Abstract
Dopaminergic degeneration is a pathologic hallmark of Parkinson's disease (PD), which can be assessed by dopamine transporter imaging such as FP-CIT SPECT. Until now, imaging has been routinely interpreted by human though it can show interobserver variability and result in inconsistent diagnosis. In this study, we developed a deep learning-based FP-CIT SPECT interpretation system to refine the imaging diagnosis of Parkinson's disease. This system trained by SPECT images of PD patients and normal controls shows high classification accuracy comparable with the experts' evaluation referring quantification results. Its high accuracy was validated in an independent cohort composed of patients with PD and nonparkinsonian tremor. In addition, we showed that some patients clinically diagnosed as PD who have scans without evidence of dopaminergic deficit (SWEDD), an atypical subgroup of PD, could be reclassified by our automated system. Our results suggested that the deep learning-based model could accurately interpret FP-CIT SPECT and overcome variability of human evaluation. It could help imaging diagnosis of patients with uncertain Parkinsonism and provide objective patient group classification, particularly for SWEDD, in further clinical studies.
Keywords: Parkinson's disease, FP-CIT, Deep learning, Deep neural network, SWEDD
Highlights
-
•
Deep learning-based FP-CIT SPECT interpretation model was developed.
-
•
Deep learning-based model could overcome interobserver variability.
-
•
Its accuracy for discriminating PD from normal was comparable to the clinical standard.
-
•
It also showed high accuracy for differentiating PD from nonparkinsonian tremor.
-
•
Clinical follow-up results showed SWEDD could be reclassified to PD by our model.
1. Introduction
Dopamine transporter (DAT) imaging such as 123I-fluoropropyl-carbomethoxyiodophenylnortropane (FP-CIT) single-photon emission computed tomography (SPECT) is one of the established tools for the diagnosis of Parkinson's disease (PD) (de la Fuente-Fernandez, 2012). In the clinical setting, visual analysis of FP-CIT SPECT has been routinely performed for determining whether a subject has dopaminergic degeneration. Currently, visual analysis combined with striatal DAT quantification is regarded as a standard practice in clinical studies (Albert et al., 2016). However, visual analysis is suboptimal because it causes interobserver variability (McKeith et al., 2007, Papathanasiou et al., 2012, Tondeur et al., 2010).
The main indication of FP-CIT SPECT is differentiating mild or uncertain Parkinsonism patients (Marshall and Grosset, 2003). However, because of uncertainty in PD classification and DAT imaging interpretation, atypical subgroup among PD patients has been consistently identified. It is scans without evidence of dopaminergic deficit (SWEDD). The term SWEDD refers to the absence of imaging abnormality in patients who are clinically diagnosed as PD. SWEDD patients are approximately 10–15% of clinically diagnosed PD patients (Group, P.S., 2000, Marek et al., 2014, Parkinson Study, G, 2002). There is growing evidence that the SWEDD is different from typical PD in terms of pathophysiology and prognosis (Fahn et al., 2004, Schwingenschuh et al., 2010). However, the determination of SWEDD is often inconsistent because of visual interpretation of DAT imaging which has high sensitivity (98%) but low specificity (67%) in early PD (de la Fuente-Fernandez, 2012).
In this study, we aimed to develop an automated FP-CIT SPECT interpretation system based on deep learning for the objective diagnosis. Recent development of deep learning is changing a variety of scientific and industrial fields (LeCun et al., 2015). Deep convolutional neural networks (CNN), a type of deep learning, have dramatically improved the performance in image classification and detection (Krizhevsky et al., 2012, LeCun et al., 2015). Recently, deep learning techniques have started to be applied to medical images for segmentation, lesion-detection, and disease classification (Choi and Jin, 2016, Ithapu et al., 2015, Moeskops et al., 2016, Pereira et al., 2016, Shen et al., 2015, Wong and Bressler, 2016, Zhang et al., 2015). Our objective in terms of clinical application was to discriminate PD among patients with uncertain Parkinsonism. In this study, the system was developed using Parkinson's Progression Markers Initiative (PPMI) database. It was further validated in an independent data acquired from Seoul National University Hospital (SNUH) that consists of patients with PD and nonparkinsonian tremor.
2. Materials and methods
2.1. Subjects
Data used in the preparation of this article were obtained from two different cohorts, the PPMI database (www.ppmi-info.org/data) and SNUH cohort. For up-to-date information of PPMI database on the study, visit www.ppmi-info.org. The subjects of the PPMI cohort in this study consisted of 431 patients with PD, 193 normal controls (NCs) and 77 patients with SWEDD. PD patients and NCs were divided into two datasets, training/validation set and test set, to develop the CNN and test its accuracy. Training/validation set consisted of 549 subjects (379 PD and 170 NCs). 75 subjects (52 PD and 23 NCs) were included in the PPMI test set to evaluate the accuracy of our framework. Training and test sets were randomly selected from the PPMI cohort. The two sets were divided so that the ratio between PD and NC was the same. SNUH cohort was applied as an independent test set from the training data. SNUH cohort included 82 patients initially suspected of PD who underwent FP-CIT SPECT from Mar 2014 to Sep 2016. FP-CIT SPECT scans were acquired to determine treatment plan and obtain accurate diagnosis.
Informed consents to clinical testing and neuroimaging prior to participation of the PPMI cohort were obtained, approved by the institutional review boards (IRB) of all participating institutions. The retrospective study using SNUH cohort was approved by IRB of our institute, and informed consent was waived due to the retrospective design. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Baseline diagnosis in the PPMI was made by clinical evaluation according to the UK PD Brain Bank criteria (Gibb and Lees, 1988). Patients with PD have had their clinical diagnosis for 2 years or less, and they were untreated status. In addition, according to the PPMI diagnosis criteria, PD was diagnosed if a patient also had imaging evidence for dopaminergic deficits interpreted by the PPMI imaging core. Thus, the gold standard of our further analysis was the clinical diagnosis and the results of visual imaging interpretation determined by the imaging core consensus of PPMI. Patients with SWEDD were clinically PD patients, but they had no evidence of dopaminergic deficit in the imaging. Motor ratings were clinically assessed with the revised Movement Disorder Society Unified Parkinson's Disease Rating Scale (MDS-UPDRS) part 3 at baseline.
