Abstract
Background
Better understanding and prediction of PD progression could improve disease management and clinical trial design. We aimed to use longitudinal clinical, molecular, and genetic data to develop predictive models, compare potential biomarkers, and identify novel predictors for motor progression in PD. We also sought to assess the use of these models in the design of treatment trials in PD.
Methods
A Bayesian multivariate predictive inference platform was applied to data from the Parkinson’s Progression Markers Initiative (PPMI) study (NCT01141023). We used genetic data and baseline molecular and clinical variables from PD patients and healthy controls to construct an ensemble of models to predict the annualised rate of the Movement Disorder Society-Unified Parkinson’s Disease Rating Scale parts II and III combined. We tested our overall explanatory power, as assessed by the coefficient of determination (R2), and replicated novel findings in an independent clinical cohort of PD patients from the Longitudinal and Biomarker Study in PD (LABS-PD; NCT00605163). The potential utility of these models for clinical trial design was quantified by comparing simulated randomized placebo-controlled trials within the out-of sample LABS-PD cohort.
Findings
A total of 117 controls and 312 PD cases were available for analysis. Our model ensemble exhibited strong performance in-cohort (5-fold cross-validated R2=41%, 95% CI: 35% – 47%) and significant, though reduced, performance out-of-cohort (R2=9%, 95% CI: 4% – 16%). Individual predictive features identified from PPMI data were confirmed in the LABS-PD cohort of 317 PD patients. These included significant replication of higher baseline motor score, male sex, and increased age, as well as a novel PD-specific epistatic interaction all indicative of faster motor progression. Genetic variation was the most useful predictive marker of motor progression (2.9%, 95%CI: 1.5–4.3%). CSF biomarkers at baseline showed a more modest (0.3%; 95%CI: 0.1–0.5%), but still significant effect on motor progression prediction. The simulations (n=5000) showed that incorporating the predicted rates of motor progression into the final models of treatment effect reduced the variability in the study outcome allowing significant differences to be detected at sample sizes up to 20% smaller than in naïve trials.
Interpretation
Our model ensemble confirmed established and identified novel predictors of PD motor progression. Improving existing prognostic models through machine learning approaches should benefit trial design and evaluation, as well as clinical disease monitoring and treatment.
Funding
Michael J. Fox Foundation for Parkinson’s Research and National Institute of Neurological Disorders and Stroke (1P20NS092529-01).
Introduction
Parkinson’s disease (PD) is a chronic, debilitating neurodegenerative disorder characterized clinically by progressive motor dysfunction and various non-motor features 1. There is substantial heterogeneity in the presentation of these symptoms and the rates of their progression among PD patients, making it difficult for care-providers to give accurate prognoses to patients and challenging for researchers to develop drugs to modify the course of disease 2. Increasing evidence supports a complex interplay between genetic, biological, and molecular abnormalities of the disease explaining this heterogeneity between patients. Understanding the etiologic and physiological factors that contribute to this variability in the evolution of PD symptoms is therefore a high priority area of PD research 2. The Parkinson’s Progression Markers Initiative (PPMI) study was initiated with the support of the Michael J. Fox Foundation for Parkinson’s Research to address this research gap by providing a uniquely comprehensive set of longitudinal clinical, imaging, and bio-sample data from de novo PD patients and controls.
Previous analyses of PD progression data have generally focused on investigating the individual associations of predictive features such as age, sex, baseline scores, clinical subtypes and varied potential biomarkers, as opposed to developing comprehensive multivariable prognostic models 3–5. There have been exceptions, however, including logistic regression and Bayesian classification models to predict cognitive impairment in PD6,7, backwards selection models to predict negative outcomes (e.g. postural instability, dementia or death)8, and machine-learning random survival forests to predict time to initiation of symptomatic treatment9. However, models that predict the rate of change of motor scores assessed using the Movement Disorder Society-Unified Parkinson’s Disease Rating Scale (MDS-UPDRS), instead of specific clinical events, remain elusive, despite that changes in this rating scale score and its subcomponents serve as the primary endpoint in many clinical trials of PD medications 10. Here, we used a novel Bayesian machine-learning platform, Reverse Engineering and Forward Simulation (REFSTM), with the PPMI compendia to identify an ensemble of predictive models, instead of a single best model, of the rate of motor progression.
This analysis is differentiated from previous studies not only in the outcome studied, but also in the scope and complexity of the predictive relationships that are explored by examining the entire complement of available genetic, molecular and clinical variables11–14. In particular, REFS allows for the detection of higher order interactions across these different data types and, consequently, identification of subpopulation-specific effects. This allows the crucial distinction between modifiers that are specific to PD cases from those that are more generally related to natural aging or other risk factors.
Taken together, these features permit three complementary objectives for this study. (1) The construction of clinically useful predictive models to identify PD patients at risk of rapid disease motor progression. (2) A comparison of the predictive utility of different types of potential biomarkers of motor progression. (3) The identification of novel progression markers for subsequent validation in an independent test sample (LABS-PD, N=317), which, notably, uncovered a significant epistatic genetic association to disease progression upon replication. We also demonstrate the utility of these models in designing PD clinical trials that aim to test potential disease-modifying therapies.
Materials and Methods
Study Population
Data and study documentation used in the preparation of this article were obtained from the PPMI database (www.ppmi-info.org/data) through the PPMI@LONI data portal December 28th 2015, and all numbers reported in this document are current as of that date. Data collection procedures have been documented previously15 and are described further in supplemental methods.
