Summary
Non-coding regions comprise most of the human genome and harbor a significant fraction of risk alleles for neuropsychiatric diseases, yet their functions remain poorly defined. We created a high-resolution map of non-coding elements involved in human cortical neurogenesis by contrasting chromatin accessibility and gene expression in the germinal zone and cortical plate of the developing cerebral cortex. We link distal regulatory elements (DREs) to their cognate gene(s) together with chromatin interaction data and show that target genes of human-gained enhancers (HGEs) regulate cortical neurogenesis and are enriched in outer radial glia, a cell-type linked to human cortical evolution. We experimentally validate the regulatory effects of predicted enhancers for FGFR2 and EOMES. We observe that common genetic variants associated with educational attainment, risk for neuropsychiatric disease, and intracranial volume are enriched within regulatory elements involved in cortical neurogenesis, demonstrating the importance of this early developmental process for adult human cognitive function.
Graphical abstract
A high-resolution map of non-coding regulatory elements driving human cortical neurogenesis reveals uniquely human enhancers linked to common genetic variants associated with cognitive function.

Introduction
The highly structured mammalian cerebral cortical wall is divided into laminae enriched for distinct cell-types and functions during cortical neurogenesis (Bystron et al., 2008; Lui et al., 2011). Decades of comparative cross-species studies reveal that cerebral cortical expansion in primates plays a major role in the evolution of human cognition (Krasnegor et al., 1997). Yet, we are just beginning to understand the molecular and cellular basis of this dramatic process (Bae et al., 2015; Lui et al., 2011; Sun and Hevner, 2014; Taverna et al., 2014). Since gene regulation plays a predominant role in human brain evolution (Varki et al., 2008), acquiring a more complete understanding of the gene regulatory mechanisms that orchestrate neurogenesis, which is central to the expansion of the primate neocortex (Rakic, 2009), will allow insights into molecular mechanisms generating human cognitive capabilities and their dysregulation in neuropsychiatric disease (Bae et al., 2015; Geschwind and Rakic, 2013).
Gene expression programs in the developing human cortex are beginning to be defined (Kang et al., 2011; Miller et al., 2014; Pollen et al., 2015). Yet, compared with mouse, non-coding regions regulating gene expression in human corticogenesis are less well defined (Gray et al., 2017; Visel et al., 2013). Characterizing these regions is of great importance since most human specific genetic variation and risk loci for neuropsychiatric diseases are found in these likely regulatory, non-coding regions (Ward and Kellis, 2012). Enhancers and promoters are often found in open or accessible regions of chromatin where DNA-binding proteins like transcription factors (TFs) preferentially bind (Nord et al., 2015). Regions of accessible chromatin in bulk human fetal and adult brain have been identified using DNase-seq (Roadmap Epigenomics et al., 2015), but not with a spatial resolution permitting identification of regulatory elements involved in human cortical neurogenesis. Chromatin regulators and TFs are key regulators of neuronal differentiation and function, further highlighting the need to define the cis-regulatory elements governing corticogenesis (Nord et al., 2015; Ronan et al., 2013).
Here, we perform ATAC-seq, a high-throughput method to profile accessible chromatin (Buenrostro et al., 2013), on the germinal zone and subplate/cortical plate in developing human brain, and by integrating these data with chromatin interaction measured via Hi-C, we identify DREs of key genes involved in cortical neurogenesis, including HGEs. We show that target genes of HGEs regulate neural progenitor biology and are enriched in genes specifically expressed in outer radial glia (oRG), a cell population that is notably increased in humans and postulated to be critical for the cerebral cortical expansion in primates and other gyrencephalic species (Lui et al., 2011; Pollen et al., 2015; Taverna et al., 2014). Our data also suggest that gene regulation in neural progenitors and early-born neurons affects individual variation in adult cognitive function and risk for neuropsychiatric disease. This firmly connects prenatal cortical development to adult cognitive function and later-onset neuropsychiatric disease risk.
Results
Identifying differentially accessible (DA) regions involved in neurogenesis
To obtain a high resolution depiction of chromatin structure (ATAC-seq) and gene expression (RNAseq) in developing human fetal cortex, we dissected post-conception week (PCW) 15-17 human neocortex (STAR*Methods) into two major anatomical divisions: (1) GZ: the neural progenitor-enriched region encompassing the ventricular zone (VZ), subventricular zone (SVZ), and intermediate zone (IZ) and (2) CP: the neuron-enriched region containing the subplate (SP), cortical plate (CP), and marginal zone (MZ) (Figure 1A; STAR*Methods). Comparison of gene expression between dissected regions with a gold standard human cortex laminar transcriptomic atlas (Miller et al., 2014) revealed that differential gene expression (DE) between GZ and CP best matched the transitions between either the VZ or SVZ and CP in vivo with very high correlation (Figures S1A-B) (SVZ vs CP; r=0.84; P<2×10-16), validating the dissection and tissue isolation strategy. Following ATAC-seq QC measures (Figures S1C-G), we took a conservative approach to peak identification and quantification (STAR*Methods, Figures 1B and S1H-L), finding high correlation between replicates and published DNAse-seq from brain (Roadmap Epigenomics et al., 2015), but less so with lung (Figure S2A-D), demonstrating tissue specificity. Samples clustered first by tissue type (GZ/CP) and next by donor, similar to clustering by gene expression (Figures 1C-D), further demonstrating that the major differences in chromatin accessibility and gene expression are driven by tissue type and that replicates are highly concordant (Figures S2A-B and S3A).
Figure 1.

Defining regions of differential chromatin accessibility in developing human neocortex.