Interpretation of FP-CIT SPECT from SNUH cohort was initially determined by concurrence of image interpretation among 3 nuclear medicine physicians. Images were visually assessed and classified into two groups, patients with preserved and reduced DAT density. To reach a consensus on the imaging interpretation, readers referred clinical symptoms, and drug response according to the clinical follow-up. Accordingly, subjects of the SNUH cohort were divided into two groups, 72 PD patients and 10 patients with nonparkinsonian tremor.
2.2. FP-CIT SPECT images
Because of different SPECT systems in different centers, PPMI used standardized imaging acquisition protocol. SPECT was performed at the screening visit. Prior to the injection of FP-CIT, subjects were pretreated with iodine solution for thyroid protection. Images were acquired within 4 ± 0.5 h after the radiotracer injection with a target dose of 111–185 MBq. SPECT data were acquired into a 128 × 128 matrix.
After the acquisition, the raw data were transferred to the PPMI imaging core and reconstructed using a hybrid ordered subset expectation maximization algorithm (Hermes Medical Solutions, Stockholm, Sweden). The subsequent processing was performed on PMOD (PMOD Technologies, Zurich, Switzerland). Attenuation correction was applied to the reconstructed data by Chang's correction. Spatial normalization into Montreal Neurological Institute (MNI) space was performed in PMOD using a template image based on a European multicenter database of healthy controls (Varrone et al., 2013). The dimension of final preprocessed images was 91 × 100 × 91, and the voxel size was 2 × 2 × 2 mm3. SPECT images of SNUH dataset were acquired by a dedicated triple-head gamma camera (TRIONIX Triad XLT 3, Trionix Research Laboratory, Inc., Twinsburg, OH, USA) with Fan-Beam collimator. Subjects were intravenously injected 185 MBq of FP-CIT 3 h before image acquisition. Images were acquired by protocols of 40 step-and-shoot for 45 s per each step. Images were reconstructed as follows: 1) 128 × 128 matrices, 2) filtered back projection, 3) Butterworth filter with high cut frequency of 0.4 and roll off degree of 5.0, 4) Chang's method for attenuation correction. Spatial normalization was performed in Statistical Parametric Mapping (SPM8, University College of London, London, UK) using an in-house template with MNI space. We checked whether normalized SPECT images using SPM8 was aligned with those of PPMI cohort normalized by PMOD. The final preprocessed images have same dimensions and voxel size with those of PPMI cohort.
2.3. Regional DAT binding ratio
Automated quantification of DAT binding ratio (BR) was performed for SPECT data as a conventional method for quantitative analysis. Each spatially normalized SPECT image was used to calculate regional BR. Mean counts of target regions were calculated. Target regions were putamen/caudate and occipital cortex. Automated anatomical labeling (AAL) template was used to segment the target regions of each SPECT image and mean counts were calculated. BR was defined as BR = (Cspecific − Cnonspecific) / Cnonspecific, where C represented mean counts of the region. Note that the counts of occipital cortex were regarded as the region of nonspecific binding.
2.4. Deep CNN architecture and training
We designed a deep CNN framework, PD Net, and the architecture is summarized in Fig. 1A. Input data were SPECT images downloaded from the PPMI database without further processing. Input values of voxels were rescaled by the range from 0 to 255, and then mean scalar value of each SPECT volume was subtracted. After this step, each 3D volume (91 × 109 × 91) was used for input argument of PD Net.
Fig. 1.
Deep convolutional neural network framework (PD Net) for interpretation of FP-CIT SPECT images. (A) A FP-CIT SPECT volume with matrix size 91 × 109 × 91 is used for an input matrix of PD Net. It consists of multiple 3-dimensional convolutional layers which learn image features from training data. Each convolutional layer is followed by ReLU activation function and max-pooling layers subsample images. The final output of PD Net has two nodes, which respectively correspond to Parkinson's disease and normal control. (B) Parameters of convolutional layers of PD Net were learned by training SPECT dataset to discriminate SPECT images of Parkinson's disease from those of normal controls. The accuracy of the classification was measured from two independent test datasets. Two expert readers interpreted same image data blinded to diagnosis. The accuracy of PD Net and the readers was compared. In addition, the classification using PD Net was tested in Parkinson's disease patients who have scans without evidence of dopaminergic deficit (SWEDD) whether PD Net interpreted those images as normal scans. (C) PPMI and SNUH cohorts were used for the PD Net training and validation. PD and NC subjects of PPMI data were randomly divided into two datasets, training/validation and test sets. SWEDD subjects of PPMI data were used for another set for testing refined diagnosis by PD Net. Another independent cohort, SNUH dataset was used for another test set for differentiating PD from nonparkinsonian tremor. For SWEDD cohorts, 2-year follow-up image and clinical diagnosis was reassessed.
Zero-padding along two dimensional axes (x- and z-axes) was applied to images have 109 × 109 × 109 matrix dimensions. The images were passed by the 3-D convolutional layer which produced 16 feature maps after the 7 × 7 × 7 convolutional filters. As a stride size of four voxels was applied, size of the feature maps was 26 × 26 × 26. After the convolutional layer, Rectified Linear Unit (ReLU) activation layer and max-pooling layer were followed. For max-pooling operation, pool size of 3 × 3 × 3 and a stride size of two voxels were applied. 3-D convolutional layers with filter size of 5 × 5 × 5 and 3 × 3 × 3 were followed. Number of filter banks of these two convolutional layers was 64 and 256, respectively. ReLU activation layers were respectively applied after these convolutional layers. Max-pooling layer was applied after the second convolutional layer. Consequently, these multiple layers produced 256 feature vectors. The 256 features were connected to two output labels (fully-connected layer), Parkinson's disease and NC. A softmax function, exponential activation function with normalized operator, was applied to discriminate two labels after the output of the fully-connected layer. The network was trained to minimize the cross entropy loss between the predicted diagnosis and the true diagnosis of the patients.
This training was conducted by stochastic gradient descent algorithm using MatConvNet deep learning library (Version 1.0-beta 20) (Vedaldi and Lenc, 2015). 90.0% of imaging data of training/validation set (494/549 subjects) were used for the training. Those 494 SPECT scans were left-right flipped for imaging data augmentation. The remaining 10.0% data of training/validation set were used for the validation which helped monitor the performance of PD Net. Therefore, the validation set was used to determine architecture and parameters including training epoch, number of nodes, layers and learning rate. PD Net was trained for 30 epochs. The momentum parameter was set to 0.9. The learning rate was initially 1 × 10− 4 and logarithmically decreased to have 1 × 10− 6 at the final epoch.
Study design.