Outcome Modeling
The MDS-UPDRS (and the original UPDRS) has been widely used for assessing the severity of motor and non-motor symptoms in PD patients, and extensively tested for its clinimetric properties16. In addition, it is responsive to therapeutic interventions, making it the standard scale for regulatory agencies. Here, the outcome studied is the annualized rate of change in the combined MDS-UPDRS Parts II (Motor Experiences of Daily Living) & III (Motor Examination) scores. The combination of UPDRS Parts II and III is a commonly used endpoint in PD clinical trials testing interventions to improve motor function17–19, is highly correlated with Hoehn & Yahr stage, and appears to be the most reliable and responsive prognostic measures of disease activity at baseline and of disease progression in early PD20. Furthermore, the motor components of the scale are convertible when measured using the UPDRS or MDS-UPDRS, whereas transformation of Parts I and IV are not possible16.
Rates of progression were estimated for each patient with at least two years of follow-up, including at minimum 3 MDS-UPDRS exams. Because treatment initiation may have a profound, if variable, effect on disease course, progression rates were estimated separately for treated and untreated periods. Individual untreated and treated progression rates were estimated from a linear mixed-effects model of the assessment scores using the R package lme421.
Genotyping
To avoid over-fitting and reduce collinearity among predictors, the ImmunoChip SNP set used for the study was further pruned (after QC described supplemental methods) based on a priori knowledge and linkage disequilibrium. 53 SNPs with established relationships to PD or PD-related traits were identified from the NHGRI GWAS catalog 22 and DisGenNet 23 and retained as potential predictors (Supplemental Table S2.a). Linkage disequilibrium (LD) pruning was applied to the remaining SNPs and identified a set of 17,403 uncorrelated SNPs. A set of 10 principal components was identified from the SNP data for evaluation of population associations.
Statistical Analysis and REFS™ Modeling Approach
Modeling was conducted using GNS Healthcare’s REFS™ platform. The predictive ensemble, consisting of 128 generalized linear models, was constructed using Markov Chain Monte Carlo sampling of the full Bayesian posterior distribution of models, given the available data; i.e. P(model|data) 11–14.
When the number of potential predictors exceeds the number of observations in a study, attempts to identify a single “best” model will inevitably lead to over-fitting; thus, our approach identifies an ensemble of models, each scored by both its goodness of fit to the observed data and its complexity. This approach allows the incorporation of prior knowledge regarding the different types of data (e.g. genetic, imaging, or molecular), including the expected relative contribution of each type into the final models. Therefore, genome-wide examination of large numbers of lower-resolution genotype variables is possible without overwhelming the signal of the fewer, though potentially more directly informative, molecular markers. This approach is well suited to small sample-high dimensionality datasets such as used here, when gradient-based learning runs into problems due to the Tanner-Donoho phase boundary.24
The 128 constituent models within the ensemble explored combinations of the available parameters, including linear additive and quadratic terms, as well as up to third-order interactions to accommodate non-linear effects within different population strata (for example treated versus untreated). To prevent over-fitting, the complexity (i.e. the number of terms) for each model in the ensemble was penalized by specifying a maximum entropy prior over the number of unique predictors selected per given variable class (see Supplemental methods for additional detail).
Modeling Results, Predictive Performance, and Variable Importance
The prospective predictive performance for each ensemble was estimated using internal five-fold cross-validation of PPMI samples, using Pearson R2 (predicted vs. observed progression rates) and root mean squared errors (Supplemental table S6). Stratified R2 values were calculated according to disease and treatment status and by follow-up time to assess the model performance across different subpopulations.
The composition of the ensembles is summarized by counting the number of times each predictor is selected into one of the 128 constituent models. A high selection frequency for a given predictor represents an increased probability of a true predictive association with the outcome 13. High confidence predictors, defined as those appearing in >5% in the constituent models of an ensemble, were evaluated to assess their relative importance towards prediction of the outcome through leave-one-out cross-validation (supplemental methods). Variable importance was calculated by determining the percent increase in mean squared error (MSE) of the predictions excluding the variable of interest over the MSE of the full ensemble, calculated from the leave-one-out predictions. Confidence intervals for variable importance were derived from a paired t-test comparing the MSE from the reduced and full ensembles.
Independent Model Validation and Variable Replication in LABS-PD
The predictive ensemble developed from the PPMI dataset was independently validated in the Longitudinal and Biomarker Study in PD (LABS-PD), which includes demographic, clinical, genetic, and dopamine transporter (DAT) imaging data in a cohort of 380 de novo PD subjects followed over 7 years25. 317 PD cases with clinical data and genotyping were available for the validation study. While the data available in the LABS-PD cohort allowed for validation of many key predictors, most importantly the SNP results, the unavailability of key predictors (including CSF biomarker data) and differences and data collection required imputation and harmonization of certain variables which is described in detail in the supplement (Supplemental methods, Supplemental Table S5).
Ensemble Prediction Evaluation
The predictive ensembles were applied to the assembled set of LABS-PD predictors and the Pearson R2 was calculated in the full replication sample and stratified by SWEDD and treatment status, and by follow-up time. Clinical and genetic features identified at greater than 5% ensemble frequency were tested in the LABS-PD dataset in both univariate and multivariate linear regression models (including model-specific significant clinical predictors). Novel findings were considered to significantly replicate if a one-sided test in the independent LABS-PD cohort exceeded an alpha level of 0.05 after Bonferroni correction for the number of novel features examined.
Complexity, cost, and success rates of clinical trials depend, in part, on the size of the population necessary to detect the sought-after treatment effect. The potential utility of these models for clinical trial design was quantified by comparing simulated randomized placebo-controlled trials within the out-of sample LABS-PD cohort with the MDS-UPDRS part II and III motor score as the primary outcome (Supplemental methods).
Role of the Funding Source
The study sponsors are listed in the acknowledgments section and played no role in the design, collection, analysis or interpretation of data, the writing of the manuscript or the decision to submit for publication.
Results
A total of 117 controls and 312 PD cases with complete genetic, molecular and 2+ years of longitudinal data were available for analysis of motor progression (Supplemental Table S1). As expected, the mean (±standard deviation) rate of motor progression was significantly higher (i.e. more severe) among PD cases compared to controls (5.05 ± 3.3 vs. −0.14 ± 0.64 points/year, p = < 2e-16), with modest but significant reductions in progression rates observed during treatment periods with symptomatic therapy among PD cases (Table S1).