(A) Left: Brightfield coronal section from PCW 15 neocortex (CTX) showing the dissected regions processed for ATAC-seq, RNA-seq, and Hi-C: cortical plate (CP) and germinal zone (GZ). Right: DAPI staining of a coronal section showing the dissected lamina: GZ encompasses the ventricular zone (VZ), inner and outer subventricular zone (I/OSVZ), and intermediate zone (IZ); and CP encompasses the subplate (SP), cortical plate (CP) and marginal zone (MZ).
(B) DA peaks (highlighted in sky blue) above coverage maps of normalized reads from ATAC-seq are shown near the PAX6 gene on chromosome 11 along with chromatin states derived from fetal tissue.
(C) Hierarchical clustering based on ATAC-seq genome-wide normalized reads within peaks shows high technical reproducibility and strong biological differences between GZ and CP.
(D) Hierarchical clustering based on RNA-seq transcriptome-wide normalized reads within genes shows similar results as chromatin accessibility clustering.
(E) Fold enrichment of DA peaks within defined regions of the genome shows strong enrichment in regions near promoters (TSS, Promoter, 5′ UTR).
(F) Fold enrichment of DA peaks in fetal brain derived chromatin states shows enrichment in enhancer and promoter regions.
(G) Correlation between effect sizes of significantly DA promoters and significantly DE exons closest to those promoters show a variable but positive correlation between chromatin accessibility at the promoter and expression for both protein-coding genes and lncRNAs. Chromatin states are defined in STAR*Methods. See also Figures S1-3 and Table S1.
To define functional regulatory elements related to neurogenesis, we identified 19,260 peaks with greater accessibility in GZ than CP (GZ>CP peaks; STAR*Methods) and 17,803 peaks with greater accessibility in CP than GZ (CP>GZ peaks; Table S1). DA peaks are significantly enriched in regions of the genome with known functionality including promoters (+/- 1kb from transcription start site [TSS]; 5′ untranslated region [UTR]), and nominally depleted in regions with poorly annotated functionality (intergenic regions; STAR*Methods) (Figures 1E and S3B). Near TSSs, DA peaks are also found at lncRNA, miRNA, and rRNA loci, in addition to protein-coding genes (Figures 1E and S3C; see Table S1 for complete list of proximal regulatory elements for ncRNAs). DA peaks are enriched in previously annotated chromatin states, including promoters and enhancers (Roadmap Epigenomics et al., 2015), and in regions containing validated human forebrain enhancers (Visel et al., 2007) (Figure 1F), but depleted heterochromatin, quiescent and actively transcribed regions of the genome (Figures 1F and S3D).
Gene expression has a variable functional relationship to accessibility at the TSS
Previous work predicts a significant relationship between chromatin accessibility and gene expression (Frank et al., 2015), although the correlations are often weak, since many additional factors, including TF binding, DNA methylation, histone tail modifications, and miRNA regulate steady-state transcript levels of a gene (Heintzman and Ren, 2007; Natarajan et al., 2012). To assess this relationship, we first visualized peaks of chromatin accessibility at genes known to be involved in neurogenesis. As expected, SOX2, a TF expressed in radial glia, has both higher chromatin accessibility at the promoter and expression in GZ, whereas HOMER1, a component of the post-synaptic density expressed in post-mitotic neurons, shows the converse pattern (Figure 2A). We observed a highly significant relationship between chromatin accessibility and gene expression across a set of marker genes known to be involved in neurodevelopment (Figure 2B), and across a less biased set of the top 240 DE genes between GZ and CP (Figure 2C; STAR*Methods). Genome-wide, we found that accessibility at the TSS has a highly significant, but not perfect, correlation with expression at protein-coding genes (r = 0.417; P=6.2×10-60) and lncRNA (r=0.454; P=0.0017) (Figure 1G; STAR*Methods), consistent with prevailing models of transcriptional regulation (Heintzman and Ren, 2007; Natarajan et al., 2012).
Figure 2.
The relationship between DA chromatin at promoter regions with gene expression and biological pathways.
(A) Chromatin accessibility and gene expression coverage maps for genes expressed in neural progenitor cells (SOX2) or differentiated neurons (HOMER1) show the expected patterns of chromatin accessibility and expression (GZ>CP for SOX2 and CP>GZ for HOMER1).
(B) Heatmaps of chromatin accessibility and expression of the exon closest to the promoter for selected genes canonically involved in neural development across samples show a variable but positive correlation between chromatin accessibility and gene expression.
(C) A similar relationship between chromatin accessibility and gene expression is observed when selecting the top 240 DE genes.
(D) Genes with DA promoters fall into known functional categories related to neural development as determined by GO analysis. See also Figure S4.
We next assessed the biological pathways revealed by DA of chromatin at protein-coding TSS, observing an enrichment of gene ontology (GO) terms related to neurogenesis and early neuronal morphogenesis in GZ>CP peaks (Figure 2D). Conversely, enrichment of terms related to neuronal physiology and synapses were found in CP>GZ peaks. Similar GO enrichment was obtained in GZ>CP and CP>GZ DE genes (Figure S4A). KEGG pathway analysis of DA sites found enrichment in Notch and Wnt signaling pathways in GZ>CP sites, in contrast with neuroactive ligand-receptor interaction and calcium signaling pathways found in CP>GZ sites (Figure S4A-B). These terms are consistent with both the expected cellular composition in regions containing neural progenitors versus neurons, as well as the correlation between chromatin accessibility and gene expression.
