Abstract
Human genetic studies implicate LRRK2 and RAB7L1 in susceptibility to Parkinson disease (PD). These two genes function in the same pathway, as knockout of Rab7L1 results in phenotypes similar to LRRK2 knockout, and studies in cells and model organisms demonstrate LRRK2 and Rab7L1 interact in the endolysosomal system. Recently, a subset of Rab proteins have been identified as LRRK2 kinase substrates. Herein, we find that Rab8, Rab10, and Rab7L1 must be membrane and GTP-bound for LRRK2 phosphorylation. LRRK2 mutations that cause PD including R1441C, Y1699C, and G2019S all increase LRRK2 phosphorylation of Rab7L1 four-fold over wild-type LRRK2 in cells, resulting in the phosphorylation of nearly one-third the available Rab7L1 protein in cells. In contrast, the most common pathogenic LRRK2 mutation, G2019S, does not upregulate LRRK2-mediated phosphorylation of Rab8 or Rab10. LRRK2 interaction with membrane and GTP-bound Rab7L1, but not Rab8 or Rab10, results in the activation of LRRK2 autophosphorylation at the serine 1292 position, required for LRRK2 toxicity. Further, Rab7L1 controls the proportion of LRRK2 that is membrane-associated, and LRRK2 mutations enhance Rab7L1-mediated recruitment of LRRK2 to the trans-Golgi network. Interaction studies with the Rab8 and Rab10 GTPase-activating protein TBC1D4/AS160 demonstrate that LRRK2 phosphorylation may block membrane and GTP-bound Rab protein interaction with effectors. These results suggest reciprocal regulation between LRRK2 and Rab protein substrates, where Rab7L1-mediated upregulation of LRRK2 kinase activity results in the stabilization of membrane and GTP-bound Rab proteins that may be unable to interact with Rab effector proteins.
Introduction
Genetic variation in the Leucine-rich repeat kinase 2 (LRRK2) gene is associated with susceptibility to Parkinson disease (PD) (1,2). There are several pathogenic missense mutations that segregate in families (1,2) and cluster in the functional kinase and GTP-binding regions of the encoded LRRK2 protein (3). Additionally, common genetic variation around the LRRK2 locus influences lifetime risk of idiopathic PD (4), possibly by influencing LRRK2 expression (5). Because of this observed association of a kinase with inherited and idiopathic disease, there is interest in the potential for targeting LRRK2 for PD therapeutics (6).
In this context, understanding LRRK2 biology is critically important for therapeutic development and in understanding potential effects of inhibiting LRRK2 activity in patients. Prior work has identified a role for LRRK2 in membrane trafficking in various contexts. LRRK2 localizes to multiple membranous compartments in cells (7,8). A major consequence of knocking out LRRK2 expression or completely inhibiting LRRK2 kinase activity is the accumulation of lysosomes and dysfunction of the autophagy-lysosomal pathway, particularly in renal endothelia in mice and rats (9–12). We have previously shown that recruitment of LRRK2 to specific membrane compartments is enhanced by binding to Rab proteins, specifically the Rab protein encoded by the PD-risk factor gene RAB7L1 (9,13), as well as other membrane adaptors. Genetic ablation of Rab7L1 phenocopies LRRK2 deficiency in mice and other model organisms, as knockouts of either gene results in these specific lysosomal defects such as the discoloration of the kidney cortex resultant from the accumulation of oxidized blood products and lipofuscins (14). These emerging results suggest that LRRK2 depends on interacting partners like Rab7L1, a group III Rab protein that is divergent from better-known group I Rab proteins like Rab8 and 10, to be positioned at intracellular membranes, and that a deficiency in this system results in dysregulation of membrane-bound vesicles in the autophagy-lysosome system.
Rab proteins are the largest subgroup of the Ras GTPase superfamily of small GTPases and regulate the transport and organization of vesicles in the autophagy-lysosome system (15). Recently, it has been shown that LRRK2 phosphorylates a subset of Rabs. LRRK2 phosphorylates at least two group I Rabs, the closely related Rab8 and Rab10 proteins (16–18). In transfected cells, pathogenic LRRK2 mutations that span across multiple domains enhance Rab8 and Rab10 phosphorylation (16). We have reported that all the pathogenic mutations in LRRK2 enhance Rab7L1-dependent effects of LRRK2 at the trans-Golgi network (TGN) (13). However, Rab7L1 was reported as a weak LRRK2 kinase substrate in vitro and not detected in in previous phosphoproteomic screens (16,19). The mechanism(s) by which LRRK2 mutations influence cellular functions, control Rab function, and by extension susceptibility to neurodegeneration and PD, has therefore not been resolved.
Rab proteins cycle between GDP-bound states and GTP and membrane-bound states. Within the Rab LRRK2 kinase substrate family that has been identified, LRRK2 phosphorylates threonine residues in conserved switch II domains to promote dissociation from GDP-dissociation inhibitors (GDIs) and therefore enhance membrane affinity together with the action of Rab-escort proteins (REPs) and geranylgeranylation (16). However, the preference of LRRK2 in phosphorylating particular Rab states, whether GTP and membrane-bound, or GDP-bound cytosolic states, could give clues as to the mechanisms underlying LRRK2 regulation of Rab GTPases, and by extension understand LRRK2 interaction with Rab7L1.
In the present study, we set out to examine the effects of pathogenic LRRK2 mutations on Rab8, Rab10, and Rab7L1 in transfected mammalian (HEK293) cells. We find that both Rab8/Rab10 and Rab7L1 are comparably phosphorylated by wild-type LRRK2. However, as opposed to Rab8/10, the G2019S-LRRK2 mutation increases phosphorylation of Rab7L1. Mechanistically, the phosphorylation of Rab7L1 depends on both LRRK2 and Rab7L1 to be in a GTP and membrane-bound form, minimizing the potential effects of LRRK2 phosphorylation on Rab GDI interactions, and instead highlighting possible blockade of GTPase-activating proteins (GAPs) that return active Rab proteins to inactive states. Further distinguishing Rab7L1 from Rab8/10, Rab7L1 activates LRRK2 autophosphorylation and the recruitment of LRRK2 to the TGN. These results provide molecular insights into the newly identified LRRK2-Rab7L1 pathway.