The strategy for the image interpretation using PD Net is summarized in Fig. 1B. PD Net was trained by 90.0% of data of training/validation set and the remaining 10.0% data were used for the validation which helped find the best model of PD Net. Since model architectures and parameters could be varied during experiments, validation dataset was used for the model optimization. Validation data were randomly selected among training/validation set so that they also have same ratio of PD to NC. The performance was independently tested by two different test sets, PPMI and SNUH dataset. An overall workflow for training and testing process of PD Net and information of two cohorts for the study are summarized in Fig. 1C.
Two readers visually reviewed images of PPMI test set blinded to the diagnosis and clinical information. Images were visually labeled with ‘normal’ and ‘abnormal’ DAT binding. The accuracy of PD Net was compared with that of readers. Additionally, PD Net classification was evaluated in SWEDD group to test whether PD Net classified SWEDD patients as normal SPECT as the visual analysis did.
Accuracy test for PD Net and comparison with conventional analysis
Sensitivity, specificity, and accuracy of PD Net were calculated for the PPMI test set and those of two readers were also obtained. As a conventional approach reflecting the clinical setting, the accuracy of the overall decision results of two readers referring DAT BR quantification was also obtained. Two readers referred DAT BR of putamen and caudate and made consensus image diagnosis for each image. The results of accuracy were statistically compared with McNemar's nonparametric test. The degree of interobserver agreement between the two readers was measured by calculating Cohen's kappa-values. The output of PD Net provided scores for the probability of PD and NC. Using the scores of PD Net, receiver operating characteristic (ROC) analysis was performed. ROC curves of two readers were drawn. In addition, a ROC curve of conventional quantification method, putaminal BR, was also drawn. The area-under-curves (AUCs) were compared by a nonparametric test of DeLong for comparison of two correlated ROC curves (DeLong et al., 1988). ROC analysis was additionally performed in SNUH test set. ROC curves were drawn for the output score of PD Net and putaminal BR.
2.5. Test for SWEDD group
As defined in the term of SWEDD, SWEDD patients had normal DAT binding according to the visual interpretation consensus. SPECT images of SWEDD were evaluated by PD Net to divide those patients into two groups, ‘normal’ and ‘abnormal DAT’. Follow-up SPECT scans after 2 years for the SWEDD patients were evaluated. Among 77 subjects, 42 subjects underwent 2-year follow-up SPECT scans. In addition, clinical follow-up diagnosis was reassessed. 56 subjects were available for 2-year follow-up clinical diagnosis data. In order to compare these two groups (PD Net normal/abnormal in SWEDD patients), BR at baseline as well as at 2-year follow-up was compared using Mann-Whitney test. Two-year follow-up visual interpretation results of the two groups were statistically compared using chi-square test. In addition, follow-up clinical diagnosis after 2 years for SWEDD patients was also assessed according to the PD Net classification.
3. Results
3.1. Accuracy for the classification between PD and NC
Clinical characteristics of the subjects are summarized in Table 1. Images of the PPMI test dataset were independently interpreted by two nuclear imaging experts. The interobserver agreement measured by kappa was 0.65 ± 0.11. Nine cases among 75 test data (12.0%) were disagreed between the readers.
Table 1.
Subjects' demographics and clinical data.
| PPMI training/validation set (n = 549) |
PPMI test set (n = 75) |
PPMI SWEDD set |
SNUH test set (n = 82) |
||||
|---|---|---|---|---|---|---|---|
| Parkinson's disease (n = 379) |
Normal control (n = 170) |
Parkinson's disease (n = 52) |
Normal control (n = 23) |
SWEDD (n = 77) |
Parkinson's disease (n = 72) |
Normal control (n = 10) |
|
| Age | 61.5 ± 9.9 | 60.9 ± 11.5 | 63.0 ± 7.7 | 58.9 ± 9.2 | 60.1 ± 10.8 | 62.5 ± 11.4 | 64.9 ± 11.4 |
| Sex (M/F) | 245/134 | 112/58 | 33/19 | 16/7 | 47/30 | 38/34 | 4/6 |
| Disease duration (months) | 6.5 ± 6.5 | 6.8 ± 6.8 | 7.4 ± 8.0 | N/A | |||
| MDS-UPDRS part III | 22.1 ± 9.9 | 19.3 ± 8.8 | 14.8 ± 10.8 | N/A | |||
| Hoehn and Yahr stage | 1.6 ± 0.5 | 1.6 ± 0.5 | 1.5 ± 0.6 | 2.9 ± 0.8 | |||
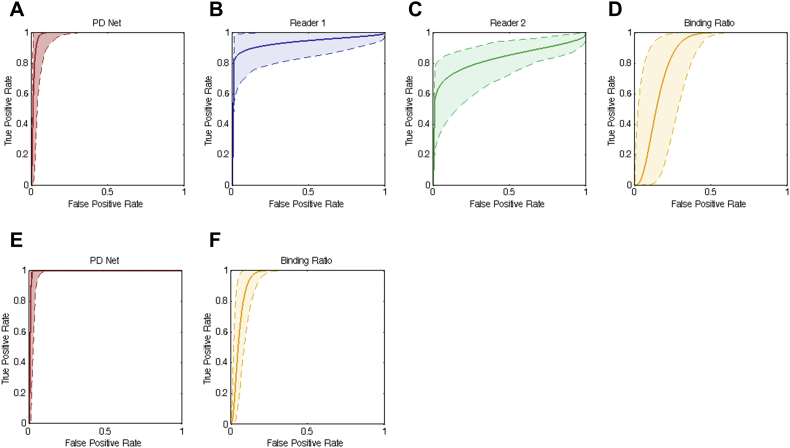
The sensitivity, specificity, and accuracy for differentiating PD from NC were evaluated (Table 2). PD Net showed 94.2% sensitivity to detect abnormal DAT which was not significantly different from the sensitivity of the two readers (98.1 and 96.2%, respectively). Specificity of PD Net was 100% and significantly higher than the two readers (73.9 and 56.5%; p = 0.030 and 0.002, respectively). Overall accuracy of PD Net was also significantly higher than that of individual readers (96.0% vs. 90.7% and 84.0%; p = 0.008 and 0.001, respectively). The accuracy of PD Net was comparable with the visual analysis referring quantitative analysis. Visual analysis combined with conventional quantification showed 96.2%, 82.6% and 92.0% for sensitivity, specificity and accuracy, respectively (Table 2). Specificity of PD Net was significantly higher than visual analysis combined with conventional quantification. The numbers of true positive, false positive, true negative and false negative were summarized in Supplementary Table 1. PD Net showed no false positive while visual analysis combined with conventional quantification showed 4 false positives. ROC curves for PD Net and the readers were drawn (Fig. 2). AUC value of PD Net was significantly higher than the individual readers as well as conventional quantification method, putaminal BR (0.988 ± 0.011, 0.860 ± 0.048, 0.763 ± 0.055 and 0.921 ± 0.034 for PD Net, reader 1, 2 and putaminal BR, respectively; p = 0.006, < 0.001 and 0.024 for PD Net vs. reader 1, vs. reader 2 and vs. putaminal BR, respectively).