Defining DREs regulating cortical neurogenesis
We next sought to define DREs involved in neurogenesis for each gene. Although regulatory elements are often assumed to regulate the closest gene, this is an overly simplifying assumption (Amano et al., 2009). Therefore, we created a mapping between chromatin accessibility peaks and their cognate genes (Figure 3A; STAR*Methods), which is predicted to represent functional enhancer-promoter interactions that regulate gene expression during corticogenesis. To avoid spurious correlations, we also required evidence of a physical interaction between the two correlated regions, as measured by chromatin capture via Hi-C from matching stages and tissue type (Won et al., 2016). Consistently, the inter-peak correlation was significantly higher between ATAC-seq peak pairs with evidence of interaction by Hi-C than in those not supported by Hi-C (P=1.1×10-100 for GZ>CP peaks with Hi-C data from GZ tissue; P=3.22×10-34 for CP>GZ peaks with Hi-C data from CP tissue) (Figure 3B). Chromatin accessibility at these DREs and gene expression of predicted targets displayed a positive correlation (r=0.372; P<5×10-324), which was substantially stronger between peaks with evidence of interaction by Hi-C (r=0.456, P=6.64×10-45; difference between correlations P=0.0017, Fisher's method), suggesting that integrating both chromatin accessibility and chromatin contact data reduces false positives when linking enhancers and their target genes (Figure 3C).
Figure 3.

Mapping DREs involved in cortical neurogenesis to their cognate genes.
(A) A schematic showing chromatin accessibility correlation and chromatin interaction assays used to map DREs to their cognate genes.
(B) Distal ATAC-seq peaks show on average a higher correlation to promoter peaks when supported by chromatin interaction derived from their cognate GZ or CP tissue. GZ>CP and CP>GZ peaks show significantly higher peak correlation when supported by GZ or CP Hi-C data as compared to the absence of Hi-C support, respectively (GZ: P=1.1×10-100, CP: P=3.22×10-34).
(C) The correlation between DREs accessibility and gene expression of their cognate gene shows a stronger relationship when the regulatory element is mapped using both chromatin accessibility correlation and interaction rather than chromatin accessibility correlation alone.
(D) DA DREs are mapped onto genes supported by both chromatin accessibility correlation and chromatin interaction that fall into known biological pathways involved in neural development. See also Table S2.
(E) An example of DRE mapping is provided at the EOMES locus, where DREs are supported by both chromatin interaction via Hi-C in GZ tissue and chromatin accessibility correlation. The location of a chromosomal breakpoint for a balanced chromosomal translocation that leads to complete absence of EOMES expression and microcephaly is shown (Baala et al., 2007).
(F) Three distinct sgRNA pairs were designed to excise the EOMES enhancer. Overlap of ATAC-seq differential peak and predicted VISTA forebrain enhancer is shown. Primers used to validate genomic deletions are represented by green half arrows.
(G) Schematic of functional validation of EOMES enhancer. sgRNA pairs flanking the EOMES enhancer were delivered along with P2A-linked GFP or RFP Cas9 via lentiviral infection into phNPCs. Following 3 wks of differentiation, cells containing both sgRNAs were selected via FACS and probed for enhancer excision and EOMES expression.
(H) Genomic PCR of the 1.8kb region containing the EOMES enhancer in control or CRISPR/Cas9 + sgRNA infected phNPCs. Red arrow points to the expected 2107bp band in controls and white arrows point to expected products following excision (see 3F).
(I) Excision of the EOMES enhancer led to a 72-77% reduction in EOMES expression as measured by qPCR in phNPCs (***P<0.0001, **P<0.001, *P<0.01, ANOVA, with Bonferroni post-hoc test, n=4, mean ± SEM shown).
(J) VISTA predicted EOMES enhancer reporter (LacZ staining) (https://enhancer.lbl.gov/) and RNA in situ hybridization for EOMES (http://developingmouse.brain-map.org/) at E11.5. Reporter signal and EOMES RNA expression display a similar pattern with enrichment in the telencephalon. See also Table S4 and Figure S5.
GO analysis demonstrated enrichment in critical processes known to drive corticogenesis in a manner consistent with the dissected regions (Figure 3D). GZ>CP distal enhancers regulate genes enriched in processes including glial cell differentiation, regulation of neuron differentiation, positive regulation of neuroblast proliferation and regulation of transcription, whereas CP>GZ distal enhancers targets are enriched in glutamate receptor activity, behavior-related categories, axon guidance and neuron projection morphogenesis (Figure 3D). Further, the expression of gene targets of GZ>CP distal enhancers were most enriched in the progenitor-enriched VZ, iSVZ, oSVZ, whereas gene targets of CP>GZ distal enhancers were enriched in the CP and MZ, as expected (Figure S5A). Together, these studies define for the first time a set of DREs, comprised primarily of enhancers (Figures 1F and S3D), which influence gene expression during human neurogenesis. We provide a list of all of the regulatory regions identified in this manner and their predicted target genes in Table S2.
To demonstrate the functional impact of these data, we identified five regulatory elements supported by both ATAC-seq and Hi-C, 264-370kb 3′ to the EOMES gene, a TF that regulates cortical neuron production and cortical thickness via effects on progenitor proliferation (Arnold et al., 2008; Sessa et al., 2008). We found a reported case of a rare homozygous chromosomal translocation 215kb upstream of the TSS of EOMES (46,XY,t(3;10)(p24;q23)2×) that causes microcephaly, polymicrogyria, and agenesis of the corpus callosum (Baala et al., 2007). When this translocation is homozygous, it results in a complete absence of EOMES expression, suggesting that regulatory elements upstream of the translocation breakpoint (chr3:2,7979,020) strongly regulate EOMES expression. Remarkably, this translocation removes all five ATAC-seq and Hi-C defined regulatory elements from chromosome 3 (Figure 3E), separating these putative enhancers from the gene. As further evidence of the enhancer functionality of these regions, one locus (chr3:28033829-28035751; hs1557) (Figure 3F) was shown to drive forebrain specific expression in mouse embryos, as predicted (Figure 3J). To experimentally confirm its enhancer activity, we engineered three partial and complete deletions of the EOMES enhancer overlapping the predicted VISTA EOMES enhancer in primary human neural progenitors (phNPCs), a well-established model system of neural development (Stein et al., 2014) (Figures 3F-G). All five putative enhancer peaks are preserved in phNPCs and display the expected pattern of decreased accessibility with differentiation, matching the in vivo GZ-CP transition and decreased EOMES expression (Figures S5B-G). We observe that all three different deletions within the predicted EOMES enhancer led to a 72-77% reduction in EOMES expression in phNPCs (Figures 3H-I), but did not change the expression of the closest genes CMC1 and AZI2, which lack combined evidence of chromatin accessibility correlation and interaction (Figure S5H and Table S2). These data provide a likely genetic mechanism for this disorder, based on inactivation of the activity of a regulatory element on its target gene, rather than deletion of the protein-coding gene itself.