Results
LRRK2 phosphorylates Rab7L1
Recent phosphoproteomic analyses have revealed that LRRK2 phosphorylates a subset of Rab proteins. Phosphorylation occurs at threonine residues in the switch II effector binding domain of the closely related Group I Rab8 and Rab10 proteins (16). The LRRK2-binding partner Rab7L1 [also known as Rab29 (9,13)] is a Group III Rab, phylogenetically distant from Rab8 and Rab10 (see Supplementary Material, Fig. S1A). To determine whether LRRK2 phosphorylates Rab7L1, we co-transfected LRRK2-encoding plasmids with eGFP-Rab plasmids into HEK293 cells (Fig. 1). Phos-tag analysis of the effects of LRRK2-mutations R1441C, Y1699C, and G2019S, together with LRRK2-kinase-dead (D1994A), demonstrated robust LRRK2 phosphorylation of Rab10 (Fig. 1A and B) and Rab8 (Supplementary Material, Fig. S2). The R1441C and Y1699C mutations enhanced phosphorylation over WT-LRRK2, but the G2019S mutation was similar to WT-LRRK2 for these substrates. Co-transfection of Rab10 or Rab8 with Y1699C-LRRK2 led to the phosphorylation of ∼30% and ∼40%, respectively, of the Rab proteins, whereas no phospho-Rab protein could be detected with kinase-dead LRRK2 co-transfections. These results indicate there are no other kinases resident in HEK293 cells that might significantly phosphorylate these Rab proteins. Co-expression of LRRK2 with Rab7L1 led to a similar level of Rab phosphorylation (Fig. 1C). In contrast to Rab8 and Rab10 substrates, G2019S-LRRK2 protein phosphorylation of Rab7L1 could not be distinguished from the effects of the Y1699C and R1441C mutations and phosphorylated ∼30% of Rab7L1 protein in cells.
Figure 1.
Pathogenic LRRK2 mutations increase the phosphorylation of GTP-bound Rabs. (A) HEK293 cells were co-transfected with plasmids expressing WT-LRRK2 or LRRK2 with pathogenic mutations R1441C, Y1699C, G2019S, or the kinase-dead D1994A (KD) together with WT-Rab10 or WT-Rab7L as indicated. Representative immunoblots are shown. Rab phosphorylation was assessed via phos-tag analysis (see Methods). The upper band (>50 kDa) represents phosphorylated Rab. (B) Calculated ratio of phosphorylated Rab10 and (C) Rab7L1. (D) R1441C-LRRK2 was co-transfected with WT, Q68L and T23N-Rab10, or WT, Q67L, T21N-Rab7L1 as indicated. Representative immunoblots are shown. Rab phosphorylation was assessed via phos-tag analysis. (E) Calculated ratio of phosphorylated Rab10 and (F) Rab7L1. (G) SH-SY5Y cells were transfected with N-terminal-eGFP-tagged WT, QL, TN-Rab10 or Rab7L1 constructs, as indicated. Representative wide-field images of live cells show typical distributions of Rab proteins 16 h after transfection, with dark phase-contrast overlays. syn bar is 25 μm. (H) R1441C-LRRK2 was co-transfected with WT, T71E and S72D Rab7L1 as indicated. Representative immunoblots are shown. Rab phosphorylation was assessed via phos-tag analysis. (I) WT, R1441C, Y1699C, G2019S-LRRK2 were co-transfected with WT or T71E Rab7L1 as indicated. Representative immunoblots are shown. Rab phosphorylation was assessed via phos-tag analysis. All data are averaged from three independent experiments or are representative of three independent experiments. Data are means ± SEM; significance assessed by one-way ANOVA with Tukey’s post hoc test. *P <0.05, **P < 0.01.
LRRK2 phosphorylation of Rab switch II domains requires Rab nucleotide binding
LRRK2 autophosphorylates its own switch-effector domains in the Rab-like ROC domain as well as in Rab8 and Rab10 (16,20–22). The switch II region is known to be relatively disordered in the inactive GDP-bound state compared with the active GTP-bound state (23), and could alter LRRK2-mediated phosphorylation. We created a panel of constructs to manipulate Rab nucleotide binding through altering intrinsic hydrolysis activity (QL mutations, see Supplementary Material, Fig. S1B) and nucleotide binding properties (TN mutations). Substitution of the P-loop threonine to asparagine (TN mutant) that shifts equilibrium towards nucleotide-free and GDP-bound states ablated LRRK2 phosphorylation (Fig. 1 and Supplementary Material, Figs S2 and S3). The TN mutations did not affect protein stability or expression compared with WT-Rab8, 10, and Rab7L1 (Fig. 1D and G and Supplementary Material, Fig. S2). The QL mutation in Rab10, designed to disrupt Rab10 GTPase activity and promote GTP-bound Rab10, resulted in significantly more LRRK2 phosphorylation activity compared with WT-Rab10 (Fig. 1E). In contrast, QL-Rab7L1 protein, which was shown to decrease GTP-binding affinity (13), cannot be phosphorylated by LRRK2 (Fig. 1F). On a subcellular level, the QL-Rab7L1 protein had a diffuse distribution different than WT-Rab7L1 and more similar to TN-Rab7L1 (Fig. 1G). These results are consistent with our past observations that the QL mutation in Rab7L1 results in a dominant-negative protein, in contrast to the equivalent mutation in Rab8/10 (13). In summary, LRRK2 phosphorylation of Rab protein substrates requires a nucleotide-bound, possibly GTP-bound, state.
LRRK2 phosphorylates Rab proteins in switch II domains. However, since Rab7L1 sequence diverges from Rab8 and Rab10, a serine residue (Ser72) in Rab7L1 is substituted for the threonine residue LRRK2 phosphorylates in the Rab8 and Rab10 switch II domains [Supplementary Material, Fig. S1A, (16)]. In Rab7L1, a threonine residue lies adjacent to the conserved phenylalanine. To clarify the site of LRRK2-mediated Rab7L1 phosphorylation, we generated an S72D and T71E mutation in Rab7L1. In contrast to S72D-Rab7L1 which showed similar LRRK2-phosphorylation as WT-Rab7L1, the T71E substitution nearly ablated LRRK2-mediated Rab7L1 phosphorylation (Fig. 1H and I). WT-LRRK2, R1441C, Y1699C, and G2019S-LRRK2 were all unable to phosphorylate T71E-Rab7L1 (Fig. 1J). These results suggest that the major LRRK2 phosphorylation site on Rab7L1 is the Thr71 residue.
LRRK2 phosphorylation of Rab substrates requires Rab prenylation
Subcellular distribution profiles of the Rab proteins suggested both cytosolic as well as membrane-associated pools of proteins (Fig. 1G). In splitting cell lysates into soluble cytosolic fractions and membrane-enriched triton-x100 extracted fractions, we could see near equal Rab protein expression between the two fractions, in contrast to the canonical proteins GAPDH that lacks membrane associations and aquaporin-1 (VDAC) that lacks a cytoplasmic component (Fig. 2A and B). Both QL and TN-Rab7L1 showed low levels of protein in the membrane-enriched fractions compared with WT-Rab7L1. Co-transfections of the QL and TN-Rab variants together with R1441C-LRRK2 demonstrated that most of the LRRK2-phosphorylated Rab proteins localized to membrane-associated fractions {[∼6: 1 Rab10, ∼10: 1 Rab8, and ∼3: 1 Rab7L1, (w/v)], Fig. 2A–D and Supplementary Material, Fig. S4}.
Figure 2.