A total of 17,499 features were included as potential predictors in the models. These included 53 a priori selected PD-related single nucleotide polymorphism (SNPs); 17,403 LD-pruned SNPs from across the genome; 7 cerebrospinal fluid (CSF) protein biomarkers; 8 DaTscan imaging variables; 10 genetic principal components; and 18 clinical and demographic variables. The included variables are described in more detail in Supplemental Tables S2.a and S2.b, and the Supplemental methods.
The final REFS ensemble is summarized as a reduced set of weighted constituent models including most of the important predictors identified from the full ensemble (N=12 weighted models; Supplemental Table S3) that optimally approximates the full ensemble of 128 total unweighted constituent models 26.
Internal and External Model Validation
The predictive ensemble explained a significant percentage of the observed variation in motor progression in 5-fold cross-validation across the full PPMI sample (Table 1). Predictive accuracy was greater among PD cases compared to controls (R2=27% vs. 1%). Within cases, a significant proportion of variability was explained by the model predictors in both patients in the earlier stage of disease (defined as < 5 years since initial PD diagnosis, [R2=29%]), and later-stage (≥ 5 years since initial PD diagnosis, [R2=19%]), and in both untreated and treated cases (R2=19% and 5%, respectively). Progression was not significantly predicted among controls, a group which had minimal variability in progression.
Table 1.
Proportion of variance explained by model in internal cross-validation (PPMI) and external validation data set (LABS-PD). The given R2 values describe the proportion of variance in the true rate of disease progression explained in a given stratum for both cohorts.
| Strata | Motor Progression | |||
|---|---|---|---|---|
| PPMI | LABS-PD | |||
| N | R2 (95% CI) | N | R2 (95% CI) | |
| All | 639 | 41% (35 – 47%) | 317 | 9% (4 – 16%) |
| PD Cases1 | 522 | 27% (21 – 34%) | 317 | 9% (4 – 16%) |
| Controls | 117 | 1% (0 – 7%) | 0 | - |
| Untreated2 | 296 | 19% (11 – 27%) | 27 | 15% (3e−5 - 45%) |
| Treated3 | 226 | 5% (1 – 12%) | 290 | 11% (5 – 18%) |
| non-SWEDD | 490 | 26% (20 – 33%) | 312 | 11% (5 – 18%) |
| SWEDD | 32 | 26% (4 – 53%) | 5 | - |
| Earlier stage4 | 421 | 29% (22 – 36%) | 15 | 0% (0 – 49%) |
| Later stage5 | 101 | 19% (7– 34%) | 302 | 10% (5 – 18%) |
Cases who contributed both treated and untreated time are included twice
progression rates in calculated for the time prior to symptomatic PD treatment
progression rates calculated for the time in which the participant was receiving symptomatic treatment.
participants with < 5 years of follow-up time since initial diagnosis of PD
participants with ≥ 5 years of follow-up time since initial diagnosis of PD
LABS-PD: Longitudinal and Biomarker Study in Parkinson’s disease; PD: Parkinson’s disease; PPMI: Parkinson’s Progression Marker Initiative; SWEDD: Scans without evidence of dopaminergic deficit
External validation of the predictive ensemble in the LABS-PD dataset overall showed reduced but significant R2 (9%; 95% CI=4–16%), despite substantial differences in the collection of progression data and the lack of data for several key predictors, including CSF biomarkers (imputed data was used, see supplemental methods for details). Evaluation of the strata-specific R2 shows significant prediction in all but the earlier-stage group (which included only 15 patients), and highest accuracy in the untreated cases.
Ensemble Summarization
Complete summarization of all identified features is provided in supplemental table S4. The ensemble models were primarily composed of expected and previously established markers of disease progression. Higher baseline motor score and PD status were predictive of faster rates of motor progression in all of the ensemble models, either as a main effect or in interaction terms, while SWEDD status (Scans without evidence of dopaminergic deficit) and presence of PD treatment suggested slower progression in all models (Table S4). Sex (defined as genetically confirmed sex, which was consistent with gender for all participants) was also an important predictive feature, appearing in >90% of the ensemble models (either by itself or interacting with PD status), with women progressing more slowly then men. Interactions were also observed, with many features having varied effects on progression between PD cases and controls (Table S4). An interaction between SWEDD status and the use of PD treatments was observed in 100% of the models. As described above, SWEDD cases and PD patients receiving treatment, independently showed slower disease progression (−3.1 points/year and −3.2 points/year respectively). However, when SWEDD patients received treatment, they demonstrated 8.3 points faster progression of motor symptoms compared to the rest of the study sample, suggesting detrimental effects of dopaminergic treatments in this population.
Genetic variants were also selected by the models, but with lower frequency than the clinical features mentioned (Table S4). The most frequently observed genetic signals, rs17710829 and rs9298897, appearing in 11.7% and 5.4% of models, respectively, demonstrated a novel PD case-only epistatic interaction in 4% of the models, with the combination of rare alleles leading to faster disease progression.
