, and the resultant OXPHOS activation in T cell activation. Altogether, our studies establish the landscapes of T cell proteome and phosphoproteome, dynamic signaling networks in T cell activation, and selective bioenergetics pathways underlying quiescence exit.
Our comprehensive pipeline allows us to integrate temporal, multi-tier omics data (e.g. transcriptome, proteome, and phosphoproteome in a time course) and various databases (e.g. PPI, TF-target and kinase-substrate) to identify core molecular circuits in complex biological systems. Compared to single-level omics analysis, this multi-tier omics pipeline shows better sensitivity (e.g. the missing data in one omics analysis can be complemented by other measurements) and reliability (multiple omics measurements improve data reliability). This strategy effectively sorts thousands of altered components into clusters and then into functional modules that are potentially controlled by a small list of interconnected key master regulators (e.g. TFs and kinases). With rapid developments in omics technologies and accumulation of large datasets, this bioinformatics pipeline provides a general platform for accelerating data interpretation and discovery.
Several unappreciated predictions about mechanisms of T cell quiescence exit were revealed. We found that TCR stimulation downregulates DNA damage responses and stress-activated MAPK signaling. Given the profound DNA and RNA synthesis associated with T cell quiescence exit, how genome integrity is maintained warrants further investigation. Also, although all MAPK subgroups (ERK, JNK and p38) are thought to be upregulated upon TCR stimulation, our temporal analysis revealed distinct patterns of ERK signaling and stress-activated JNK and p38 activation. We also found TCR stimulation mediates induction of protein degradation, which has been shown to supply needed amino acids to support robust protein synthesis for maintaining cellular homeostasis (Suraweera et al., 2012). Further studies perturbing these pathways can further test their functional importance.
Despite the increasing appreciation of mitochondrial functions in T cell responses (Chang et al., 2013; Sena et al., 2013), the specific mitochondrial pathways and upstream signals remain largely unexplored. mTORC1 has been shown to promote mitochondrial function by activating PCG-1α (Cunningham et al., 2007). In the immune compartment, however, a dispensable role of mTORC1 in mitochondrial function is observed in effector cytotoxic T lymphocytes (Hukelmann et al., 2016) and dendritic cells (Amiel et al., 2014). For T cell exit from quiescence, we showed activation of mTORC1-dependent mitochondrial biogenesis, mitoribosome expression, one-carbon metabolism and complex IV components. Direct comparison of the proteome between rapamycin-treated CTLs (Hukelmann et al., 2016) and TCR-stimulated Rptor-deficient naïve T cells revealed both overlapping and distinct effects (data not shown), further highlighting context-dependent regulation of mitochondrial activity by mTORC1.
T cell activation and differentiation require metabolic rewiring, but the functional effects of distinct metabolic programs remain a topic of interest and debate. In particular, it is often difficult to elucidate whether an observed metabolic shift is the cause or consequence of a change in the cellular phenotype. Indeed, despite the profound upregulation of HK2 after TCR stimulation, loss of HK2 or substantial reduction of glycolysis is compatible with T cell activation. In contrast, inhibition of mitoribosome biogenesis and disruption of COX10 revealed the crucial roles of OXPHOS in T cell exit from quiescence. Our data further established mTORC1-dependent mitoribosome biogenesis and COX10-mediated complex IV activity as the signal-dependent mitochondrial pathways engaged upon T cell activation. Nonetheless, Cox10-deficient cells showed no obvious defects in IL-2 production or upregulation of activation markers, which was different from deficiency of complex III (Sena et al., 2013). Furthermore, Cox10-deficient T cells could differentiation into Treg cells, but were defective in developing into inflammatory Th1 cells. Thus, T cells can rely on metabolic adaptation to engage certain activation events and maintain short-term survival, but cell cycle entry and Th1 cell differentiation are more metabolically demanding and require activation of mitochondrial OXPHOS.
In summary, we have established the landscapes of T cell proteome and phosphoproteome and the dynamic functional modules and signaling networks that orchestrate T cell activation. We have also identified bioenergetics pathways and their functional involvement in T cell quiescence exit. These molecular circuits identified in quiescence exit may also be implicated in the maintenance of naïve T cell quiescence and survival (Yang et al., 2011), re-establishment of T cell quiescence during memory formation (Shrestha et al., 2014), and lymphocyte transformation.
EXPERIMENTAL PROCEDURES
Mice
C57BL/6, Rag1−/−, CD45.1+, Cox10fl/fl and Hk2fl/fl mice were purchased from the Jackson Laboratory or EMMA. Rptorfl/fl and Cd4-Cre mice have been described (Yang et al., 2013). Mice were used at 8–10-weeks old unless otherwise noted. All mice were kept in a specific pathogen-free facility in the Animal Resource Center at St. Jude Children’s Research Hospital, and animal protocols were approved by the Institutional Animal Care and Use Committee.