LRRK2 requires Rab prenylation and membrane localization for phosphorylation. HEK293 cells were co-transfected with WT, R1441C, Y1699C, G2019S-LRRK2 and (A) Rab7L1or (B) Rab10. Lysates were sequentially separated into soluble cytosolic-enriched and detergent-extracted membrane-enriched fractions, as indicated. Representative immunoblots are shown. Rab10 and Rab7L1 phosphorylation was assessed via phos-tag analysis. The transmembrane marker VDAC and cytosolic enzyme GAPDH are shown as markers for membrane-enriched fractions and cytosolic fractions, respectively. (C) Calculated ratios of pRab7L1 and (D) pRab10 as a percent of total Rab protein. (E) Lysates from HEK293 cells co-transfected with WT-LRRK2 and WT, QL, or TN-Rab7L1 or (F) Rab10 constructs, and separated into soluble cytosolic-enriched and detergent-extracted membrane-enriched fractions as indicated. Representative immunoblots are shown. (G) Calculated ratios of pRab10 and (H) pRab7L1 as a percent of total Rab protein. (I) Representative live cell wide-field fluorescence imaging of transfected SH-SY5Y cells expressing eGFP-WT and (J) CC-Rab10, and (K) WT-Rab7L1 and (L) CC-Rab7L1 16 h after transfection. Dark phase-contrast overlays are included. Scale bar is 25 μM. (M) HEK293 cells co-transfected with WT-LRRK2 and WT or CC-Rab7L1 or (N) Rab10 constructs, treated with or without MLi2 (100 nM, 2 h) as indicated. (O) Calculated ratios of pRab10 and (P) pRab7L1 as a percent of total Rab protein. All data are averaged from three independent experiments or are representative of three independent experiments. Data are means ± SEM; significance assessed by one-way ANOVA with Tukey’s post hoc test. **P < 0.01, ***P < 0.001.
Pathogenic LRRK2 mutations may enhance the proportion of membrane-bound and kinase activity LRRK2 protein (13,24). We split cell lysates from cells co-expressing WT-Rab10 or Rab7L1 together with WT, R1441C, Y1699C and G2019S-LRRK2 into soluble cytosolic fractions and membrane-enriched fractions. All the LRRK2 mutations preferentially phosphorylated membrane-localized WT-Rab10 and Rab7L1 (Fig. 2E–H). Prenylation of C-terminal cysteine residues in Rabs is known to increase the affinity of the Rabs to membranes and is required for Rab function (25). We mutated the residues required for prenylation (Supplementary Material, Fig. S1B) and verified in vivo that GTP and GDP-binding of the resultant CC-Rab proteins was undisturbed (Supplementary Material, Fig. S5). Subcellular localization of the CC-Rab proteins revealed a diffuse distribution compared with the WT-Rab proteins, consistent with the intended effects of the CC mutations in blocking prenylation (Fig. 2I–L). In co-transfection experiments, LRRK2 was largely unable to phosphorylate CC-Rab mutants (Fig. 2M–P), as only slight LRRK2-phosphorylation in CC-Rab7L1 or CC-Rab10 could be detected above background in the cytoplasmic fraction and none in the membrane-enriched fraction. As expected, the overall CC-Rab concentration in the membrane-enriched fraction was also diminished.
Rab7L1 activates LRRK2 autophosphorylation
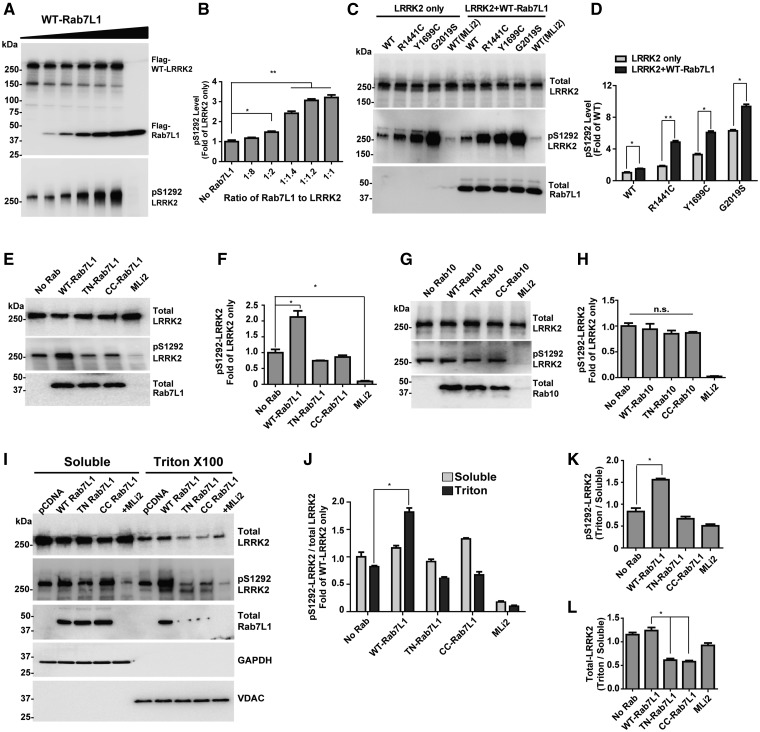
From our results, LRRK2 phosphorylation of other Rab proteins may require the Rab substrate to be membrane and GTP-bound. As LRRK2 autophosphorylates its own Rab-like ROC GTPase domain, and membrane-associated LRRK2 shows higher kinase activity than cytosolic LRRK2 (24,26), we next investigated whether Rab proteins might affect LRRK2 autophosphorylation. In comparing the intensity of FLAG-tagged WT-LRRK2 and FLAG-tagged WT-Rab7L1, we could determine that increasing levels of Rab7L1 led to increasing the proportion of LRRK2 autophosphorylated at Ser1292. At an equimolar stoichiometry, Rab7L1 enhanced pSer1292 levels in WT-LRRK2 to 3fold compared with cells that lacked FLAG-Rab7L1 expression (Fig. 3A and B). In contrast, WT-LRRK2 could only phosphorylate Rab7L1 to 4% when LRRK2 outnumbers Rab7L1 molecules 8: 1 (Supplementary Material, Fig. S6).
Figure 3.
Rab7L1 regulates membrane-associated LRRK2 kinase activity. (A) HEK293 cells were co-transfected with increasing concentrations of FLAG-WT-Rab7L1 with FLAG-WT-LRRK2. Representative immunoblots are shown. (B) Fold of increases of pS1292-LRRK2 to total LRRK2 protein with respect to no Rab7L1 expression were calculated. (C) HEK293 cells were transfected with WT, R1441C, Y1699C, and G2019S-LRRK2, with or without WT-Rab7L1. The LRRK2 kinase inhibitor MLi2 (100 nM) with WT-LRRK2 conditions, with or without Rab7L1, were included as controls for LRRK2 kinase activity. Representative immunoblots are shown. (D) Normalized pS1292-LRRK2 levels (to total LRRK2 expression) were calculated and shown as fold of WT-LRRK2 without Rab7L1. (E) Cells were co-transfected with WT-LRRK2 and WT, TN, or CC-Rab10, or empty vector (No Rab lane) as indicated. MLi2 (100 nM) is indicated. Representative immunoblots are shown. (F) Normalized pS1292-LRRK2 levels (to total LRRK2 expression) were calculated. (G) Rab7L1 constructs were combined with WT-LRRK2 as in (E). (H) Normalized pS1292-LRRK2 levels (to total LRRK2 expression) were calculated. (I) Lysates from LRRK2 and Rab7L1-expressing (or pCDNA empty vector) cells were sequentially fractionated to soluble and triton-x100 fractions. Representative immunoblots are shown. (J) Normalized pS1292-LRRK2 levels (to total LRRK2 expression) were calculated. (K) pS1292-LRRK2 levels and L) total LRRK2 protein in the triton-x100 fraction were normalized to the levels in the soluble fractions. All data are averaged from three independent experiments. Data are means ± SEM; significance is assessed by one-way ANOVA with Tukey’s post hoc test. *P < 0.01, **P < 0.001.