Comparison of Variable Importance of Potential Disease Markers
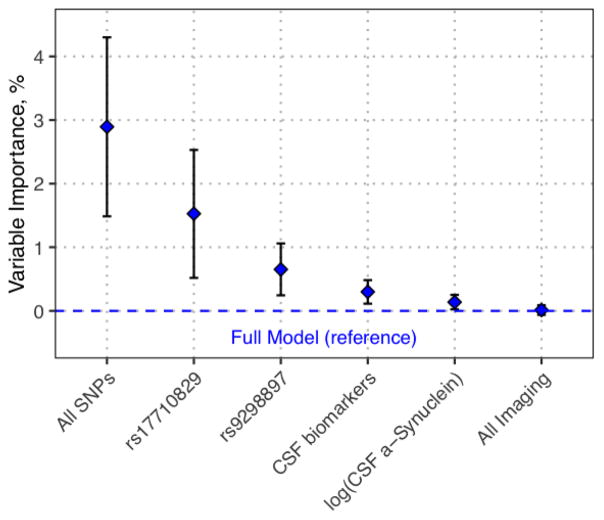
Shown in Figure 1, the relative contribution of the set of imaging, CSF, and genetic markers varied greatly. As a set, genetic variation showed the greatest importance (Figure 1) in the prediction of rate of motor progression (2.9%, 95%CI: 1.5–4.3%). The two SNPs selected most frequently by the ensemble, rs17710829 and rs9298897, both have significant effects on model predictions as well. CSF biomarkers at baseline showed a more modest (0.3%; 95%CI: 0.1–0.5%), but still significant effect on motor progression prediction, with CSF alpha-synuclein levels primarily driving the CSF effect (0.14%; 95%CI: 0.1 – 0.3%). Imaging data did not show a significant effect on motor progression prediction.
Fig. 1. Variable importance of model predictors in motor progression.
The relative contribution to the overall explanatory power for individual and/or sets of features is shown. The variable importance of the feature(s) is expressed as a percent increase in the mean squared error in leave-one-out cross-validation with each feature plotted in descending order of importance. Mean and 95% confidence intervals are indicated. The dashed blue line represents the full model without excluding any features.
Independent Replication of Candidate PD Progression Biomarkers in LABS-PD
In order to accurately assess the significance and magnitude of the effect of specific features identified in the model ensembles an independent test set (LABS-PD) was used. Several features appearing at high frequency in the ensemble showed similar effects in the LABS-PD cohort. Baseline motor score (beta=0.04, p=9e-6) and age (beta=0.05, p=3e-7) were both strongly associated with rate of motor progression. Sex also showed a significant effect on motor progression, with women progressing at a slower rate than men (beta=−0.45, p=0.009).
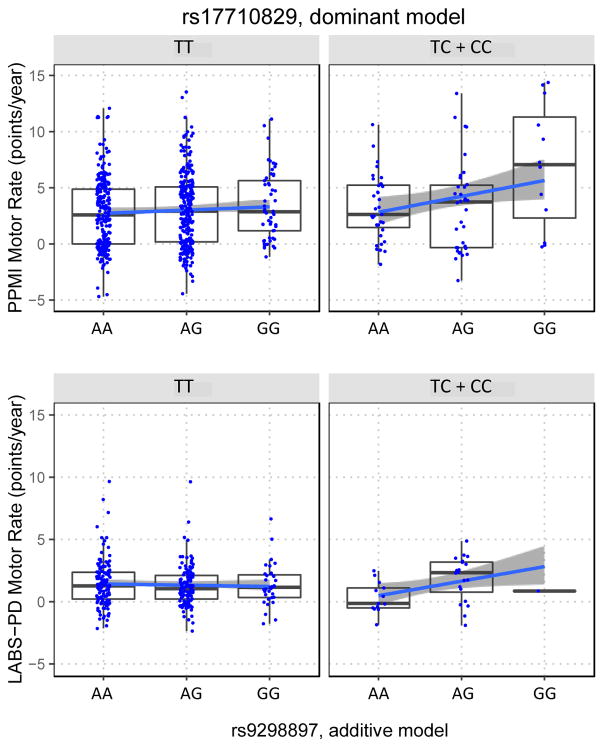
Treatment, SWEDD status, and the identified genetic principal components, features that had substantial differences in distribution between cohorts (Supplemental table S5), as well as their interacting terms, were not replicated. While the sex by treatment interaction term identified in the PPMI sample showed nominal association (beta=−2.4, p=0.023), it was not significant after accounting for multiple comparisons. In contrast, the novel epistatic interaction between the SNPs rs9298897 and rs17710829 demonstrated a significant association with a consistent direction in the LABS-PD sample (beta=1.2, p=0.011; Figure 2).
Fig. 2. Replication of PD-specific SNP interaction affecting motor progression rates.
Stratified plots of Motor progression rates vs. rs17710829 and rs9298897 genotypes for PD cases in PPMI (upper panels) and LABS-PD (lower panels). Note, dominant genetic modeling (combing the TC and CC genotypes) was used for rs17710829 due to its low minor allele frequency (C allele frequency=6%) while the more common rs9298897 (G allele frequency =35%) was modeled additively.
Predictive Utility of Model Ensemble in an Independent Sample
The true observed motor scores (median, −/+ 95% confidence intervals) over the follow-up period (which began between 3 and 4 years after baseline enrollment) for the predicted slow, moderate, and fast tertiles are shown in Figure 3. Although the actual rates of progression in the LABS-PD cohort tended to be slower overall than their predicted rates, the ordering was consistent, with significant separation in median motor scores observed between the slowest and fastest predicted progression groups across all time points. The moderate progression group also shows significant separation from the fast progression group. The slow and moderate groups were not as strongly differentiated, showing significant separation only in year 6.
Fig. 3.
LABS-PD Motor Scores by Predicted Progression Group. Median (95% CI) MDS-UPDRS motor scores parts II and III, beginning with the first follow-up exam (starting at either 3 or 4 years after baseline) are shown for cases predicted to be slow, moderate or fast progressors at study baseline.
Simulations (N=5000) show that incorporating the predicted rates of motor progression into the final models of treatment effect reduces the variability in the study outcome allowing significant differences to be detected at samples sizes up to 20% smaller than in naïve trials. The reduced set of weighted constituent models (Supplemental Table S3), incorporating far fewer features, resulted in nearly equivalent sample reduction of 19%.