Table 2.
Accuracy of PD Net and visual interpretation for discriminating Parkinson's disease from normal control (PPMI cohort) and from nonparkinsonian tremor (SNUH cohort).
| PPMI test set |
p-Value⁎ (for comparison with PD Net) |
SNUH test set |
||||||
|---|---|---|---|---|---|---|---|---|
| Rater 1 | Rater 2 | Visual + conventional quantification | PD Net | vs. Rater 1 | vs. Rater 2 | vs. visual + conventional quantification | PD Net | |
| Sensitivity | 98.1% | 96.2% | 96.2% | 94.2% | n.s. | n.s. | n.s. | 98.6% |
| Specificity | 73.9% | 56.5% | 82.6% | 100% | 0.03 | 0.002 | 0.05 | 100% |
| Accuracy | 90.7% | 84.0% | 92.0% | 96.0% | 0.008 | 0.001 | n.s. (0.06) | 98.8% |
p-Value was uncorrected for multiple comparison.
Fig. 2.
Receiver operating characteristic (ROC) curves for PD Net, human readers and conventional quantification. ROC curves are drawn for PD Net, the readers and putaminal binding ratio (BR) using PPMI test set data (Red line: PD Net, Blue line: reader 1, Green line: reader 2, Orange line: putaminal BR). Color shading shows the ROC curves of 95% CI. Area under curves were 0.988 ± 0.011, 0.860 ± 0.048, 0.763 ± 0.055 and 0.921 ± 0.034 for PD Net (A), reader 1 (B), reader 2 (C) and putaminal BR (D), respectively. ROC curves were also drawn for SNUH test set (E, F). Area under curves were 0.997 ± 0.003 and 0.968 ± 0.017 for PD Net and putaminal BR, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.2. PD Net for discriminating PD from nonparkinsonian tremor: independent test set
To validate PD Net, the performance was tested in an independent dataset acquired from SNUH. The PD Net was used to differentiate PD from patients with nonparkinsonian tremor. Accuracy of PD Net in this dataset was comparable with that in the PPMI dataset. Sensitivity, specificity and accuracy of PD Net for discriminating PD were 98.6%, 100% and 98.8%, respectively. ROC analysis revealed a trend of higher AUC value of PD Net than that of quantitative analysis using putaminal BR (0.997 ± 0.003 for PD Net and 0.968 ± 0.017 for putaminal BR; p = 0.081).
3.3. PD Net for SWEDD classification
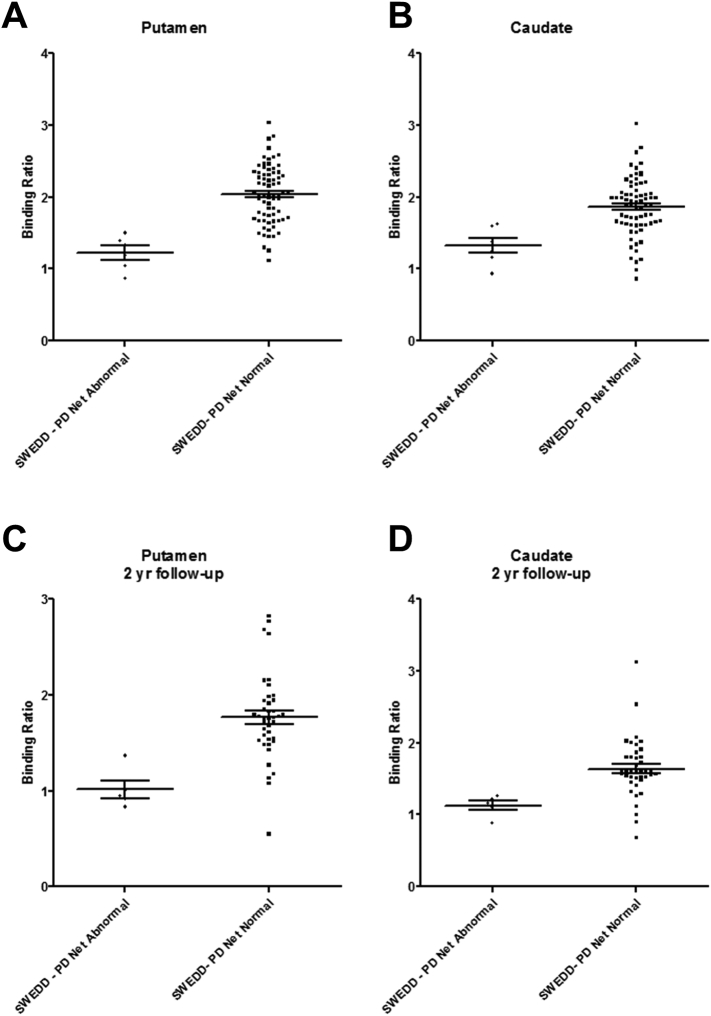
All the scans of SWEDD patients were classified as ‘normal scan’ according to the consensus of PPMI visual interpretation. Among 77 patients, 6 patients (7.8%) revealed dopaminergic deficit when PD Net analyzed the SPECT images. They showed significantly lower DAT binding ratio (BR) of putamen and caudate nuclei than SWEDD patients with normal DAT according to the PD Net analysis (Putaminal BR: 1.22 ± 0.24 vs. 2.03 ± 0.40; Caudate BR: 1.33 ± 0.27 vs. 1.86 ± 0.40; p = 0.0002 and 0.002, respectively) (Table 3, Fig. 3). The follow-up assessment including FP-CIT SPECT was performed at 2 years after the baseline study. Follow-up SPECT scans were also classified by the consensus of PPMI visual interpretation. 42 among 77 subjects underwent 2-year follow-up SPECT as well as baseline. The follow-up visual interpretation was changed in 80.0% (4/5) subjects of SWEDD patients who showed abnormal DAT in PD Net at baseline and underwent follow-up SPECT scans. Those 4 subjects were clinically PD according to the follow-up diagnosis and a subject who showed normal SPECT in follow-up scan was a patient with Alzheimer's dementia. On the other hand, 5.4% (2/37) subjects of SWEDD patients who showed normal DAT in PD Net and underwent follow-up SPECT became positive in 2-year follow-up. So, the conversion of imaging diagnosis into abnormal DAT in 2-year follow-up was significantly more in subjects who showed abnormal DAT in baseline PD Net than those who showed normal in PD Net (p = 0.0001, Table 3). According to the clinical follow-up diagnosis, it was revealed that 76.5% (39/51) subjects of SWEDD with normal PD Net had nonparkinsonian tremor including essential tremor and psychogenic illness. Only 23.5% (12/51) of them were still clinically PD in follow-up exams. Among 12 clinical PD, 9 subjects had 2-year follow-up SPECT. 2 subjects were abnormal DAT in 2-year follow-up scan while seven subjects showed still normal DAT in the follow-up (Table 3).