Co-transfection experiments with WT-Rab7L1 expressed together with WT, R1441C, Y1699C, or G2019S-LRRK2 showed that pSer1292 can be simulated by Rab7L1 irrespective of LRRK2 mutation status (Fig. 3C and D). To further study the mechanism of Rab7L1-mediated LRRK2 kinase activation, we co-transfected R1441C-LRRK2, which shows the most robust Rab7L1 activation (Fig. 3C and D), together with WT, TN, and CC-Rab7L1. The TN-Rab7L1 protein that LRRK2 cannot phosphorylate could not enhance pS1292-LRRK2 levels. Likewise, expression of the CC-Rab7L1 mutant that holds Rab7L1 cytoplasmic had no effect on pS1292-LRRK2 levels (Fig. 3E and F). These results suggest that Rab7L1 must be GTP and membrane-bound to stimulate LRRK2 autophosphorylation. In contrast to Rab7L1, co-expression of LRRK2 together with WT, TN, or CC-Rab10 did not affect pS1292-LRRK2 levels (Fig. 3G and H).
In parsing out pS1292-LRRK2 protein into cytosolic soluble fractions and triton-x100 extracted membrane-enriched fractions, we found that nearly all of the increased levels of pS1292-LRRK2 caused by Rab7L1 co-expression were occurring at the membrane (Fig. 3I and J). While the proportion of pS1292-LRRK2 in the membrane-enriched fraction is increased in the presence of WT-Rab7L1 (Fig. 3K), expression of TN or CC-Rab7L1 diminished total LRRK2 protein in the membrane-enriched fraction relative to the cytosolic fraction (Fig. 3L). As expected, the pS1292-LRRK2 signal was not present in a S1292A-LRRK2 mutant control construct, whether or not Rab7L1 was expressed (Supplementary Material, Fig. S7). These results suggest that WT-Rab7L1, but not mutant forms of Rab7L1 that lack membrane and/or nucleotide binding, activate LRRK2 autophosphorylation in membrane-enriched fractions.
Rab7L1 promotes LRRK2 recruitment to the trans-Golgi network
Previously, we demonstrated that LRRK2 and Rab7L1 lead to the clearance of Golgi-derived vesicles through endolysosomal pathways (9,13). We tested whether the increase in LRRK2 protein recruited to membrane-enriched fractions caused by WT-Rab7L1 expression is due to enhanced LRRK2 relocalization to the trans-Golgi network. We imaged the co-localization of LRRK2 and Rab7L1 (Fig. 4A) or TN-Rab7L1 (Fig. 4B) together with the trans-Golgi marker TGN46 using an automated high content imaging assay previously described (37). All three pathogenic LRRK2 mutations R1441C, Y1699C, and G2019S increased LRRK2 recruitment to the TGN and co-localization with Rab7L1 (Fig. 4A). Conversely, TN-Rab7L1 expression did not allow for LRRK2 localization to the TGN (Fig. 4B), consistent with measurements from membrane-enriched fractions (Fig. 3). Therefore, blocking the ability of either LRRK2 or Rab7L1 to bind GDP/GTP (T1348N-LRRK2, or TN-Rab7L1, respectively) abolishes LRRK2 co-localization with TGN46, and all pathogenic LRRK2 mutations increase localization to the TGN (Fig. 3C).
Figure 4.

Pathogenic mutations in LRRK2 increase relocalization to the TGN. (A) Confocal images of HEK293 cells stained for an endogenous marker of the trans-Golgi network (TGN, TNG46, red) transfected WT-Rab7L1 (cyan) or (B) TN-Rab7L1 (cyan) together with WT-LRRK2 or the indicated mutant LRRK2 (green). White arrows indicate co-localization of the three proteins (LRRK2, Rab7L1, TGN46) within a complex in the same cell. Scale bar is 10 μm. (C) To quantify these results, a high content imaging platform was used to count the number of cells with a white-spot that corresponds to the co-localization of LRRK2, Rab7L1 and TGN26 within an individual cell, in the indicated condition. The percent of cells that show co-localization are plotted. Data are from over 1000 cells assessed from six tissue culture wells from at least two independent experiments. Means are plotted with error bars showing ± SEM; significance was assessed by a one-way ANOVA with Tukey’s post hoc test. **P < 0.01, **P < 0.001.
Rab proteins function as poor LRRK2 kinase substrates in vitro
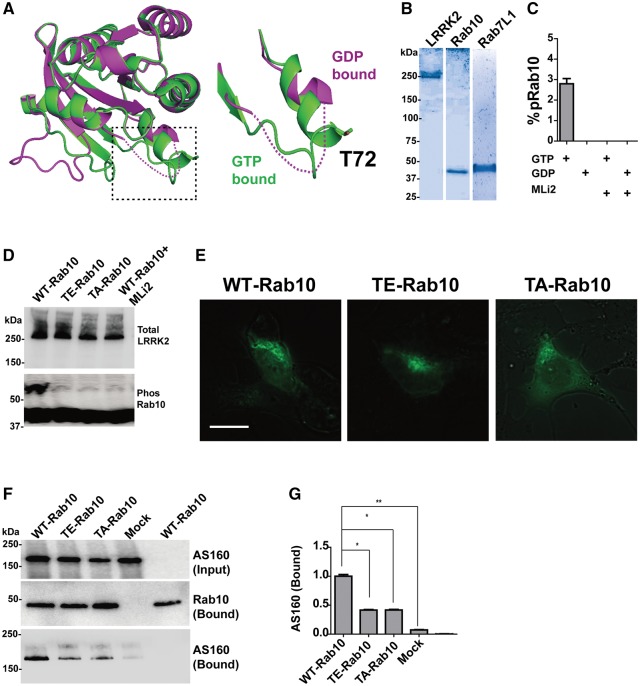
Recent work has demonstrated that Rab8/10 proteins, purified from e. coli., serve as exceptional LRRK2 substrates in vitro (16). While the crystal structures of Rab7L1 and GDP-bound Rab10 are not known, both GDP-bound (PDB ID: 4LHV) and GTP-bound Rab8 (PDB ID: 4LHW) crystal structures are solved (27). Structural alignment showed the switch II motif, where the LRRK2 dependent phosphorylation site Thr72 resides (Supplementary Material, Fig. S1), is highly unstructured in the GDP-bound form (Fig. 5A). In reasoning that an unstructured domain may poorly satisfy conformational requirements for efficient LRRK2 phosphorylation, we purified Rab10 and Rab7L1 from mammalian cells and bound the protein to a solid surface (FLAG-resin). In combining this form of Rab protein with recombinant full-length G2019S-LRRK2 protein, we could measure limited LRRK2-mediated phosphorylation of GTP-bound Rab10 and not GDP-Rab10 (Fig. 5B and C and Supplementary Material, Fig. S8). Only a small fraction of GTP-Rab10 could be phosphorylated by G2019S-LRRK2 (∼3%) in these conditions, and we were unable to visualize any phosphorylation of GTP or GDP-bound FLAG-Rab7L1 in vitro. These results highlight the possible requirements we demonstrated in cells of Rab prenylation and membrane interaction, as well as GTP-binding, for physiologically meaningful interaction with LRRK2.