Discussion
Using a hypothesis-free machine learning ensemble approach suitable for large-scale multivariate modeling, we have developed predictive models of motor symptom progression in early-stage Parkinson’s disease cases and age- and gender-matched controls. We have further identified the relative contribution of individual patient factors and sets of factors, and replicated several specific associations in an independent PD cohort, including a novel epistatic interaction. While previous progression modeling efforts have often focused on specific binary clinical outcomes (such as initiation of dopaminergic treatment or development of MCI), we used linear mixed effects models to estimate the continuous treated and untreated rates of clinical progression for each individual. Looking at strata-specific model performance, the predictive accuracy of the models was unsurprisingly greater among the PD cases, as there was very little variability in motor symptoms over time in the controls. Drug-naïve progression was similarly modeled more accurately than treated progression both in and out of sample, potentially due to heterogeneity in treatment regimens among those receiving PD medications. Additionally, it is well-recognized that there is large inter-individual variability in response to anti-Parkinsonian treatments, with this variability becoming more prominent over time as disease progresses 27,28.
While a substantial drop-off in the amount of variability explained was observed in the LABS-PD test set, this was expected, as several predictors were unavailable in LABS-PD (including CSF biomarkers and the baseline SCOPA exam). The required conversion of UPDRS to MDS-UPDRS scores for the outcome data also likely introduced significant variability. Furthermore, among cases, predictive accuracy for motor progression was greater in the earlier stages of disease among the PPMI cohort. In contrast, prediction in later-stage patients was only significant in the LABS-PD validation cohort. These discrepancies in the accuracy of predictions were likely due to differences in the composition of cohorts, as the large majority of the sample in LABS-PD (95%) was followed for longer than 5 years as opposed to a minority in PPMI (19%). As the models assume a linear trend in progression, another explanation may be that progression in later disease stages is associated with the same factors as early-stage, but the relationships are non-linear over time 4,28. Although these limitations in comparability between the test and training cohorts resulted in reduced (but still significant) R2, they also provide an important demonstration of the robustness of the ensemble modeling to diverse cohorts and data missingness. We can see that despite the differences from the training sample, the models demonstrate an unequivocal ability to prospectively differentiate between patients in the test set who would manifest slow or moderate progression and those whose condition would deteriorate more rapidly (Figure 3).
Variable importance measures determined using leave-one-out cross validation provided insight into the comparative predictive value of three different general types of potential predictive markers or tests available to clinicians (i.e. genetic variation, CSF biomarkers, and DaTscan imaging). The relatively limited prognostic utility of the DaTscan imaging data was in contrast to a previous study showing association between baseline imaging measures and various long-term PD outcomes 29. As our study is focused on early-stage PD progression, as opposed to long-term outcomes, this is intriguingly suggestive of distinctions between the factors determining disease progression at different disease stages. Of the CSF biomarkers, alpha synuclein levels, which have previously been inversely associated with motor symptom severity30, were the most common predictor of motor progression, with higher levels similarly predictive of slower progression.
Examination of the implicated features available for study in the LABS-PD cohort replicated a majority of clinical features, including sex, which showed women having a slower rate of decline in both cohorts. Sex differences are consistently observed in PD, most notably in prevalence, with men being more commonly affected, but also in age of onset and disease presentation 31. An interaction between sex and treatment was also observed, but only reached nominal significance in the replication cohort. Treatment and SWEDD status, both as main effects and in interaction, failed to replicate, likely as a consequence of the significant differences in prevalence of each factor in the two cohorts.
An interaction between two SNPs identified at high frequency in the motor progression ensemble was replicated in the LABS-PD cohort. As shown in Figure 2, cases who carry minor alleles for both SNPs have on average a substantially faster rate of motor decline in both the LABS-PD (1.2 points/per year faster) and PPMI (2.4 points/year). Neither SNP was among the a priori selected PD-related SNPS identified from GWAS, but intriguingly, rs9298897 is located in the fifth intron of the gene leucine rich repeat and Ig domain containing 2 (LINGO2). LINGO2 and its paralog LINGO1 encode for type I leucine-rich transmembrane proteins exclusively expressed in the central nervous system32,33. Like LRRK2, which is also of the leucine-rich repeat protein family and is the greatest known genetic contributor to PD, LINGO1 and LINGO2 are thought to function in the regulation of cell signaling related to neuronal survival and growth as well as glial function34–37. In experimental models of PD, increased expression of LINGO-1 were found in animal models of PD after parkinsonism-inducing neurotoxic lesions, while the absence or inhibition of LINGO-1 resulted in increased dopamine neuron protection35. Polymorphisms at these genes have been previously implicated in risk of PD and essential tremor (ET)38 but these findings have not been conclusive 39. The interacting SNP rs17710829 is located in an intergenic region on 2q14.1. The nearest gene dipeptidyl peptidase like 10 (DPP10) has not been linked to neurodegenerative disease in GWAS or other studies of genetic variation, however abnormal expression of the encoded protein, DPP10, has been observed in Alzheimer’s disease and other neurodegenerative diseases 40. Additional replication of this finding is warranted, as the functional link between these two genes is unclear, but further understanding of this interaction may lead to important mechanistic insights in the disease process.
Despite stringent sample inclusion criteria, robust data pre-processing, and penalization of model complexity via Bayesian ensemble inference, the generality of findings reported here are subject to several limitations associated with PPMI and LABS-PD study designs. The PPMI profiles used to develop the models are principally restricted to short-term follow-up after enrollment (median < 4 years), which may limit predictive accuracy when evaluated over longer periods of disease progression. Notably, in the LABS-PD cohort with average follow-up times exceeding 6 years, we see slower overall rates of progression (Supplemental Table S5) with a narrower range (Figure 2) suggestive of a potential plateau or other non-linear treads in progression. Additional characterization of the natural disease trajectories for each assessment domain, including more sophisticated incorporation of site and treatment effects may refine the clinically reliable window for linear rate model predictions beyond 5 years 28,41–43. Independent validation and variable replication findings reported herein may be additionally biased by variation in follow-up time distribution, distinct outcome assessment tools, and missing covariates between discovery and validation cohorts.