Table 3.
Reclassification of SWEDD patients according to the results of PD Net.
| SWEDD patients | Baseline SPECT (n = 77) |
2-years follow-up SPECT (n = 42) |
Clinical follow-up diagnosis (n = 56) | ||||
|---|---|---|---|---|---|---|---|
| n | Putamen BR | Caudate BR | PPMI visual consensus (abnormal:normal) | Putamen BR | Caudate BR | ||
| PPMI visual consensus normal/PD Net abnormal | 6 | 1.22 ± 0.24 | 1.33 ± 0.27 | 4:1 (80.0%)a | 1.01 ± 0.21 | 1.12 ± 0.15 | 4: PD 1: Alzheimer's dementia |
| PPMI visual consensus normal/PD Net normal | 71 | 2.03 ± 0.40 | 1.86 ± 0.40 | 2:35 (5.4%)b | 1.77 ± 0.45 | 1.64 ± 0.41 | 12: PDc 10: Essential tremor 10: No neurologic disease 6: Psychogenic illness 13: Other types of nonparkinsonian tremor |
| p-Value | 0.0002 | 0.002 | 0.0001 | 0.001 | 0.002 | ||
No follow-up imaging and clinical diagnosis data for a subject.
No follow-up imaging data for 34 subjects and no follow-up clinical diagnosis data for 20 subjects.
Among 12 subjects, two showed abnormal follow-up SPECT and 7 subjects showed normal follow-up scan, still clinically SWEDD. The others did not undergo 2-years follow-up SPECT.
Fig. 3.
Binding ratio (BR) of SWEDD patients according to the PD Net classification. Baseline putaminal (A) and caudate (B) BR of SWEDD patients who showed decreased dopamine transporter (DAT) in PD Net analysis were significantly lower than those of SWEDD patients who showed normal DAT in PD Net (Putaminal BR: 1.22 ± 0.24 vs. 2.03 ± 0.40; Caudate BR: 1.33 ± 0.27 vs. 1.86 ± 0.40; p = 0.0002 and 0.002, respectively). BR was calculated in 2-years follow-up scans (C, D). Follow-up putaminal (C) and caudate (D) BR were also significantly lower in SWEDD patients with baseline abnormal PD Net than SWEDD patients with baseline normal PD Net (Putaminal BR: 1.01 ± 0.21 vs. 1.77 ± 0.45; Caudate BR: 1.12 ± 0.15 vs. 1.64 ± 0.41; p = 0.001 and 0.002, respectively).
Additionally, DAT BR of putamen and caudate nuclei in 2-year follow-up scans also showed significant difference between two groups (Putaminal BR: 1.01 ± 0.21 vs. 1.77 ± 0.45; Caudate BR: 1.12 ± 0.15 vs. 1.64 ± 0.41; p = 0.001 and 0.002, respectively) (Fig. 3). Representative cases of refined SWEDD diagnosis were presented in Fig. 4. Though two baseline SPECT images were classified as normal according to the visual interpretation consensus, the PD Net classified one as normal and the other as abnormal. The subject who had abnormal DAT on PD Net analysis showed abnormal DAT at 2-year follow-up even in visual analysis, while the other subject with normal DAT on PD Net analysis stayed normal in the follow-up scan.
Fig. 4.
Refining SWEDD classification using PD Net analysis. Representative two cases show different image diagnosis analyzed by PD Net. Two subjects had normal DAT according to the visual interpretation consensus, while PD Net revealed that a subject (above) had reduced DAT in the striatum. The 2-years follow-up SPECT of the subject was abnormal according to the visual interpretation consensus. However, a SWEDD subject (below) who also showed normal DAT in PD Net persistently has normal DAT in the follow-up scan.
4. Discussion
We showed that the deep learning-based FP-CIT SPECT interpretation system could accurately and objectively determine dopaminergic degeneration and refine diagnosis. The accuracy of PD Net was comparable with experts' reading referring conventional quantitative analysis which has been regarded as a clinical standard. Our approach was validated in the independent SNUH SPECT dataset for discriminating PD from nonparkinsonian tremor patients. In addition, some of SWEDD patients, a heterogeneous group that could be inconsistently classified according to the clinical studies, had dopaminergic degeneration in PD Net analysis. It was revealed that those patients eventually had dopaminergic degeneration in follow-up study, which implied they could be initially misclassified as SWEDD. As complicated image feature selection was not required and provided objective classification of SPECT images, PD Net was practical to use in the clinical setting.
The main advantage of PD Net is in its objectiveness and high accuracy. It could overcome interobserver variability of visual interpretation which has been routinely performed in FP-CIT SPECT analysis (McKeith et al., 2007, Papathanasiou et al., 2012, Tondeur et al., 2010). In our study, Cohen's kappa of two independent readers was 0.65 ± 0.11, and the interpretation of 12.0% cases of the test dataset was disagreed. Such interobserver variability in image interpretation could affect treatment plan as well as clinical diagnosis. Moreover, overall accuracy of PD Net for discriminating PD was significantly higher than that of conventional quantification method, putaminal BR, as well as visual interpretation. Accuracy of PD Net was comparable with a clinical standard of image diagnosis made by multiple experts' reading referring quantification results (Albert et al., 2016). Moreover, specificity of PD Net was significantly higher than this conventional analysis method. Because of its high accuracy and objective results, PD Net could have clinical impacts on the diagnosis of PD.