Figure 5.
LRRK2 phosphorylation of GTP-bound Rab10 may decrease interaction with the GAP protein AS160. (A) Structural alignment of Rab8-GTP (Green) and Rab8-GDP (magenta). The unstructured switch II motif in the GDP-bound form is shown as dashed line. The Thr72 side chain is shown as sticks. (B) Assessment of purity of recombinant proteins isolated from HEK293 cells, for G2019S-LRRK2, WT-Rab10, and WT-Rab7L1, by SDS-PAGE followed by coomassie staining. All proteins showed >95% purity. (C) Calculated ratios of pRab10 from in vitro kinase assays in the presence of 500 μM of GTP, or GDP, with MLi2 (100 nM), as indicated. (D) Lysates from HEK293 cells co-transfected with LRRK2 and WT, TA, TD-Rab10, as indicated. MLi2 was included at 100 nM as indicated. Representative phos-tag analysis and immunoblots are shown. (E) Live cell imaging of SH-SY5Y cells. Wide-field fluorescence of the eGFP-tagged Rab10 construct is shown on a dark phase-contrast background, 16 h after transfection. (F) Cells over-expressing the Rab10 GAP AS160 or mock (empty vector) controls were lysed and incubated with bead-immobilized WT, TA or TD-Rab10. Complexes were analysed by immunoblots as shown and quantified (G) as column graphs depicting Rab10 interaction with Rab10 variants, relative to WT-Rab10. All data are averaged from three independent experiments or are representative of three independent experiments. Data are means ± SEM; significance assessed by one-way ANOVA with Tukey’s post hoc test. *P < 0.01, **P < 0.001.
LRRK2 phosphorylation of Rab10 blocks interaction with TBC1D4/AS160
As LRRK2 requires Rab8, Rab10, and Rab7L1 substrates to be membrane and GTP-bound for phosphorylation in cells, we next tested whether LRRK2-mediated Rab phosphorylation may affect interaction with GAP proteins that revert the GTP-membrane-bound state back to an inactive GDP-bound state. Although the identities of GAP proteins for Rab7L1 have not yet been described, TBC1D4/AS160 is a known critical GAP for Rab8 and Rab10 function (28–30). We first confirmed that all the LRRK2-mediated phosphorylation we could visualize in Rab10 was localized to the threonine 73 position as described (16). Accordingly, substitution of the LRRK2 phosphorylation site residue (Thr73) to either alanine (TA) or glutamic acid (TE) largely blocked LRRK2 phosphorylation, similar to the effects of the LRRK2 kinase inhibitor MLi2 (Fig. 5D). The phospho-mimic mutation T72E but not T72A increases the perinuclear localization of Rab10, suggesting phosphorylation could potentially change the localization of Rab10 (Fig. 5E). However, immunoprecipitation assays demonstrated that substitution of the switch II LRRK2 phosphorylation site blocked the interaction between Rab10 and TBC1D4/AS160 (Fig. 5F and G). These results suggest that LRRK2 modification of the critical Thr-switch II residue may block interaction with TBC1D4/AS160 and thus prolong a Rab membrane and GTP-bound state, with altered interaction with effector proteins.
Discussion
The impact of our results center on three novel observations. First, we define Rab7L1 protein as a LRRK2 kinase substrate in cells. All the pathogenic LRRK2 mutations we studied increased the ability of LRRK2 to phosphorylate the Thr71 residue in cells. The inclusion of Rab7L1 as one of the Rab proteins LRRK2 can phosphorylate is notable, because of the strong genetic association between the Rab7L1 gene locus and PD susceptibility. Second, using novel panels of mutated Rabs, we can show that LRRK2 cannot phosphorylate Rab GTPases in an inactive and cytosolic state, but phosphorylates them in a membrane and GTP-bound state. This has important implications because LRRK2 modification of switch II domains in Rab substrates can have opposing effects on Rab function due to modifying both GDI and GAP protein interactions. Since LRRK2 phosphorylates only the membrane and GTP-bound state, inhibition of GAP interaction would lead to the promotion of GTP-bound Rab protein. Third, we find that Rab7L1 promotes mutant LRRK2 recruitment to the trans-Golgi network, and in the process, promotes LRRK2 autophosphorylation. Proteins that regulate PD-linked LRRK2 kinase activity, especially those already linked to PD susceptibility through genetic studies, are of particular interest for therapeutic approaches.
Mutations in the LRRK2 gene represent one of the most common known heritable causes of neurodegeneration (31), and pathogenic mutations appear to increase LRRK2 kinase activities in both animal models and clinical samples (32–35). Early studies demonstrated that neurotoxicity requires LRRK2 kinase activity (36,37) and LRRK2 kinase activity results in the autophosphorylation of the LRRK2 Rab-like ROC domain (20,21,38). Mutating the most abundant LRRK2 autophosphorylation site, pSer1292, to an alanine reduces LRRK2 neurotoxicity, implicating autophosphorylation activities in LRRK2-linked pathogenic mechanisms (33). Herein, we show that Rab7L1 is a robust co-factor in regulating pS1292-LRRK2 protein levels. Rab7L1 can increase pS1292-LRRK2 whereas inactivated Rab7L1 can reduce membrane-bound pS1292-LRRK2 levels. Noticeably, WT-LRRK2 can only phosphorylate ∼4% Rab7L1 in our overexpression system. One possibility is that Rab7L1 is predominantly an upstream regulator of WT-LRRK2, with this paradigm partially reversing with the pathogenic LRRK2 mutations that affect a much larger proportion of Rab7L1 (up to ∼30% in our system). It will be important to further explore this relationship in different model systems as well as clinical samples and patient-derived cells. Blocking Rab7L1 expression, or expressing dominant-negative inactivated Rab7L1, may potently dampen LRRK2 kinase activities associated with pathogenic LRRK2 mutations, thereby presenting a novel strategy to block LRRK2 kinase activity.
Rab proteins are well known to regulate a diversity of membrane functions and vesicle trafficking activities. Few studies have explored the effects of phosphorylation modifications on Rab proteins. If LRRK2-mediated phosphorylation of Rab proteins begins with Rabs that are membrane-bound and GTP-activated, modification of the switch II domain that binds GAPs that inactivate the Rabs suggests that LRRK2 stabilizes membrane and GTP-bound Rabs (Fig. 6). However, the phosphorylation likely alters interaction with other effector proteins, opening the possibility GTP-bound inactive Rabs.
Figure 6.