In summary, this analysis highlights the ability of ensemble modeling to capture the complex interplay of clinical, genetic, and molecular predictors of the highly heterogeneous PD progression phenotype. Bayesian model inference using REFS identified a combination of established and novel patient factors predictive of PD motor progression, particularly in the earlier stages of disease. The ability of the Bayesian model selection process to integrate genetic data with CSF-based biomarkers is of notable clinical relevance, as these data classes showed the greatest predictive impact in our models. Further, the quantification of the comparative predictive importance of the different data types, particularly the limited predictive utility of the DaTscan imaging in early-stage progression, provides guidance to the effective deployment of clinical and research resources in longitudinal patient evaluations.
The ability of the ensembles to prospectively identify patients most likely to have rapid progression of symptoms at the earliest stage of the disease has immediate significance towards enabling more effective trial recruitment and clinical disease management. The identification and independent confirmation of predictive factors also provides potential mechanistic insight into the disease process. Identification of a PD-specific novel genetic interaction between an intronic LINGO2 (9p21.1) SNP rs9298897 and the 2q14.1 variant rs17710829 was made possible through the multi-dimensional, hypothesis-free methodologies implemented here. Taken together these results advance the goal of establishing validated biomarkers of PD progression rate and improving existing prognostic models to the benefit of trial design and evaluation, as well as clinical disease monitoring and treatment.
Supplementary Material
Table S1. PPMI Sample demographics.
Table S2. Predictors included for study. Includes a) List of included known PD SNPs and b) List of included non-genetic variables
Table S3. Weighted summary Networks.
Table S4. Full list of Motor progression features with ensemble frequencies.
Table S5. Comparison of LABS-PD and PPMI demographics
Research in context.
Evidence before this study
We searched PubMed for articles including the terms “Parkinson’s progression”, “Parkinson’s prognostic”, or “Parkinson’s predictive” up until December 15, 2016. We examined both studies that conducted purely associative analyses as well as those that presented prognostic models, focusing on models that included some combination of additional molecular, genetic, or imaging data in addition to baseline clinical assessments and demographics. While several studies were found that quantify associations to motor progression and predictive models for given clinical event such as onset of dementia or initiation of treatment, we did not identify models simultaneously incorporating broad genetic, molecular and imaging data predictive of the rate of change of motor score.
Added value of this study
Use of a Bayesian machine learning method enabled us to examine complex interactions across data types, resulting in the identification and replication of a novel gene by gene interaction. The ensemble framework also allowed for comparison of the relative importance of different disease markers toward predicting disease progression. Through simulations we show that these predictive models have the potential to reduce cost and increase the efficiency of clinical trials.
Implications of all the available evidence
The results of this study demonstrate the benefits of a unified analysis incorporating the full complement of data types, increasingly becoming available in large longitudinal disease cohorts such as PPMI.
Acknowledgments
Funding: PPMI – a public-private partnership – is funded by the Michael J. Fox Foundation for Parkinson’s Research and funding partners, including Abbvie, Avid Radiopharmaceuticals, Biogen, Bristol-Myers Squibb, Covance, GE Healthcare, Genentech, GlaxoSmithKline, Lilly, Lundbeck, Merck, MesoScaleDiscovery, Pfizer, Piramal, Roche, Servier, UCB, and Golub Capital.
This work was supported by grants from the Michael J. Fox Foundation for Parkinson’s Research and National Institute of Neurological Disorders and Stroke (1P20NS092529-01)
Glossary
- Bayesian machine learning
Statistical methods focused on making explicit the prior assumptions about the model or the data that are used to build the model
- REFS
Bayesian machine-learning platform, Reverse Engineering and Forward Simulation (REFS), used to construct an ensemble of regularized generalized linear models. As applied here, REFS takes into account model complexity and model composition in terms of variable classes (i.e. demographic, clinical, genetic) and class sizes, effectively incorporating multiple testing correction into the model fitting step.
- Ensemble
a set of models, each individually scored by its goodness of fit to the observed data. By combining predictions from each of constituent models, the ensemble incorporates the natural heterogeneity in the predictor-outcomes relationships and reduces overfitting.
- Compressed Ensemble
To aid interpretation, the full ensemble, which can include hundreds of constituent models, can be compressed by clustering similar constituent models to provide a reduced set of representative models. The models in the reduced set are weighted to indicate the proportion of the full ensemble represented by each.
- Variable Importance
a measure of the usefulness of a given predictor (or set of predictors) to predicting an outcome based on the reduction of predictive accuracy (here quantified by change in mean squared error) when the predictor is removed from the ensemble.
Footnotes
Author contributions: JCL, MTB, TCH, DMW, IGK, BH, CSV conceived and designed the study. JCL, MTB analyzed the data. REM, JNO, MPV contributed analysis tools. JCL wrote the first draft of the manuscript. JCL, MTB, REM, JNO, MPV, DMW, BH, CSV Contributed to the writing of the manuscript: JCL, MTB, TCH, REM, JNO, MPV, DMW, IGK, BH, CSV Agree with the manuscript’s results and conclusions.
Competing interests: JCL, MTB, TCH, REM, JNO, MPV, DMW, IGK, BH, are currently (JCL, TCH, REM, DMW, IGK, BH) or were at time of study (MTB, JNO, MPV) employees of GNS Healthcare.