Mass spectrometry and data analysis
Detailed analyses were described in Extended Experimental Procedures. In brief, cell pellets were lysed, digested and labeled with TMT tags respectively. Equal amount of peptides from each sample were pooled and fractionated by off-line basic pH reverse phase liquid chromatography (LC). Fractionated peptides were analyzed by LC-MS/MS. Phosphopeptides were enriched (Tan et al., 2015) before LC-MS/MS analysis. Using the JUMP software (Li et al., 2016; Wang et al., 2014), spectra were searched against the Uniprot mouse database (52,490 protein entries; downloaded in February 2015) and filtered to achieve 1% FDR at either unique protein (for whole proteome) or peptide (for phosphoproteome) level. To minimize redundancy, protein identifications from shared peptide sequences (Data S6) were grouped into unique proteins according to the principal of parsimony. TMT intensities were extracted, filtered, normalized and summarized into peptide and protein quantification. For T cell activation analysis, one-way ANOVA was used to identify DE events, and p values were corrected for multiple testing using the method of Benjamini-Hochberg. To define Raptor-dependent events, z-score based DE analysis was performed, in which proteins with significant z scores (1.96 as cutoff) were overlapped in two experiments. Principal component analysis and hierarchical clustering were performed using R (version 3.0.1).
Network analysis
Co-expression clusters were identified by the WGCNA R package (Langfelder et al., 2008) and superimposed onto a composite PPI database of STRING (http://string-db.org/) and BioPlex (http://wren.hms.harvard.edu/bioplex/). Kinase activity was inferred based on known substrates in the PhosphoSitePlus database (Hornbeck et al., 2015) using the IKAP algorithm (Mischnik et al., 2016). TF activity was inferred based on target expression levels (Gerstein et al., 2012). Kinase-TF network were constructed by superimposing activity-altered kinases and TFs onto networks of known kinase-substrate (Hornbeck et al., 2015) and TF-target relationships (Neph et al., 2012).
Listeria monocytogenes infection
Mice were intravenously infected with 3 × 104 colony-forming units (CFUs) of L. monocytogenes expressing the chicken ovalbumin (LM-OVA). At seven days post infection, lymphocytes were isolated from the spleen and liver and stained with OVA peptide (comprised of amino acids Ser-Ile-Ile-Asn-Phe-Glu-Lys-Leu (SIINFEKL)-loaded mouse H-2Kb tetramer (H-2Kb-OVA; Baylor Tetramer Production Facility). To examine cytokine production, splenocytes were stimulated with OVA257–264 (1 μM) or LLO189–201 (1 μM) for 5 h in the presence of monensin before fixation, permeabilization and intracellular staining of IFNγ and TNFα.
Statistical analysis for small-scale immunologic experiments
P values were calculated by Mann-Whitney test or two-tailed unpaired Student’s t test using GraphPad Prism, unless otherwise noted. P values of less than 0.05 were considered as significant. All error bars represent the s.e.m.
Supplementary Material
Acknowledgments
The authors thank for all other members in the Peng and Chi labs and in St. Jude Proteomics Facility for insightful discussion, and Anil KC and Melissa Hendren for technical assistance. This work was partially supported by National Institutes of Health grant AG047928 (J.P.), GM114260 (J.P.), AI101407 (H.C.), CA176624 (H.C.), AI105887 (H.C.), NS064599 (H.C.), American Asthma Foundation, and ALSAC (American Lebanese Syrian Associated Charities). The MS analysis was performed in the St. Jude Proteomics Facility, partially supported by NIH P30CA021765. The authors declare no conflicts of interest.