Identifying TFs driving neurogenesis
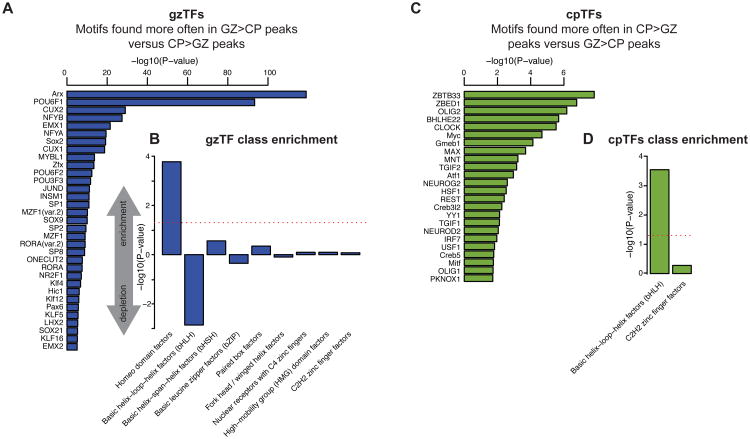
Since TFs are key drivers of gene expression and cell fate determination (Guillemot, 2007; Nord et al., 2015), we hypothesized that TF motifs found more often in GZ accessible regions would be enriched in TFs known to promote neural progenitor maintenance and proliferation (gzTFs), whilst those motifs found more often in CP accessible regions would be enriched in TFs involved in neurogenesis and neuronal maturation (cpTFs). Observing such coordination would permit identification of TFs driving human cortical neurogenesis, while providing validation of the functional relevance of chromatin accessibility. As a proof of principle, we conducted a differential motif enrichment analysis between fibroblasts and human embryonic stem cells (hESCs) using a logistic regression framework (STAR*Methods), identifying the major known pluripotency-associated transcription factors (Buganim et al., 2013), demonstrating the validity of this approach (Figure S4C).
We then applied this analysis to our data, identifying 97 gzTFs and 26 cpTFs (FDR adjusted P<0.05; Figure 4; Table S3). gzTFs included SOX2 and PAX6, canonical markers of radial glia known to regulate neural stem cell self-renewal (Estivill-Torrus et al., 2002; Graham et al., 2003) and several TFs known to be required for neural progenitor expansion or proliferation in developing cortex, including ARX, EMX1/2, LHX2, KLF4, NR2F1 and SP2 (Liang et al., 2013; Qin and Zhang, 2012; Rubenstein and Rakic, 2013) (Figure 4A). Notably, gzTFs as a group were enriched in the homeodomain class of TF, which are implicated in neural stem cell patterning and neural progenitor fate specification (Guillemot, 2007) (Figure 4B).
Figure 4.
Predicting TFs involved in neural progenitor proliferation and neurogenesis.
(A,C) TFs with significant differential enrichment of conserved motifs in DA peaks. The statistical test identifies TFs likely involved in neural progenitor proliferation and maintenance (gzTFs; A) or neurogenesis and maturation (cpTFs; C). An abridged list is shown here for clarity, and the full list can be found in Table S3.
(B,D) Enrichment of TF classes for gzTFs (B) or cpTFs (D). See also Figure S4.
Conversely, cpTFs were enriched in pro-neural basic helix-loop-helix (bHLH) factors including NEUROG2 and NEUROD2, which promote neuronal differentiation (Guillemot, 2007), whereas gzTFs were depleted in this class (Figures 4C-D). We also identified REST, a canonical repressor of neuronal gene transcription (Ballas et al., 2005). Of all identified TFs, 68% of gzTFs and 88% of cpTFs were DE between GZ and CP, confirming their dynamic regulation (FDR adjusted P<0.05). Together, these results demonstrate that differential motif enrichment analysis in DA chromatin identifies TFs known to be involved in neural stem cell proliferation and neural differentiation and dozens of additional TFs (Table S3) that were previously unknown to be involved in these processes.