Data and materials availability: PPMI data is available at PPMI website: http://www.ppmi-info.org/access-data-specimens/download-data/
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Notes
- 1.Kalia LV, Lang AE. Parkinson’s disease. Lancet Lond Engl. 2015;386:896–912. doi: 10.1016/S0140-6736(14)61393-3. [DOI] [PubMed] [Google Scholar]
- 2.Sieber B-A, Landis S, Koroshetz W, et al. Prioritized research recommendations from the National Institute of Neurological Disorders and Stroke Parkinson’s Disease 2014 conference. Ann Neurol. 2014;76:469–72. doi: 10.1002/ana.24261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Post B, Merkus MP, de Haan RJ, Speelman JD CARPA Study Group. Prognostic factors for the progression of Parkinson’s disease: a systematic review. Mov Disord Off J Mov Disord Soc. 2007;22:1839–51. doi: 10.1002/mds.21537. quiz 1988. [DOI] [PubMed] [Google Scholar]
- 4.Reinoso G, Allen JC, Au W-L, Seah S-H, Tay K-Y, Tan LCS. Clinical evolution of Parkinson’s disease and prognostic factors affecting motor progression: 9-year follow-up study. Eur J Neurol. 2015;22:457–63. doi: 10.1111/ene.12476. [DOI] [PubMed] [Google Scholar]
- 5.Terrelonge M, Marder KS, Weintraub D, Alcalay RN. CSF β-Amyloid 1–42 Predicts Progression to Cognitive Impairment in Newly Diagnosed Parkinson Disease. J Mol Neurosci MN. 2016;58:88–92. doi: 10.1007/s12031-015-0647-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morales DA, Vives-Gilabert Y, Gómez-Ansón B, et al. Predicting dementia development in Parkinson’s disease using Bayesian network classifiers. Psychiatry Res. 2013;213:92–8. doi: 10.1016/j.pscychresns.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 7.Schrag A, Siddiqui UF, Anastasiou Z, Weintraub D, Schott JM. Clinical variables and biomarkers in prediction of cognitive impairment in patients with newly diagnosed Parkinson’s disease: a cohort study. Lancet Neurol. 2017;16:66–75. doi: 10.1016/S1474-4422(16)30328-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Velseboer DC, de Bie RMA, Wieske L, et al. Development and external validation of a prognostic model in newly diagnosed Parkinson disease. Neurology. 2016;86:986–93. doi: 10.1212/WNL.0000000000002437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simuni T, Long JD, Caspell-Garcia C, et al. Predictors of time to initiation of symptomatic therapy in early Parkinson’s disease. Ann Clin Transl Neurol. 2016;3:482–94. doi: 10.1002/acn3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhattaram VA, Siddiqui O, Kapcala LP, Gobburu JVS. Endpoints and Analyses to Discern Disease-Modifying Drug Effects in Early Parkinson’s Disease. AAPS J. 2009:11. doi: 10.1208/s12248-009-9123-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedman N, Koller D. Being Bayesian About Network Structure. A Bayesian Approach to Structure Discovery in Bayesian Networks. Mach Learn. 2003;50:95–125. [Google Scholar]
- 12.Xing H, McDonagh PD, Bienkowska J, et al. Causal modeling using network ensemble simulations of genetic and gene expression data predicts genes involved in rheumatoid arthritis. PLoS Comput Biol. 2011;7:e1001105. doi: 10.1371/journal.pcbi.1001105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson JP, Parikh JR, Shenfeld DK, et al. Reverse Engineering and Evaluation of Prediction Models for Progression to Type 2 Diabetes: An Application of Machine Learning Using Electronic Health Records. J Diabetes Sci Technol. 2016;10:6–18. doi: 10.1177/1932296815620200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steinberg GB, Church BW, McCall CJ, Scott AB, Kalis BP. Novel predictive models for metabolic syndrome risk: a ‘big data’ analytic approach. Am J Manag Care. 2014;20:e221–8. [PubMed] [Google Scholar]
- 15.Marek K, Jennings D, Lasch S, et al. The Parkinson Progression Marker Initiative (PPMI) Prog Neurobiol. 2011;95:629–35. doi: 10.1016/j.pneurobio.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goetz CG, Tilley BC, Shaftman SR, et al. Movement Disorder Society-sponsored revision of the Unified Parkinson’s Disease Rating Scale (MDS-UPDRS): scale presentation and clinimetric testing results. Mov Disord Off J Mov Disord Soc. 2008;23:2129–70. doi: 10.1002/mds.22340. [DOI] [PubMed] [Google Scholar]
- 17.Pahwa R, Lyons KE, Hauser RA, et al. Randomized trial of IPX066, carbidopa/levodopa extended release, in early Parkinson’s disease. Parkinsonism Relat Disord. 2014;20:142–8. doi: 10.1016/j.parkreldis.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 18.Hauser RA, Panisset M, Abbruzzese G, et al. Double-blind trial of levodopa/carbidopa/entacapone versus levodopa/carbidopa in early Parkinson’s disease. Mov Disord Off J Mov Disord Soc. 2009;24:541–50. doi: 10.1002/mds.22343. [DOI] [PubMed] [Google Scholar]
- 19.Storch A, Jost WH, Vieregge P, et al. Randomized, double-blind, placebo-controlled trial on symptomatic effects of coenzyme Q(10) in Parkinson disease. Arch Neurol. 2007;64:938–44. doi: 10.1001/archneur.64.7.nct60005. [DOI] [PubMed] [Google Scholar]
- 20.Parashos SA, Luo S, Biglan KM, et al. Measuring disease progression in early Parkinson disease: the National Institutes of Health Exploratory Trials in Parkinson Disease (NET-PD) experience. JAMA Neurol. 2014;71:710–6. doi: 10.1001/jamaneurol.2014.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bates DM. lme4: Mixed-effects modeling with R. [accessed March 17, 2014];URL Httplme4 R-Forge R-Proj Orgbook. 2010 http://lme4.r-forge.r-project.org/lMMwR/lrgprt.pdf.