Model for LRRK2 interaction with Rab protein substrates. An inactive GDP-bound Rab interacts with a Rab-escort protein (REP) that critically increases interaction with a RabGGTase resulting in prenylation (specifically geranylation) at one or two C-terminal cysteines, and increases membrane interaction. GEFs (guanine nucleotide exchange factors) facilitate the exchange of GDP for GTP, a process that would otherwise be very slow in most Rabs. In the GTP-bound active state on the membrane, the LRRK2 dimer is recruited and LRRK2 can then phosphorylate the Rab proteins. LRRK2-mediated phosphorylation may block the interaction of GAPs (GTPase-activating proteins) that would otherwise facilitate GTPase activity in GTP-bound Rabs. Without GAPs, intrinsic Rab GTPase activity is usually very slow. Therefore, LRRK2 potentiates the membrane and GTP-bound state of Rab substrates. In this process, LRRK2 is recruited to the membrane for interaction, such as the trans-Golgi network (TGN), and LRRK2 autophosphorylation activity is reciprocally stimulated, potentially affecting the phosphorylation of other membrane-bound Rabs.
Notably, intrinsic GTPase activity can vary orders-of-magnitude within the Rab family (22), so the effects of LRRK2 phosphorylation on different Rabs could likewise vary from negligible to critical depending on the Rab substrate. Theoretically, in Rab proteins that show higher-intrinsic GTPase activity, LRRK2 phosphorylation may block GDI-mediated membrane extraction. In a transient GDP-bound membrane state, further LRRK2 Rab phosphorylation would not be possible and equilibrium with phosphatases should eventually return the Rab to a GDI-bound cytosolic state. This scenario is compatible with our observation that we did not detect any robust depletions in the cytosolic pool of WT or even QL-mutated Rab proteins due to LRRK2 expression, suggesting LRRK2 is unlikely to cause the accumulation of membrane-bound inactive Rab8/10 and Rab7L1. However, this relationship between LRRK2 and Rab proteins could change depending on the abundance of GAPs, GDIs, phosphatases, and Rab7L1 levels that regulate LRRK2 kinase activity.
Despite LRRK2 interaction with numerous Rab protein substrates, through our comparative studies of the group I Rab proteins 8 and 10 with the group III Rab protein Rab7L1, we noticed a pronounced difference in the effect of the most common LRRK2 mutation G2019S. We could not detect significant difference between the pathogenic G2019S-LRRK2 protein from WT-LRRK2 protein in phosphorylating Rab8 and Rab10, whereas with Rab7L1 the increases were very clear. In addition, Rab7L1, but not Rab8 or Rab10, can activate LRRK2 protein and recruit LRRK2 to the TGN. Further, all pathogenic LRRK2 mutations increase the proportion of LRRK2 localized to the TGN. As LRRK2 protein kinase activity is now convincingly linked to PD (3), and numerous components of the TGN are implicated in PD (13), data overwhelmingly favor closer examinations of Rab7L1 in LRRK2-linked neurodegeneration.
Materials and Methods
DNA plasmids
N-terminal Myc-tagged LRRK2 constructs have been previously described (36). The LRRK2 mutation construct G2019S-S1292A was generated using Phusion Site-Directed Mutagenesis Kit (Life Technologies). To generate Rab10 expression plasmids, WT, Rab10 DNA sequence were synthesized and sub-cloned into pCDNA3.1GFP-N1 plasmid or pCDNA3.1-Flag plasmids from Genscript. Rab8 constructs are obtained from Addgene (Plasmids 86075, 86076, 86077) courtesy of the Lei Lu laboratory. pEGFP-Rab7L1 plasmids are generated as previously described (9). Novel mutations in the Rab expression constructs were all made with the Phusion Site-Directed Mutagenesis Kit (Life Technologies). The N-terminal Flag-tagged AS160 expression plasmid was a kind gift from Gustav Lienhard. All plasmids were purified with Qiagen Maxi-prep kits following manufacturer’s instructions.
Cell culture
HEK 293 (clone FT, Invitrogen) and SH-SY5Y (clone CRL-2266, ATCC) cells were maintained in 10% fetal bovine serum and DMEM. All cells were seeded at ∼60% confluency 24-h prior to transfections of plasmids using polyethylenimine at a 3: 2 ratio (w/v) with plasmids. Cells were harvested or imaged 16-h post-transfection. Cell imaging was accomplished with Zen Blue software (Carl Zeiss) on a Cell Observer with incubation and cool LED imaging. To separate lysates into soluble cytosolic fractions and membrane-enriched fractions, cells were scraped into lysisbuffer containing 150 mM NaCl, 50 mM Tris-HCl, pH 7.4, 10mM MgCl2 1x PhosSTOP and Protease inhibitor cocktails (Roche) followed by ultracentrifugation at 150, 000 x g for 20 min. Pellets were next resuspended into the same lysis buffer supplemented with 1% triton-x100.
Immunoblotting
Proteins and lysates were analysed via SDS-PAGE followed by transfer to PVDF membranes and analyses with Classico ECL reagent (Millipore) and signal recorded digitally on a Chemidoc Touch (BioRad). Intensities were calculated with the latest version of ImageLab software. The following antibodies were used: N241A/34 anti-LRRK2 (Antibodies Inc), MJFR-19–7-8 anti-pS1292-LRRK2 (Abcam), anti-eGFP antibody (Abcam), anti-FLAG M2 (Sigma), VDAC, β-actin, and GAPDH (Santa Cruz).
Phos-tag gel
Phosphoproteins were separated using a ‘phos-tag’ approach, as described (39) with acrylamide-gel matrices supplemented with 100 µM MnCl2 and 50 µM of Phos-Tag reagent (Wako Chemicals). For separation of GFP-Rabs and phospho-GFP-Rabs, 5% phos-tag gels were used. For separation of FLAG-Rab10 and phospho-FLAG-Rab10, 10% gel was used. Gels were run at 100 V for 1.5 h in 25 mM Tris, 192 mM glycine, 0.1% SDS running buffer, followed by 2x washing in running buffer supplemented with 5 mM EDTA before transfer to PVDF (Immobilon-FL) membrane at 35 V for 10 h.
Protein purification
N-terminal Flag-Rab10 and Flag-Rab7L1 plasmids were transfected to HEK293 cells as described and harvested into lysis buffer (described above) supplemented with 100 μM GDP (Sigma). Lysates were cleared by centrifugation at 50,000 × g and incubated with M1-Flag-resin (Sigma). Resins were washed five times with lysis buffer and eluted with the same lysis buffer supplemented with 100 μg ml−1 3x Flag-peptide (Sigma). Recombinant full-length N-terminal FLAG-LRRK2 protein was purchased from Invitrogen. All protein purities were assessed by coomassie blue staining PAGE analysis and were all above >95% purity as assessed by ImageLab software (BioRad).
Immunoprecipitations
For Rab10-GAP interaction experiments, anti-GFP antibody resin loaded with eGFP-tagged WT, T73A, and T73D-Rab10 were incubated with lysates containing AS160 protein. After incubation for 4 h at 4 °C, resins were washed five times in lysis buffer and eluate analysed via immunoblot as above. In all immunoprecipitation experiments, anti-eGFP resin with immobilized Rab10 or Rab7L1 were used as negative controls to assess non-specific binding.