In the clinical setting, FP-CIT SPECT is mainly performed to discriminate neurodegenerative Parkinsonism from nonparkinsonian tremor. On the other hand, PD Net was trained to discriminate between PD patients and controls. It is not regarded as a common clinical indication for FP-CIT SPECT because initial diagnosis of PD was made by clinical examination (Marshall and Grosset, 2003). In our study, using SNUH dataset, we showed that PD Net could differentiate PD from nonparkinsonian tremor with high accuracy. It suggested a feasibility of application of PD Net to differentiating neurodegenerative Parkinsonism from clinically ambiguous patients. Nevertheless, this test dataset was retrospectively collected and hardly reflected the performance for patients with mild or uncertain Parkinsonism. Therefore, further prospective study of the application of PD Net to validating clinical usefulness for patients with uncertain Parkinsonism will be needed.
In addition, our approach could be used to refine diagnostic subgroups in clinical trials by objective identification of SWEDD participants. According to the result, PD Net identified abnormal DAT in 6 (7.8%) SWEDD patients and most of them (80.0%) eventually showed abnormal DAT in longitudinal follow-up visual interpretation. However, only two subjects among SWEDD patients who were also baseline normal DAT in PD Net analysis were changed to abnormal DAT in the follow-up interpretation . SWEDD patients are different from PD as previous studies showed poor responsiveness to levodopa, first-line drug for the management of PD (Fahn et al., 2004). Of note, DAT of SWEDD patients is mostly remained normal in long-term follow-up (Marek et al., 2005, Marek et al., 2014). SWEDD patients who had normal DAT in PD Net analysis mostly remained normal DAT after 2 years (94.6%). It suggested SWEDD patients who showed abnormal DAT in PD Net might be resulted from misclassification. Furthermore, DAT BR of SWEDD patients who showed abnormal DAT in PD Net was significantly lower than that of SWEDD patients who showed normal DAT. Our results also imply that some patients with PD could be misclassified as SWEDD in several clinical trials as the imaging diagnosis has been made by visual interpretation. A recent retrospective study also showed that a large proportion of SWEDD population was due to SPECT misinterpretation (Nicastro et al., 2016). Moreover, a systematic review related to SWEDD revealed that SWEDD patients were heterogeneous and mostly due to a clinical misdiagnosis of PD (Erro et al., 2016). Our results corresponded to this review as most (76.5%) SWEDD patients with normal PD Net result had nonparkinsonian tremor in long term follow-up. This misclassification issue might influence the result of therapeutic interventions in clinical trials. An important advance in the application of PD Net to clinical studies could be an objective identification of dopaminergic degeneration, which results in refining subgroup classification of PD patients, particularly for SWEDD group.
According to the PD Net analysis, three PD subjects of PPMI data were misclassified as normal. Among them, two subjects were also misclassified by experts' reading referring conventional quantitative analysis. They were relatively early PD (UPDRS part 3 score was 8 and 17). For another misclassified subject, the output score of PD Net was 0.42. This value was the highest in NCs. It suggests that decision criteria using a different threshold value for the PD Net output score could improve the diagnosis for clinical settings. Thus, we also used ROC analysis, which revealed that AUC of PD Net was higher than conventional methods.
PD Net is superior to other automated methods in terms of ease of application and performance. Recently, other machine learning methods using quantitative parameters of FP-CIT SPECT combined with or without clinical factors showed good accuracy (90–96%) for the diagnosis of PD (Huertas-Fernandez et al., 2015, Illan et al., 2012, Prashanth et al., 2014). Though these methods also showed high accuracy, there are limitations in generalization and clinical implementation. They used imaging features such as striatal BR rather than images themselves. The feature selection procedures are not standardized as quantification of striatal BR could be affected by image processing steps such as normalization and selection of nonspecific regions (Brahim et al., 2015, Tossici-Bolt et al., 2006). PD Net directly analyzed all input voxels and automatically found patterns of them, which resulted in high accuracy without striatal BR calculation. Moreover, generalized application of PD Net was validated by an independent cohort of SNUH.
In spite of high accuracy of PD Net, it was tested by discriminating PD patients from NCs. In the clinical setting, FP-CIT SPECT scans were acquired for patients with atypical Parkinsonism and SWEDD as well as PD. Thus, the accuracy of PD Net did not reflect the patient characteristics in the clinic and could be overestimated. In addition, PD Net training relied on the gold standard diagnosis of PPMI cohort, which was the clinical diagnosis combined with the visual imaging interpretation instead of pure clinical diagnosis independent from the image interpretation. Nonetheless, the strength of PD Net is less interobserver variability which could provide consistent interpretation results. Because of this strength, it can be used in clinical trials which require objective biomarkers. As another limitation in the study design, PD Net ignored patients' characteristics such as age. Because DAT is influenced by aging (Pirker et al., 2000), PD Net could not differentiate age-related degeneration from PD-related degeneration. In the future, modified designs of deep neural network which considers clinical variables could improve diagnostic performance in the clinical setting. The independent test set, SNUH test set, includes relatively small number of patients with nonparkinsonian tremor. Furthermore, the gold standard diagnosis for SNUH test set was made by visual interpretation results considering clinical information. Therefore, PD Net should be validated in a larger prospective cohort that includes patients with several movement disorders and uncertain Parkinsonism with clinically follow-up diagnosis.
5. Conclusion
We designed a deep CNN model, PD Net, for FP-CIT SPECT interpretation. Its accuracy for discriminating PD from NCs was comparable to that of the clinical standard, experts' visual interpretation combined with quantification. Our approach was also validated for discriminating PD from nonparkinsonian tremor using independent SPECT data. As an automated system, it could overcome interobserver variability which might result in misclassification of subject groups. Accordingly, a promising application of PD Net will be an objective diagnosis for patients with clinically uncertain Parkinsonism who showed ambiguous FP-CIT SPECT results. Furthermore, it will apply to reclassification of SWEDD group. In the future, the application will be extended to imaging interpretation in various diseases and development of imaging biomarkers.
The following is the supplementary data related to this article.
Number of true positive, false positive, true negative and false negative for the PPMI test set.