HGEs regulate neural progenitor biology
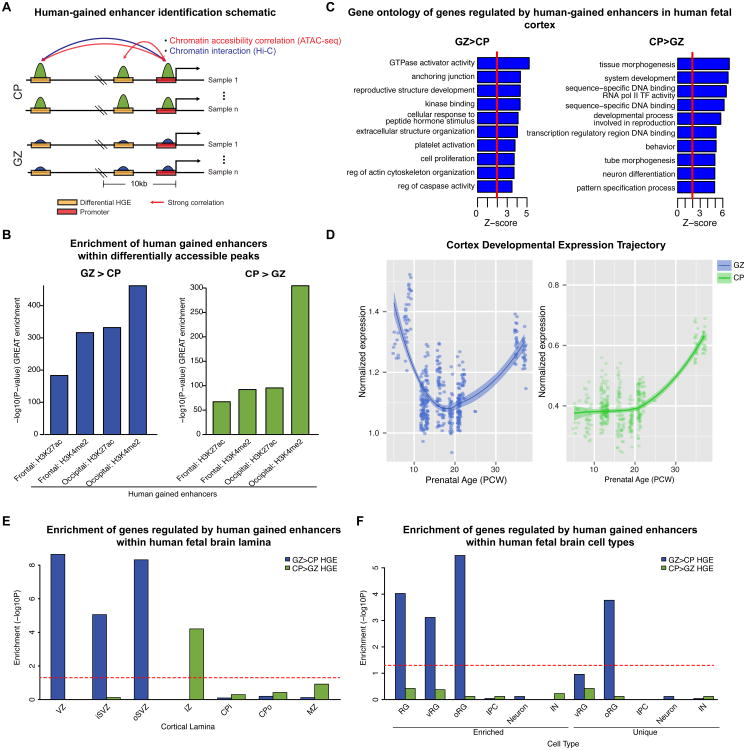
The expansion of the primate cerebral cortex is one of foundations on which the evolution of human cognition is based (Geschwind and Rakic, 2013) and differences in neural progenitor cell-types and their proliferation are predicted to play a major role (Lui et al., 2011; Taverna et al., 2014). We reasoned that these chromatin accessibility data provide a unique opportunity to identify human-specific regulatory elements driving cortical neurogenesis and their predicted target genes. We obtained a list of HGEs that are preferentially active in developing human brain, as compared to Rhesus macaque and mice (Reilly et al., 2015). Next, we identified the genes regulated by these HGEs using chromatin accessibility correlation and chromatin interaction (Figure 5A; STAR*Methods), resulting in high-confidence sets of 347 (GZ) and 246 (CP) candidate genes regulated by HGEs. HGEs fall within GZ/CP DA peaks significantly more often than would be expected by chance, suggesting key roles in cortical neurogenesis by virtue of their dynamic regulation during this period (Figure 5B).
Figure 5.
Gene expression, biological pathways, and cell-types impacted by HGEs.
(A) schematic showing chromatin accessibility correlation and chromatin interaction are used to nominate genes regulated by DREs.
(B) HGEs are strongly enriched in DA peaks.
(C) GO analysis of genes regulated by HGEs within GZ>CP DA peaks.
(D) Gene expression profiles throughout prenatal human cortical development (Kang et al., 2011) are shown for genes regulated by GZ>CP (GZ) or CP>GZ (CP) HGEs.
(E) Enrichment of genes regulated by HGEs within specific human fetal cortical laminae (Miller et al., 2014) (VZ: ventricular zone; i/oSVZ: inner/outer subventricular zone; IZ: intermediate zone; CPi/o: inner/outer cortical plate; MZ: marginal zone). Enrichment of GZ HGEs targets is observed in the progenitor-enriched VZ, iSVZ and oSVZ, whereas CP HGEs targets show enrichment in the IZ enriched in migrating neurons.
(F) Enrichment of genes regulated by HGEs within specific cell types present in human fetal cortex (Pollen et al., 2015) (RG: radial glia, IPC: intermediate progenitor cell; IN: inhibitory neuron; v/oRG: ventricular or outer radial glia). Enrichment of GZ HGEs targets is observed in vRGs and oRGs and oRG-unique genes. Cell type gene sets enriched or unique (no overlapping genes between cell types) are shown.
Genes targeted by GZ>CP HGEs were enriched in the modulation of GTPase activity, cell proliferation, caspase activity and the actin cytoskeleton, as well as in components of anchoring junctions and the extracellular matrix, processes that have been linked to human neural stem cell biology and cortical expansion (Pollen et al., 2015; Sun and Hevner, 2014; Taverna et al., 2014) (Figure 5C). GZ>CP HGE genes also showed a declining expression trajectory during early neurogenesis, followed by an increase at the onset of gliogenesis (Figure 5D), a pattern observed with genes enriched in neural progenitors and glia (Kang et al., 2011; Stein et al., 2014). These genes were highly expressed in the human VZ and oSVZ (Figure 5E). Since the oSVZ is significantly expanded in humans (Lui et al., 2011), our data suggested that these HGEs may be acting preferentially within cell-types predominately found within this human-expanded region.
To test this, we analyzed the overlap between the high confidence set of HGE targets with a single cell atlas of human fetal brain gene expression (Pollen et al., 2015). Remarkably, we found that genes targeted by the HGEs in GZ were preferentially expressed in oRG (FDR-corrected P=3.4×10-6), and to a lesser degree in vRG (FDR-corrected P=7.6×10-4) (Figure 5F). This enrichment is also observed when comparing to a list of genes uniquely expressed in oRG (FDR-corrected P=1.7×10-4) (Figure 5F), but not in vRG, suggesting a critical role of these regulatory elements in oRG biology. This analysis connects the seminal observations indicating that the oRG play an important role in elaboration of the human cerebral cortex (Lui et al., 2011; Taverna et al., 2014), with the putative transcriptional regulatory elements driving this process. Further, the enrichment of enhancers gained on the human lineage in this specific cell population provides unbiased, genome-wide evidence supporting the contention that oRG play a significant role in human brain evolution (Lui et al., 2011).