In vitro kinase assay
25 nM full-length G2019S-LRRK2 protein (Life Technologies) was incubated with 100 nM recombinant Flag-Rab10 or Flag-Rab7L1 in a kinase reaction buffer containing 150 mM NaCl, 50 mM Tris-HCl, pH 7.4, 10 mM MgCl2, and 1 mM ATP, in the presence of 500 μM of GTP or GDP as indicated, with or without MLi2 compound at 100 nM (synthesized in-house). Reactions were allowed to proceed for 1 h at 30 °C prior to termination.
Trans-Golgi localization assay
Rab7L1-induced LRRK2 recruitment to the trans-Golgi network was measured using an automated assay as previously described (40). Briefly, transiently 3x-flag LRRK2/2x-myc Rab7L1 transfected HEK293 cells were fixed and stained 24-h post-transfection for FLAG (LRRK2), Myc (Rab7L1) and TGN46. Transfected cells were imaged and quantified on a Cellomics VTI high content imager using Spot Detector bioapplication for % of cells with LRRK2 and TGN46 positive spots from total number of LRRK2 transfected cells. At least 1000 cells in each of 6-independent wells were measured for each condition, and this was repeated at least twice. Cells were also imaged on a Zeiss LSM880 confocal microscope.
Statistics
Analyses were performed in GraphPad Prism 5.0. Graphs were generated in GraphPad Prism 5.0 and arranged in Adobe Illustrator 9.0. All statistical tests are reported in figure legends.
Supplementary Material
Supplementary Material is available at HMG online.
Conflict of Interest statement. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could potentially cause any conflict of interest.
Funding
Michael J. Fox Foundation for Parkinson’s Disease research, National Institutes of Health/National Institute of Neurological Disorders and Stroke grants [R01 NS064934, R21 NS097643], Intramural Research Program of the National Institutes of Health, National Institute on Aging [AG000948, AG000937].
Supplementary Material
References
- 1. Paisan-Ruiz C., Jain S., Evans E.W., Gilks W.P., Simon J., van der Brug M., Lopez de Munain A., Aparicio S., Gil A.M., Khan N. (2004) Cloning of the gene containing mutations that cause PARK8-linked Parkinson's disease. Neuron, 44, 595–600. [DOI] [PubMed] [Google Scholar]
- 2. Zimprich A., Biskup S., Leitner P., Lichtner P., Farrer M., Lincoln S., Kachergus J., Hulihan M., Uitti R.J., Calne D.B.. et al. (2004) Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron, 44, 601–607. 10.1016/j.neuron.2004.11.005 [DOI] [PubMed] [Google Scholar]
- 3. West A.B. (2017) Achieving neuroprotection with LRRK2 kinase inhibitors in Parkinson disease. Exp Neurol, 298(Pt B), 236–245. 10.1016/j.expneurol.2017.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nalls M.A., Pankratz N., Lill C.M., Do C.B., Hernandez D.G., Saad M., DeStefano A.L., Kara E., Bras J., Sharma M.. et al. (2014) Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson's disease. Nat. Genet., 46, 989–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Trabzuni D., Ryten M., Emmett W., Ramasamy A., Lackner K.J., Zeller T., Walker R., Smith C., Lewis P.A., Mamais A.. et al. (2013) Fine-mapping, gene expression and splicing analysis of the disease associated LRRK2 locus. PLoS One, 8, e70724.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. West A.B. (2015) Ten years and counting: moving leucine-rich repeat kinase 2 inhibitors to the clinic. Mov. Disord., 30, 180–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Alegre-Abarrategui J., Christian H., Lufino M.M., Mutihac R., Venda L.L., Ansorge O., Wade-Martins R. (2009) LRRK2 regulates autophagic activity and localizes to specific membrane microdomains in a novel human genomic reporter cellular model. Hum. Mol. Genet., 18, 4022–4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Biskup S., Moore D.J., Celsi F., Higashi S., West A.B., Andrabi S.A., Kurkinen K., Yu S.W., Savitt J.M., Waldvogel H.J.. et al. (2006) Localization of LRRK2 to membranous and vesicular structures in mammalian brain. Ann. Neurol., 60, 557–569. [DOI] [PubMed] [Google Scholar]
- 9. MacLeod D.A., Rhinn H., Kuwahara T., Zolin A., Di Paolo G., McCabe B.D., Marder K.S., Honig L.S., Clark L.N., Small S.A.. et al. (2013) RAB7L1 interacts with LRRK2 to modify intraneuronal protein sorting and Parkinson's disease risk. Neuron, 77, 425–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boddu R., Hull T.D., Bolisetty S., Hu X., Moehle M.S., Daher J.P., Kamal A.I., Joseph R., George J.F., Agarwal A.. et al. (2015) Leucine-rich repeat kinase 2 deficiency is protective in rhabdomyolysis-induced kidney injury. Hum. Mol. Genet., 24, 4078–4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Herzig M.C., Kolly C., Persohn E., Theil D., Schweizer T., Hafner T., Stemmelen C., Troxler T.J., Schmid P., Danner S.. et al. (2011) LRRK2 protein levels are determined by kinase function and are crucial for kidney and lung homeostasis in mice. Hum. Mol. Genet., 20, 4209–4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tong Y., Yamaguchi H., Giaime E., Boyle S., Kopan R., Kelleher R.J. 3rd, Shen J. (2010) Loss of leucine-rich repeat kinase 2 causes impairment of protein degradation pathways, accumulation of alpha-synuclein, and apoptotic cell death in aged mice. Proc. Natl Acad. Sci. U S A, 107, 9879–9884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Beilina A., Rudenko I.N., Kaganovich A., Civiero L., Chau H., Kalia S.K., Kalia L.V., Lobbestael E., Chia R., Ndukwe K.. et al. (2014) Unbiased screen for interactors of leucine-rich repeat kinase 2 supports a common pathway for sporadic and familial Parkinson disease. Proc. Natl Acad. Sci. U S A, 111, 2626–2631. 10.1073/pnas.1318306111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kuwahara T., Inoue K., D’Agati V.D., Fujimoto T., Eguchi T., Saha S., Wolozin B., Iwatsubo T., Abeliovich A. (2016) LRRK2 and RAB7L1 coordinately regulate axonal morphology and lysosome integrity in diverse cellular contexts. Sci. Rep., 6, 29945.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hutagalung A.H., Novick P.J. (2011) Role of Rab GTPases in membrane traffic and cell physiology. Physiol. Rev., 91, 119–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Steger M., Tonelli F., Ito G., Davies P., Trost M., Vetter M., Wachter S., Lorentzen E., Duddy G., Wilson S.. et al. (2016) Phosphoproteomics reveals that Parkinson's disease kinase LRRK2 regulates a subset of Rab GTPases. Elife, 29, 5, pii: e12813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Thirstrup K., Dachsel J.C., Oppermann F.S., Williamson D.S., Smith G.P., Fog K., Christensen K.V. (2017) Selective LRRK2 kinase inhibition reduces phosphorylation of endogenous Rab10 and Rab12 in human peripheral mononuclear blood cells. Sci. Rep., 7, 10300.