Footnotes
AUTHOR CONTRIBUTIONS
J.P., H.C., K.Y., Y.L. and H.T. designed the research; H.T. performed proteomics experiments; K.Y., Y.W. and D.B.B. performed mouse, culture and immunoblot experiments; Y.L., T.I.S., X.W., J.H.C. and H.W. implemented the pipeline for computational analysis; S.R. performed immunoblot experiments; C.G. performed imaging experiments; and K.Y., H.T., Y.L., T.I.S., J.P. and H.C. prepared the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amiel E, Everts B, Fritz D, Beauchamp S, Ge B, Pearce EL, Pearce EJ. Mechanistic target of rapamycin inhibition extends cellular lifespan in dendritic cells by preserving mitochondrial function. J Immunol. 2014;193:2821–2830. doi: 10.4049/jimmunol.1302498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amit I, Regev A, Hacohen N. Strategies to discover regulatory circuits of the mammalian immune system. Nat Rev Immunol. 2011;11:873–880. doi: 10.1038/nri3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck MD, O’Sullivan D, Pearce EL. T cell metabolism drives immunity. J Exp Med. 2015;212:1345–1360. doi: 10.1084/jem.20151159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo SE, Clauser KR, Mootha VK. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2016;44:D1251–1257. doi: 10.1093/nar/gkv1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CH, Curtis JD, Maggi LB, Jr, Faubert B, Villarino AV, O’Sullivan D, Huang SC, van der Windt GJ, Blagih J, Qiu J, et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013;153:1239–1251. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi H. Regulation and function of mTOR signalling in T cell fate decisions. Nat Rev Immunol. 2012;12:325–338. doi: 10.1038/nri3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450:736–740. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilardi F, Migliavacca E, Naldi A, Baruchet M, Canella D, Le Martelot G, Guex N, Desvergne B, Cycli XC. Genome-wide analysis of SREBP1 activity around the clock reveals its combined dependency on nutrient and circadian signals. PLoS Genet. 2014;10:e1004155. doi: 10.1371/journal.pgen.1004155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton SE, Jameson SC. CD8 T cell quiescence revisited. Trends Immunol. 2012;33:224–230. doi: 10.1016/j.it.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43:D512–520. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hukelmann JL, Anderson KE, Sinclair LV, Grzes KM, Murillo AB, Hawkins PT, Stephens LR, Lamond AI, Cantrell DA. The cytotoxic T cell proteome and its shaping by the kinase mTOR. Nat Immunol. 2016;17:104–112. doi: 10.1038/ni.3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R, Greenbaum D, Gerstein M. Relating whole-genome expression data with protein-protein interactions. Genome Res. 2002;12:37–46. doi: 10.1101/gr.205602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidani Y, Elsaesser H, Hock MB, Vergnes L, Williams KJ, Argus JP, Marbois BN, Komisopoulou E, Wilson EB, Osborne TF, et al. Sterol regulatory element-binding proteins are essential for the metabolic programming of effector T cells and adaptive immunity. Nat Immunol. 2013;14:489–499. doi: 10.1038/ni.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Zhang B, Horvath S. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics. 2008;24:719–720. doi: 10.1093/bioinformatics/btm563. [DOI] [PubMed] [Google Scholar]
- Li F, Wang Y, Zeller KI, Potter JJ, Wonsey DR, O’Donnell KA, Kim JW, Yustein JT, Lee LA, Dang CV. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol Cell Biol. 2005;25:6225–6234. doi: 10.1128/MCB.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JJ, Biggin MD. Gene expression. Statistics requantitates the central dogma. Science. 2015;347:1066–1067. doi: 10.1126/science.aaa8332. [DOI] [PubMed] [Google Scholar]
- Li Y, Wang X, Cho JH, Shaw TI, Wu Z, Bai B, Wang H, Zhou S, Beach TG, Wu G, et al. JUMPg: An Integrative Proteogenomics Pipeline Identifying Unannotated Proteins in Human Brain and Cancer Cells. J Proteome Res. 2016;15:11. doi: 10.1021/acs.jproteome.6b00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Beyer A, Aebersold R. On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell. 2016;165:535–550. doi: 10.1016/j.cell.2016.03.014. [DOI] [PubMed] [Google Scholar]
- MacIver NJ, Michalek RD, Rathmell JC. Metabolic regulation of T lymphocytes. Annu Rev Immunol. 2013;31:259–283. doi: 10.1146/annurev-immunol-032712-095956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann M, Kulak NA, Nagaraj N, Cox J. The coming age of complete, accurate, and ubiquitous proteomes. Mol Cell. 2013;49:583–590. doi: 10.1016/j.molcel.2013.01.029. [DOI] [PubMed] [Google Scholar]