- 22.Hindorff LA, Sethupathy P, Junkins HA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–7. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piñero J, Queralt-Rosinach N, Bravo À, et al. DisGeNET: a discovery platform for the dynamical exploration of human diseases and their genes. Database J Biol Databases Curation. 2015;2015:bav028. doi: 10.1093/database/bav028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Donoho D, Tanner J. Observed universality of phase transitions in high-dimensional geometry, with implications for modern data analysis and signal processing. Philos Trans R Soc Lond Math Phys Eng Sci. 2009;367:4273–93. doi: 10.1098/rsta.2009.0152. [DOI] [PubMed] [Google Scholar]
- 25.Ravina B, Tanner C, Dieuliis D, et al. A longitudinal program for biomarker development in Parkinson’s disease: a feasibility study. Mov Disord Off J Mov Disord Soc. 2009;24:2081–90. doi: 10.1002/mds.22690. [DOI] [PubMed] [Google Scholar]
- 26.Hayete B, Valko M, Greenfield A, Yan R. MDL-motivated compression of GLM ensembles increases interpretability and retains predictive power. [accessed Dec 21, 2016];ArXiv161106800 Stat. 2016 published online Nov 21. http://arxiv.org/abs/1611.06800.
- 27.Nutt JG, Holford NH. The response to levodopa in Parkinson’s disease: imposing pharmacological law and order. Ann Neurol. 1996;39:561–73. doi: 10.1002/ana.410390504. [DOI] [PubMed] [Google Scholar]
- 28.Venuto CS, Potter NB, Ray Dorsey E, Kieburtz K. A review of disease progression models of Parkinson’s disease and applications in clinical trials. Mov Disord Off J Mov Disord Soc. 2016;31:947–56. doi: 10.1002/mds.26644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ravina B, Marek K, Eberly S, et al. Dopamine transporter imaging is associated with long-term outcomes in Parkinson’s disease. Mov Disord Off J Mov Disord Soc. 2012;27:1392–7. doi: 10.1002/mds.25157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang J-H, Irwin DJ, Chen-Plotkin AS, et al. Association of cerebrospinal fluid β-amyloid 1-42, T-tau, P-tau181, and α-synuclein levels with clinical features of drug-naive patients with early Parkinson disease. JAMA Neurol. 2013;70:1277–87. doi: 10.1001/jamaneurol.2013.3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gillies GE, Pienaar IS, Vohra S, Qamhawi Z. Sex differences in Parkinson’s disease. Front Neuroendocrinol. 2014;35:370–84. doi: 10.1016/j.yfrne.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carim-Todd L, Escarceller M, Estivill X, Sumoy L. LRRN6A/LERN1 (leucine-rich repeat neuronal protein 1), a novel gene with enriched expression in limbic system and neocortex. Eur J Neurosci. 2003;18:3167–82. doi: 10.1111/j.1460-9568.2003.03003.x. [DOI] [PubMed] [Google Scholar]
- 33.Laurén J, Airaksinen MS, Saarma M, Timmusk T. A novel gene family encoding leucine-rich repeat transmembrane proteins differentially expressed in the nervous system. Genomics. 2003;81:411–21. doi: 10.1016/s0888-7543(03)00030-2. [DOI] [PubMed] [Google Scholar]
- 34.MacLeod D, Dowman J, Hammond R, Leete T, Inoue K, Abeliovich A. The familial Parkinsonism gene LRRK2 regulates neurite process morphology. Neuron. 2006;52:587–93. doi: 10.1016/j.neuron.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 35.Inoue H, Lin L, Lee X, et al. Inhibition of the leucine-rich repeat protein LINGO-1 enhances survival, structure, and function of dopaminergic neurons in Parkinson’s disease models. Proc Natl Acad Sci U S A. 2007;104:14430–5. doi: 10.1073/pnas.0700901104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mi S, Miller RH, Lee X, et al. LINGO-1 negatively regulates myelination by oligodendrocytes. Nat Neurosci. 2005;8:745–51. doi: 10.1038/nn1460. [DOI] [PubMed] [Google Scholar]
- 37.Li W, Walus L, Rabacchi SA, et al. A neutralizing anti-Nogo66 receptor monoclonal antibody reverses inhibition of neurite outgrowth by central nervous system myelin. J Biol Chem. 2004;279:43780–8. doi: 10.1074/jbc.M401803200. [DOI] [PubMed] [Google Scholar]
- 38.Vilariño-Güell C, Wider C, Ross OA, et al. LINGO1 and LINGO2 variants are associated with essential tremor and Parkinson disease. Neurogenetics. 2010;11:401–8. doi: 10.1007/s10048-010-0241-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nalls MA, Pankratz N, Lill CM, et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat Genet. 2014;46:989–93. doi: 10.1038/ng.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen T, Gai W-P, Abbott CA. Dipeptidyl peptidase 10 (DPP10(789)): a voltage gated potassium channel associated protein is abnormally expressed in Alzheimer’s and other neurodegenerative diseases. BioMed Res Int. 2014;2014:209398. doi: 10.1155/2014/209398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo S, Wang J. Bayesian hierarchical model for multiple repeated measures and survival data: an application to Parkinson’s disease. Stat Med. 2014;33:4279–91. doi: 10.1002/sim.6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van den Hout A, Matthews FE. Estimating dementia-free life expectancy for Parkinson’s patients using Bayesian inference and microsimulation. Biostat Oxf Engl. 2009;10:729–43. doi: 10.1093/biostatistics/kxp027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee JY, Gobburu JVS. Bayesian quantitative disease-drug-trial models for Parkinson’s disease to guide early drug development. AAPS J. 2011;13:508–18. doi: 10.1208/s12248-011-9293-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. PPMI Sample demographics.
Table S2. Predictors included for study. Includes a) List of included known PD SNPs and b) List of included non-genetic variables
Table S3. Weighted summary Networks.
Table S4. Full list of Motor progression features with ensemble frequencies.
Table S5. Comparison of LABS-PD and PPMI demographics