Several genes predicted to be regulated by GZ>CP HGEs are required for normal human brain development and cognition, including: 1) seven genes with human-specific developmental cortical expression trajectories, UTRN, MAK, TMEM134, PDLIM2, EPB41L3, PHLDB1, and TBC1D2B (Bakken et al., 2016) suggesting that modulation by HGEs may underlie their species-specific expression pattern; 2) GLI3 implicated in macrocephaly (Jamsheer et al., 2012); 3) seven genes in which deleterious, de novo loss of function mutations have been identified in ASD probands, but not in controls, MAD1L1, DDX60, DNAH5, MAK, DVL3, SCN7A, RAPGEF3 (De Rubeis et al., 2014; Iossifov et al., 2014); and 4) eight genes implicated in intellectual disability (ID) (Parikshak et al., 2013), including the oRG-enriched genes GLI3, GFAP, and AASS (Pollen et al., 2015).
Among the genes implicated in ID and predicted to be regulated by GZ>CP HGEs is FGFR2 (Figure 6A), a receptor for fibroblast growth factor family members (FGF). In mice, FGFR2 is expressed in telencephalic radial glia of the anterior cortex and its loss leads to a decrease in cortical neural progenitor self-renewal and excitatory projection neurons (Stevens et al., 2010). Predicted gain of function mutations in FGFR2 also cause Apert syndrome, which can present with megalencephaly and atypical gyrification (Cohen and Kreiborg, 1990; Sun and Hevner, 2014). We identified a GZ>CP HGE ∼550 kb away from the TSS of FGFR2 predicted to functionally interact with the FGFR2 promoter based on chromatin accessibility correlation and confirmed by Hi-C-defined physical chromatin interactions in brain (Figure 6A; STAR*Methods).
Figure 6.

FGFR2 is a target of a HGE impacting human cortical neurogenesis.
(A) An HGE within a DA peak interacts with the FGFR2 promoter with evidence from both chromatin accessibility correlation and chromatin interaction.
(B) Seven sgRNA pairs were used to excise the DA peak and/or the overlapping HGE peak measured via H3K27ac ChIP-seq. Primers used to validate genomic deletions are represented by green half arrows.
(C) Genomic PCR of the 3.7kb region containing the FGFR2 HGE in control or CRISPR/Cas9 + sgRNA infected phNPCs. Red arrow points to the expected 2107bp band in controls and white arrows point to expected products following excision (see 6B). Asterisks indicate background PCR amplicons.
(D) Excision of the FGFR2 HGE led to a 19-40% reduction in FGFR2 expression as measured by qPCR in phNPCs (***P<0.0001, **P<0.001, *P<0.01, ANOVA, with Bonferroni post-hoc test, n=4, mean ± SEM shown).
(E) Excision of the FGFR2 HGE led to an increase in the number of neurons generated after two weeks of differentiation. phNPCs infected with sgRNA pairs designed to excise the FGFR2 enhancer were stained with antibodies to GFP, tRFP, and the early neuron marker Tuj1. The percent of dual GFP/RFP+ cells positive for Tuj1 (arrows) was quantified. Co-infection with any of the three sgRNA pairs led to an increase in Tuj1+ positive cells compared to control (P=1.31×10-7, sgRNA -127/+1067; P=2.38×10-8, sgRNA -1213/+1067; P=1.44×10-8, sgRNA - 1213/+20; n=4 independent infections with 30 image fields/infection, mean ± SEM shown). Scale bar = 50μm. See also Table S4 and Figure S6.
We hypothesized that an active HGE involved in human cortical expansion should promote FGFR2 expression, thus stimulating neural progenitor proliferation, and conversely, its removal would lead to diminished FGFR2 levels (Raballo et al., 2000; Stevens et al., 2010). To test this, we first confirmed that the enhancer peak was present in undifferentiated phNPCs (Figure S6A), and engineered seven different partial and complete deletions of the FGFR2 enhancer (Figure 6B-C). All seven deletions led to a 19-40% reduction in FGFR2 expression in phNPCs (Figure 6D). Excision of the FGFR2 enhancer did not change the expression of the closest gene WDR11, which does not show evidence of chromatin accessibility correlation or interaction (Figure 6A and S6B). We next assessed whether ablation of this enhancer affects neurogenesis, observing that the percentage of neurons generated in deletion lines increased, consistent with decreased self-renewal of neural progenitors (Kang et al., 2009) (Figure 6E). These data validate the distal regulation of FGFR2 by an HGE and provide a mechanism by which its expression could contribute to human cortical expansion by promoting neural progenitor proliferation, consistent with prevailing models of cortical evolution (Geschwind and Rakic, 2013; Lui et al., 2011).
Common genetic variants within DA peaks regulate human brain volume and cognition
Thus far, most genetic variants associated with common disease are found in non-coding intergenic or intronic regions of the genome limiting their functional interpretation (Ward and Kellis, 2012). Thus, we next determined if common genetic variation within the functionally annotated non-coding regions identified here, which regulate neurogenesis, influence complex human traits including cognition and ICV. Using a partitioned heritability approach based on LD score regression (Finucane et al., 2015) (Figure 7A), we find that genetic variants associated with risk for schizophrenia (Schizophrenia Working Group of the Psychiatric Genomics, 2014), ADHD (Demontis et al., 2017), depressive symptoms (Okbay et al., 2016a), neuroticism (Okbay et al., 2016a), educational attainment (Okbay et al., 2016b) and ICV (Adams et al., 2016) were all significantly enriched in GZ>CP peaks, and much less, or not at all in CP>GZ peaks (Figures 7B-C and S7A). As a negative control, we tested if genetic variants associated with inflammatory bowel disease (IBD), identified by well-powered GWAS (Jostins et al., 2012) and finger whorls (Ho et al., 2016), were enriched in differential accessibility peaks. We did not observe enrichment of IBD or finger whorl variants (Figures 7B-C) in DA peaks, highlighting the importance of tissue-specific gene regulation.
Figure 7.
Common genetic variation within DA peaks affects adult cognition, risk for neuropsychiatric disease, and global brain size.