Acknowledgments
Acknowledgments
Data used in the preparation of this article were obtained from the Parkinson's Progression Markers Initiative database (www.ppmi-info.org/data). For up-to-date information on the study, visit www.ppmi-info.org. PPMI – a public-private partnership (http://www.ppmi-info.org/) – is funded by the Michael J. Fox Foundation for Parkinson's Research and funding partners, including Abbvie, Avid Radiopharmaceuticals, Biogen Idec, Bristol-Myers Squibb, Covance, Eli Lilly & Co, F Hoff man-La Roche, GE Healthcare, Genentech, GlaxoSmithKline, Lundbeck, Merck, MesoScale, Piramal, Pfizer, and UCB. This research was supported by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (HI14C0466), and funded by the Ministry of Health & Welfare, Republic of Korea (HI14C3344), and funded by the Ministry of Health & Welfare, Republic of Korea (HI14C1277), and the Technology Innovation Program (10052749), and supported by the National Research Foundation of Korea (NRF) Grant funded by the Korean Government (MSIP) (2017M3C7A1048079). This study was also supported by the Korea Institute of Planning & Evaluation for Technology in Food, Agriculture, Forestry, and Fisheries, Republic of Korea (311011-05-3-SB020) by the Korea Healthcare Technology R&D Project funded by Ministry of Health & Welfare, Republic of Korea (HI11C21100200) and by the Technology Innovation Program (10050154, Business Model Development for Personalized Medicine Based on Integrated Genome and Clinical Information) funded by the Ministry of Trade, Industry & Energy (MI, Korea) and by the Bio & Medical Technology Development Program of the NRF funded by the Korean government, MSIP (2015M3C7A1028926) and by the National Research Foundation of Korea Grant Funded by the Ministry of Science and ICT (NRF-2017M3C7A1047392).
Conflict of interest
None.
Author contributions
D.S.L. and S.H.P. designed the study. H.C. and S.H. analyzed the data and designed the framework. H.J.I. contributed to image processing. S.H.P. performed clinical study for SNUH cohort. H.C., S.H. and D.S.L. wrote this manuscript mainly and all other authors also wrote down their own part of this manuscript according to their own specialties.
Contributor Information
Sun Ha Paek, Email: paeksh@snu.ac.kr.
Dong Soo Lee, Email: dsl@plaza.snu.ac.kr.
References
- Albert N.L., Unterrainer M., Diemling M., Xiong G., Bartenstein P., Koch W., Varrone A., Dickson J.C., Tossici-Bolt L., Sera T., Asenbaum S., Booij J., Kapucu L.O., Kluge A., Ziebell M., Darcourt J., Nobili F., Pagani M., Sabri O., Hesse S., Borght T.V., Van Laere K., Tatsch K., la Fougere C. Implementation of the European multicentre database of healthy controls for [(123)I]FP-CIT SPECT increases diagnostic accuracy in patients with clinically uncertain parkinsonian syndromes. Eur. J. Nucl. Med. Mol. Imaging. 2016;43:1315–1322. doi: 10.1007/s00259-015-3304-2. [DOI] [PubMed] [Google Scholar]
- Brahim A., Ramirez J., Gorriz J.M., Khedher L., Salas-Gonzalez D. Comparison between different intensity normalization methods in 123I-Ioflupane imaging for the automatic detection of parkinsonism. PLoS One. 2015;10 doi: 10.1371/journal.pone.0130274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H., Jin K.H. Fast and robust segmentation of the striatum using deep convolutional neural networks. J. Neurosci. Methods. 2016;274:146–153. doi: 10.1016/j.jneumeth.2016.10.007. [DOI] [PubMed] [Google Scholar]
- DeLong E.R., DeLong D.M., Clarke-Pearson D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- Erro R., Schneider S.A., Stamelou M., Quinn N.P., Bhatia K.P. What do patients with scans without evidence of dopaminergic deficit (SWEDD) have? New evidence and continuing controversies. J. Neurol. Neurosurg. Psychiatry. 2016;87:319–323. doi: 10.1136/jnnp-2014-310256. [DOI] [PubMed] [Google Scholar]
- Fahn S., Oakes D., Shoulson I., Kieburtz K., Rudolph A., Lang A., Olanow C.W., Tanner C., Marek K., Parkinson Study, G Levodopa and the progression of Parkinson's disease. N. Engl. J. Med. 2004;351:2498–2508. doi: 10.1056/NEJMoa033447. [DOI] [PubMed] [Google Scholar]
- de la Fuente-Fernandez R. Role of DaTSCAN and clinical diagnosis in Parkinson disease. Neurology. 2012;78:696–701. doi: 10.1212/WNL.0b013e318248e520. [DOI] [PubMed] [Google Scholar]
- Gibb W.R., Lees A.J. The relevance of the Lewy body to the pathogenesis of idiopathic Parkinson's disease. J. Neurol. Neurosurg. Psychiatry. 1988;51:745–752. doi: 10.1136/jnnp.51.6.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Group, P.S. A randomized controlled trial comparing pramipexole with levodopa in early Parkinson's disease: design and methods of the CALM-PD Study. Parkinson Study Group. Clin. Neuropharmacol. 2000;23:34–44. doi: 10.1097/00002826-200001000-00007. [DOI] [PubMed] [Google Scholar]
- Huertas-Fernandez I., Garcia-Gomez F.J., Garcia-Solis D., Benitez-Rivero S., Marin-Oyaga V.A., Jesus S., Caceres-Redondo M.T., Lojo J.A., Martin-Rodriguez J.F., Carrillo F., Mir P. Machine learning models for the differential diagnosis of vascular parkinsonism and Parkinson's disease using [(123)I]FP-CIT SPECT. Eur. J. Nucl. Med. Mol. Imaging. 2015;42:112–119. doi: 10.1007/s00259-014-2882-8. [DOI] [PubMed] [Google Scholar]
- Illan I.A., Gorrz J.M., Ramirez J., Segovia F., Jimenez-Hoyuela J.M., Ortega Lozano S.J. Automatic assistance to Parkinson's disease diagnosis in DaTSCAN SPECT imaging. Med. Phys. 2012;39:5971–5980. doi: 10.1118/1.4742055. [DOI] [PubMed] [Google Scholar]