- Mischnik M, Sacco F, Cox J, Schneider HC, Schafer M, Hendlich M, Crowther D, Mann M, Klabunde T. IKAP: A heuristic framework for inference of kinase activities from Phosphoproteomics data. Bioinformatics. 2016;32:424–431. doi: 10.1093/bioinformatics/btv699. [DOI] [PubMed] [Google Scholar]
- Neph S, Stergachis AB, Reynolds A, Sandstrom R, Borenstein E, Stamatoyannopoulos JA. Circuitry and dynamics of human transcription factor regulatory networks. Cell. 2012;150:1274–1286. doi: 10.1016/j.cell.2012.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron-Harel N, Santos D, Ghergurovich JM, Sage PT, Reddy A, Lovitch SB, Dephoure N, Satterstrom FK, Sheffer M, Spinelli JB, et al. Mitochondrial Biogenesis and Proteome Remodeling Promote One-Carbon Metabolism for T Cell Activation. Cell Metab. 2016;24:104–117. doi: 10.1016/j.cmet.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Sena LA, Li S, Jairaman A, Prakriya M, Ezponda T, Hildeman DA, Wang CR, Schumacker PT, Licht JD, Perlman H, et al. Mitochondria are required for antigen-specific T cell activation through reactive oxygen species signaling. Immunity. 2013;38:225–236. doi: 10.1016/j.immuni.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha S, Yang K, Wei J, Karmaus PW, Neale G, Chi H. Tsc1 promotes the differentiation of memory CD8+ T cells via orchestrating the transcriptional and metabolic programs. Proc Natl Acad Sci U S A. 2014;111:14858–14863. doi: 10.1073/pnas.1404264111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrtic M, Sriskanthadevan S, Jhas B, Gebbia M, Wang X, Wang Z, Hurren R, Jitkova Y, Gronda M, Maclean N, et al. Inhibition of mitochondrial translation as a therapeutic strategy for human acute myeloid leukemia. Cancer Cell. 2011;20:674–688. doi: 10.1016/j.ccr.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suraweera A, Munch C, Hanssum A, Bertolotti A. Failure of amino acid homeostasis causes cell death following proteasome inhibition. Mol Cell. 2012;48:242–253. doi: 10.1016/j.molcel.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan H, Wu Z, Wang H, Bai B, Li Y, Wang X, Zhai B, Beach TG, Peng J. Refined phosphopeptide enrichment by phosphate additive and the analysis of human brain phosphoproteome. Proteomics. 2015;15:500–507. doi: 10.1002/pmic.201400171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton TM, Delgado P, Chen L, Salas B, Krementsov D, Fernandez M, Vernia S, Davis RJ, Heimann R, Teuscher C, et al. Inactivation of nuclear GSK3beta by Ser(389) phosphorylation promotes lymphocyte fitness during DNA double-strand break response. Nat Commun. 2016;7:10553. doi: 10.1038/ncomms10553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang Y, Li Y, Bai B, Wang X, Tan H, Liu T, Beach TG, Peng J, Wu Z. Systematic optimization of long gradient chromatography mass spectrometry for deep analysis of brain proteome. J Proteome Res. 2015;14:829–838. doi: 10.1021/pr500882h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Li Y, Wu Z, Wang H, Tan H, Peng J. JUMP: a tag-based database search tool for peptide identification with high sensitivity and accuracy. Mol Cell Proteomics. 2014;13:3663–3673. doi: 10.1074/mcp.O114.039586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R, Dephoure N, Haas W, Huttlin EL, Zhai B, Sowa ME, Gygi SP. Correct interpretation of comprehensive phosphorylation dynamics requires normalization by protein expression changes. Mol Cell Proteomics. 2011;10:M111009654. doi: 10.1074/mcp.M111.009654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue HH, Bollenbacher J, Rovella V, Tripuraneni R, Du YB, Liu CY, Williams A, McCoy JP, Leonard WJ. GA binding protein regulates interleukin 7 receptor alpha-chain gene expression in T cells. Nat Immunol. 2004;5:1036–1044. doi: 10.1038/ni1117. [DOI] [PubMed] [Google Scholar]
- Yang K, Neale G, Green DR, He W, Chi H. The tumor suppressor Tsc1 enforces quiescence of naive T cells to promote immune homeostasis and function. Nat Immunol. 2011;12:888–897. doi: 10.1038/ni.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Shrestha S, Zeng H, Karmaus PW, Neale G, Vogel P, Guertin DA, Lamb RF, Chi H. T cell exit from quiescence and differentiation into Th2 cells depend on Raptor-mTORC1-mediated metabolic reprogramming. Immunity. 2013;39:1043–1056. doi: 10.1016/j.immuni.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZF, Drumea K, Mott S, Wang J, Rosmarin AG. GABP transcription factor (nuclear respiratory factor 2) is required for mitochondrial biogenesis. Mol Cell Biol. 2014;34:3194–3201. doi: 10.1128/MCB.00492-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Nicholatos J, Dreier JR, Ricoult SJ, Widenmaier SB, Hotamisligil GS, Kwiatkowski DJ, Manning BD. Coordinated regulation of protein synthesis and degradation by mTORC1. Nature. 2014;513:440–443. doi: 10.1038/nature13492. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.