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ito G., Katsemonova K., Tonelli F., Lis P., Baptista M.A., Shpiro N., Duddy G., Wilson S., Ho P.W., Ho S.L.. et al. (2016) Phos-tag analysis of Rab10 phosphorylation by LRRK2: a powerful assay for assessing kinase function and inhibitors. Biochem. J., 473, 2671–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jaleel M., Nichols R.J., Deak M., Campbell D.G., Gillardon F., Knebel A., Alessi D.R. (2007) LRRK2 phosphorylates moesin at threonine-558: characterization of how Parkinson's disease mutants affect kinase activity. Biochem. J., 405, 307–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Greggio E., Taymans J.M., Zhen E.Y., Ryder J., Vancraenenbroeck R., Beilina A., Sun P., Deng J., Jaffe H., Baekelandt V.. et al. (2009) The Parkinson's disease kinase LRRK2 autophosphorylates its GTPase domain at multiple sites. Biochem. Biophys. Res. Commun., 389, 449–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Webber P.J., Smith A.D., Sen S., Renfrow M.B., Mobley J.A., West A.B. (2011) Autophosphorylation in the leucine-rich repeat kinase 2 (LRRK2) GTPase domain modifies kinase and GTP-binding activities. J. Mol. Biol., 412, 94–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu Z., Mobley J.A., DeLucas L.J., Kahn R.A., West A.B. (2016) LRRK2 autophosphorylation enhances its GTPase activity. faseb J., 30, 336–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pfeffer S.R. (2005) Structural clues to Rab GTPase functional diversity. J. Biol. Chem., 280, 15485–15488. [DOI] [PubMed] [Google Scholar]
- 24. Berger Z., Smith K.A., Lavoie M.J. (2010) Membrane localization of LRRK2 is associated with increased formation of the highly active LRRK2 dimer and changes in its phosphorylation. Biochemistry, 49, 5511–5523. 10.1021/bi100157u [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee M.T., Mishra A., Lambright D.G. (2009) Structural mechanisms for regulation of membrane traffic by rab GTPases. Traffic, 10, 1377–1389. 10.1111/j.1600-0854.2009.00942.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schapansky J., Nardozzi J.D., Felizia F., LaVoie M.J. (2014) Membrane recruitment of endogenous LRRK2 precedes its potent regulation of autophagy. Hum. Mol. Genet., 23, 4201–4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guo Z., Hou X., Goody R.S., Itzen A. (2013) Intermediates in the guanine nucleotide exchange reaction of Rab8 protein catalyzed by guanine nucleotide exchange factors Rabin8 and GRAB. J. Biol. Chem., 288, 32466–32474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sano H., Eguez L., Teruel M.N., Fukuda M., Chuang T.D., Chavez J.A., Lienhard G.E., McGraw T.E. (2007) Rab10, a target of the AS160 Rab GAP, is required for insulin-stimulated translocation of GLUT4 to the adipocyte plasma membrane. Cell Metab., 5, 293–303. [DOI] [PubMed] [Google Scholar]
- 29. Ishikura S., Bilan P.J., Klip A. (2007) Rabs 8A and 14 are targets of the insulin-regulated Rab-GAP AS160 regulating GLUT4 traffic in muscle cells. Biochem. Biophys. Res. Commun., 353, 1074–1079. [DOI] [PubMed] [Google Scholar]
- 30. Larance M., Ramm G., Stockli J., van Dam E.M., Winata S., Wasinger V., Simpson F., Graham M., Junutula J.R., Guilhaus M.. et al. (2005) Characterization of the role of the Rab GTPase-activating protein AS160 in insulin-regulated GLUT4 trafficking. J. Biol. Chem., 280, 37803–37813. [DOI] [PubMed] [Google Scholar]
- 31. Trinh J., Guella I., Farrer M.J. (2014) Disease penetrance of late-onset parkinsonism: a meta-analysis. JAMA Neurol., 71, 1535–1539. [DOI] [PubMed] [Google Scholar]
- 32. Fraser K.B., Moehle M.S., Alcalay R.N., West A.B. and Consortium, L.C. (2016) Urinary LRRK2 phosphorylation predicts parkinsonian phenotypes in G2019S LRRK2 carriers. Neurology, 86, 994–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sheng Z., Zhang S., Bustos D., Kleinheinz T., Le Pichon C.E., Dominguez S.L., Solanoy H.O., Drummond J., Zhang X., Ding X.. et al. (2012) Ser1292 autophosphorylation is an indicator of LRRK2 kinase activity and contributes to the cellular effects of PD mutations. Sci. Transl. Med., 4, 164ra161.. [DOI] [PubMed] [Google Scholar]
- 34. West A.B., Moore D.J., Choi C., Andrabi S.A., Li X., Dikeman D., Biskup S., Zhang Z., Lim K.L., Dawson V.L.. et al. (2007) Parkinson's disease-associated mutations in LRRK2 link enhanced GTP-binding and kinase activities to neuronal toxicity. Hum. Mol. Genet., 16, 223–232. [DOI] [PubMed] [Google Scholar]
- 35. West A.B., Moore D.J., Biskup S., Bugayenko A., Smith W.W., Ross C.A., Dawson V.L., Dawson T.M. (2005) Parkinson's disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc. Natl. Acad. Sci. U S A, 102, 16842–16847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Greggio E., Jain S., Kingsbury A., Bandopadhyay R., Lewis P., Kaganovich A., van der Brug M.P., Beilina A., Blackinton J., Thomas K.J.. et al. (2006) Kinase activity is required for the toxic effects of mutant LRRK2/dardarin. Neurobiol. Dis., 23, 329–341. [DOI] [PubMed] [Google Scholar]
- 37. MacLeod D., Dowman J., Hammond R., Leete T., Inoue K., Abeliovich A. (2006) The familial Parkinsonism gene LRRK2 regulates neurite process morphology. Neuron, 52, 587–593. [DOI] [PubMed] [Google Scholar]
- 38. Gloeckner C.J., Boldt K., von Zweydorf F., Helm S., Wiesent L., Sarioglu H., Ueffing M. (2010) Phosphopeptide analysis reveals two discrete clusters of phosphorylation in the N-terminus and the Roc domain of the Parkinson-disease associated protein kinase LRRK2. J. Proteome Res., 9, 1738–1745. [DOI] [PubMed] [Google Scholar]
- 39. Kinoshita E., Kinoshita-Kikuta E., Takiyama K., Koike T. (2006) Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol. Cell Proteomics, 5, 749–757. [DOI] [PubMed] [Google Scholar]
- 40. Chia R., Haddock S., Beilina A., Rudenko I.N., Mamais A., Kaganovich A., Li Y., Kumaran R., Nalls M.A., Cookson M.R. (2014) Phosphorylation of LRRK2 by casein kinase 1alpha regulates trans-Golgi clustering via differential interaction with ARHGEF7. Nat. Commun., 5, 5827.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.