- Ithapu V.K., Singh V., Okonkwo O.C., Chappell R.J., Dowling N.M., Johnson S.C., Alzheimer's Disease Neuroimaging I. Imaging-based enrichment criteria using deep learning algorithms for efficient clinical trials in mild cognitive impairment. Alzheimers Dement. 2015;11:1489–1499. doi: 10.1016/j.jalz.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizhevsky A., Sutskever I., Hinton G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Proces. Syst. 2012:1097–1105. [Google Scholar]
- LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- Marek K., Jennings D., Seibyl J. Long-term follow-up of patients with scans without evidence of dopaminergic deficit (SWEDD) in the ELLDOPA study. Neurology. 2005 (A274-A274) [Google Scholar]
- Marek K., Seibyl J., Eberly S., Oakes D., Shoulson I., Lang A.E., Hyson C., Jennings D., Parkinson Study Group, P.I Longitudinal follow-up of SWEDD subjects in the PRECEPT Study. Neurology. 2014;82:1791–1797. doi: 10.1212/WNL.0000000000000424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall V., Grosset D. Role of dopamine transporter imaging in routine clinical practice. Mov. Disord. 2003;18:1415–1423. doi: 10.1002/mds.10592. [DOI] [PubMed] [Google Scholar]
- McKeith I., O'Brien J., Walker Z., Tatsch K., Booij J., Darcourt J., Padovani A., Giubbini R., Bonuccelli U., Volterrani D., Holmes C., Kemp P., Tabet N., Meyer I., Reininger C., Group, D.L.B.S. Sensitivity and specificity of dopamine transporter imaging with 123I-FP-CIT SPECT in dementia with Lewy bodies: a phase III, multicentre study. Lancet Neurol. 2007;6:305–313. doi: 10.1016/S1474-4422(07)70057-1. [DOI] [PubMed] [Google Scholar]
- Moeskops P., Viergever M.A., Mendrik A.M., de Vries L.S., Benders M.J., Isgum I. Automatic segmentation of MR brain images with a convolutional neural network. IEEE Trans. Med. Imaging. 2016 doi: 10.1109/TMI.2016.2548501. [DOI] [PubMed] [Google Scholar]
- Nicastro N., Garibotto V., Badoud S., Burkhard P.R. Scan without evidence of dopaminergic deficit: a 10-year retrospective study. Parkinsonism Relat. Disord. 2016;31:53–58. doi: 10.1016/j.parkreldis.2016.07.002. [DOI] [PubMed] [Google Scholar]
- Papathanasiou N., Rondogianni P., Chroni P., Themistocleous M., Boviatsis E., Pedeli X., Sakas D., Datseris I. Interobserver variability, and visual and quantitative parameters of (123)I-FP-CIT SPECT (DaTSCAN) studies. Ann. Nucl. Med. 2012;26:234–240. doi: 10.1007/s12149-011-0564-1. [DOI] [PubMed] [Google Scholar]
- Parkinson Study, G Dopamine transporter brain imaging to assess the effects of pramipexole vs levodopa on Parkinson disease progression. JAMA. 2002;287:1653–1661. doi: 10.1001/jama.287.13.1653. [DOI] [PubMed] [Google Scholar]
- Pereira S., Pinto A., Alves V., Silva C.A. Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Trans. Med. Imaging. 2016 doi: 10.1109/TMI.2016.2538465. [DOI] [PubMed] [Google Scholar]
- Pirker W., Asenbaum S., Hauk M., Kandlhofer S., Tauscher J., Willeit M., Neumeister A., Praschak-Rieder N., Angelberger P., Brucke T. Imaging serotonin and dopamine transporters with 123I-beta-CIT SPECT: binding kinetics and effects of normal aging. J. Nucl. Med. 2000;41:36–44. [PubMed] [Google Scholar]
- Prashanth R., Roy S.D., Mandal P.K., Ghosh S. Automatic classification and prediction models for early Parkinson's disease diagnosis from SPECT imaging. Expert Syst. Appl. 2014;41:3333–3342. [Google Scholar]
- Schwingenschuh P., Ruge D., Edwards M.J., Terranova C., Katschnig P., Carrillo F., Silveira-Moriyama L., Schneider S.A., Kagi G., Palomar F.J., Talelli P., Dickson J., Lees A.J., Quinn N., Mir P., Rothwell J.C., Bhatia K.P. Distinguishing SWEDDs patients with asymmetric resting tremor from Parkinson's disease: a clinical and electrophysiological study. Mov. Disord. 2010;25:560–569. doi: 10.1002/mds.23019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W., Zhou M., Yang F., Yang C., Tian J. Multi-scale convolutional neural networks for lung nodule classification. Inf. Process. Med. Imaging. 2015;24:588–599. doi: 10.1007/978-3-319-19992-4_46. [DOI] [PubMed] [Google Scholar]
- Tondeur M.C., Hambye A.S., Dethy S., Ham H.R. Interobserver reproducibility of the interpretation of I-123 FP-CIT single-photon emission computed tomography. Nucl. Med. Commun. 2010;31:717–725. doi: 10.1097/mnm.0b013e32833b7ea4. [DOI] [PubMed] [Google Scholar]
- Tossici-Bolt L., Hoffmann S.M., Kemp P.M., Mehta R.L., Fleming J.S. Quantification of [123I]FP-CIT SPECT brain images: an accurate technique for measurement of the specific binding ratio. Eur. J. Nucl. Med. Mol. Imaging. 2006;33:1491–1499. doi: 10.1007/s00259-006-0155-x. [DOI] [PubMed] [Google Scholar]
- Varrone A., Dickson J.C., Tossici-Bolt L., Sera T., Asenbaum S., Booij J., Kapucu O.L., Kluge A., Knudsen G.M., Koulibaly P.M., Nobili F., Pagani M., Sabri O., Vander Borght T., Van Laere K., Tatsch K. European multicentre database of healthy controls for [123I]FP-CIT SPECT (ENC-DAT): age-related effects, gender differences and evaluation of different methods of analysis. Eur. J. Nucl. Med. Mol. Imaging. 2013;40:213–227. doi: 10.1007/s00259-012-2276-8. [DOI] [PubMed] [Google Scholar]
- Vedaldi A., Lenc K. Proceedings of the 23rd Annual ACM Conference on Multimedia Conference. ACM. 2015. MatConvNet: convolutional neural networks for matlab; pp. 689–692. [Google Scholar]
- Wong T.Y., Bressler N.M. Artificial intelligence with deep learning technology looks into diabetic retinopathy screening. JAMA. 2016;316:2366–2367. doi: 10.1001/jama.2016.17563. [DOI] [PubMed] [Google Scholar]
- Zhang W., Li R., Deng H., Wang L., Lin W., Ji S., Shen D. Deep convolutional neural networks for multi-modality isointense infant brain image segmentation. NeuroImage. 2015;108:214–224. doi: 10.1016/j.neuroimage.2014.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Number of true positive, false positive, true negative and false negative for the PPMI test set.