(A) A schematic demonstrating the partitioned heritability approach via LD (linkage disequilibrium)-score regression implemented to determine enrichment of association statistics within DA peaks.
(B) The FDR-corrected significance of partitioned heritability enrichment demonstrates a significant enrichment of heritability in specific brain traits. References for each GWAS is found in the STAR*Methods.
(C) Partitioned heritability enrichment across multiple GWAS. Error bars represent standard error. See also Figure S7.
To explore temporal specificity, we identified peaks in adult cortical ATAC-seq data from the PsychENCODE project (STAR*Methods) and found no significant enrichment of genetic variants associated with depressive symptoms, ADHD, neuroticism, or ICV in adult cortical peaks, whereas variants associated with schizophrenia and educational attainment were enriched in both fetal and adult open chromatin regions (Figure S7B). These data suggest that variation in gene regulation within neural progenitors and early-born neurons in fetal stages has a more substantial effect on later occurring, adolescent phenotypes than variation occurring in post-mitotic neurons within the fetal CP.
Discussion
Here we have performed a comprehensive assessment of chromatin accessibility during human cortical neurogenesis and integrated it with gene expression and chromatin interaction (Hi-C) data from the same tissue and developmental period. We illustrate how to leverage these data to define functional cis-regulatory elements involved in human corticogenesis and their target genes, identify candidate TFs regulating these genes, and show how these data inform our understanding of human genetic disorders and brain evolution. In doing so, we identify a human specific FGFR2 enhancer that likely plays a role in human cortical expansion and brain evolution. We additionally highlight potential mechanisms of a rare genetic disorder involving disruption of gene regulatory elements by structural chromosomal variation, rather than disruption or deletion of the coding region. By integrating these data with GWAS, we also identify regulatory mechanisms by which non-coding genetic variation likely influences neuropsychiatric disease, brain volume and cognition.
Our analysis also has fundamental implications for understanding the pathophysiology of neuropsychiatric disease, guiding future genetic discovery and disease modeling. Functional annotation in non-coding regions is critical to a mechanistic understanding of genetic disease-associations (Roadmap Epigenomics et al., 2015; Won et al., 2016). The power of this is two-fold, 1) to annotate functionality of sequence alterations within cis-regulatory elements, and 2) to define the target gene likely to be affected by variants, which is of critical importance, since gene regulation is often long-range (GTEx, 2015; Won et al., 2016). Moreover, multiple lines of evidence support a neurodevelopmental basis for ASD and schizophrenia (de la Torre-Ubieta et al., 2016; Parikshak et al., 2015). Thus, these data provide a necessary link to interpret the impact of genetic variation during a critical period for susceptibility to neuropsychiatric disease risk. Similarly, that genetic variation regulating ICV and educational attainment is enriched in regions preferentially accessible in the GZ provides strong genetic evidence that differences in neural progenitor biology have substantial influence on brain size and cognitive performance later in life (Adams et al., 2016; Okbay et al., 2016b).
Identification of functional regulatory elements in cortical neurogenesis also increases power for genetic discovery. Here we provide annotations of the genome relevant to a key biological process implicated in neuropsychiatric disorders, which may be used as an annotation feature in the burden test for whole genome sequencing studies. Regulatory regions defined here may also be used to create custom capture libraries for efficient next-generation sequencing similar to previous “HAR-ome” studies, which focused on another class of functionally relevant human-evolved regulatory sequences (Doan et al., 2016). Additionally, endogenous enhancer regions for marker genes can also be used to more faithfully drive expression for cell-type specific study of pathways involved in disease (Dimidschstein et al., 2016).
Often TF prediction efforts to define gene regulatory networks rely exclusively in motif prediction, which are substantially strengthened by incorporating chromatin accessibility and interaction (Rackham et al., 2016). Our analysis identifies many TFs known to orchestrate neurogenesis and differentiation, including Sox, homeodomain, and bHLH family members, among others (Guillemot, 2007). Based on these data we predict roles in cortical neurogenesis for several dozen other TFs not previously implicated in this process (Table S3). Follow-up ChIP-seq experiments will be necessary to assess individual TF binding in progenitors and neurons, but they rely on highly specific antibodies for native in vivo validation, which are not yet available for many of these TFs (Johnson et al., 2007).
It will clearly be important to expand this analysis to other brain regions, specific cell types, and developmental periods. Genes regulating progenitors and glia are overlapping (Kriegstein and Alvarez-Buylla, 2009; Stein et al., 2014), thus it is possible that HGEs are also regulating glial-specific genes, which will require chromatin accessibility data from a later time period to directly test this hypothesis. We also found that genetic variants associated with ICV (Adams et al., 2016) were not enriched in open chromatin regions in adult brain, in contrast to our findings in fetal brain, which not only helps define the critical epoch when these variants are likely acting, but emphasizes the need for stage and tissue specific maps of gene regulation. Finally, we identified GZ/CP DA putative regulatory elements for ncRNAs that will be important to characterize in the future. This is of particular interest given the growing role of lncRNAs and miRNAs in regulating cortical neurogenesis (Briggs et al., 2015; Sun and Hevner, 2014), and the paucity of knowledge surrounding their spatial and temporal regulation.
STAR*Methods
Contact for Reagent and Resource Sharing
Please contact D.H.G. (dhg@mednet.ucla.edu) for reagents and resources generated in this study.
Experimental Model and Subject Details
Developing Human Brain Samples
Fetal tissue was obtained from the UCLA Gene and Cell Therapy Core according to IRB guidelines from 3 donors (post-conception weeks 15:male, 16:female, 17: female) following voluntary termination of pregnancy (sex determined via homozygosity on the X chromosome). This study was performed under the auspices of the UCLA Office of Human Research Protection, which determined that it was exempt because samples are anonymous pathological specimens. Full informed consent was obtained from all of the parent donors. Coronal sections were taken from dissociated tissue that visually appeared cortical, which was confirmed via matching RNA-seq data to Allen Institute Human Fetal Brain microdissected tissue using Transition Mapping (Stein et al., 2014) (Figures S1A and S1B).
Primary Human Neuronal Culture
Primary human neural progenitor cells were previously isolated from two donors (both 16 PCW: male), which were subsequently differentiated and cultured for up to 8 weeks following previously described culture protocols (Stein et al., 2014). In detail, after establishing monolayer cultures, cells were expanded by plating on polyornithine/fibronectin coated plates with proliferation media [Neurobasal A (Invitrogen) supplemented with 10% BIT 9500 (Stem Cell Technologies), Antibiotics and Antimycotics (Gibco), GlutaMAX (Gibco), and heparin (1 μg/mL; Sigma)] with freshly added EGF, FGF2, PDGF (each at 20 ng/mL; Peprotech), and LIF (2 ng/mL; EMD-Millipore) and passaged when confluent. For differentiation, cells were plated at 2×104 cells/cm² on polyornithine/laminin coated plastic plates (Figures 3G-I, S5, 6B-D, and S6) or acid-washed coverslips (Figure 6E), and then switched to differentiation media [Neurobasal A (Invitrogen) supplemented with B27 (Gibco), GlutaMAX (Gibco), and Antibiotics and Antimycotics (Gibco)] as well as Retinoic Acid (500 ng/mL; Sigma-Aldrich), Forskolin (10 μM; Sigma-Aldrich), BDNF (10 ng/mL; Peprotech), NT-3 (10 ng/mL; Peprotech), and KCl (10mM). Half of the media was replaced three times per week for the duration of the differentiation. All cells were incubated at 37°C and 5% CO 2. Using unbiased quantitative approaches these cultures have been previously shown to: 1) match mid-fetal in vivo cortical development to a high degree, 2) best match cortical neuroanatomical identity, 3) are enriched in all of the major cortical markers, including lamina-specific markers and excitatory cell markers [BCL11, SOX5, TBR1, FEZF2, SATB2, POU3F2, EMX1, NEUROD2, NEUROD6, and SLC17A7, and 4) remarkably preserve the gene-co-expression architecture observed in vivo. Undifferentiated phNPCs are enriched in the dorsal telencephalic markers FOXG1, PAX6 and SOX2. Upon differentiation, progenitors gradually differentiate into MAP2-positive neurons (up to 40% MAP2 positive at 8 weeks), express cortical laminar markers including BCL11B (50% of the cells) and POU3F2 (35% of the cells), and undergo stereotypic neuronal morphogenesis including axo-dendritic polarization and more complex dendritic arborization with maturation (Stein et al., 2014).
Method Details
Tissue dissection
Under a dissection microscope, the coronal sections were cut with a razor blade along the less dense intermediate zone to divide the tissue into two segments from each donor (1) GZ: the neural progenitor rich region encompassing the ventricular zone, subventricular zone, and intermediate zone and (2) CP: the neuron rich region encompassing the subplate, cortical plate, and marginal zone (Figure 1A). For each donor and lamina (GZ or CP), 3-4 replicates were processed for ATAC-seq and RNA-seq.
ATAC-seq library preparation
ATAC-seq processing followed the original protocol (Buenrostro et al., 2013), which involves enzymatic dissociation of tissue using papain inactivated by ovomucoid (Worthington) followed by aliquoting of 50,000 cells, nuclear isolation using an IGEPAL solution, transposition using Nextera Tn5 transposase (Illumina), barcoding and amplification (8-11 cycles), quality control via gel electrophoresis, quantification via qPCR with DNA Standards (Kapa Biosystems), and massively parallel 50 bp paired end (PE) sequencing on an Illumina HiSeq2000 or HiSeq2500 to acquire ∼25M fragments per sample. For each donor, GZ and CP samples were dissected, library prepped, and sequenced within the same batch to prevent batch effects correlated with the biological condition of interest. Following preparation of a homogeneous suspension of cells isolated from the GZ or CP, 3-4 technical replicates consisting of independent nuclear isolations, Tn5 tagmentation, and library preparation were generated. phNPC samples were processed for ATAC-seq library preparation at 0 wks (3 replicates - independent nuclear isolations, Tn5 tagmentation, and library preparation from the same differentiation), 2 wks (3 replicates), 4 wks (2 replicates), and 8 wks (3 replicates) post differentiation as above.
RNA extraction and library preparation
RNA was extracted from samples using the Qiagen miRNeasy Mini Kit. Concentration and purity were evaluated with a NanoDrop spectrophotomoter. RNA Integrity Number was evaluated for each sample on an Agilent BioAnalyzer (mean±s.d. RIN = 8.7±0.6). Library preparation was completed using the Illumina Stranded TruSeq kit with RiboZero Gold ribosomal RNA depletion. Massively parallel 50 bp PE sequencing was completed on an Illumina HiSeq2500 to acquire ∼50M fragments per sample. For each donor, RNA extraction was completed on the same day for GZ and CP samples to prevent batch effects. Additionally, samples were pseudo-randomly assigned to a library preparation batch and sequencing lane as part of a larger experiment that reduced correlation between any covariate of interest (RIN, sample purity, biological condition, gestation week) and batch.
Functional validation of DREs
To rule out potential off-target effects from non-specific guide targeting, we designed multiple combinations of sgRNA pairs flanking the EOMES and FGFR2 enhancers (Figures 3F and 6B). sgRNAs were designed using the Benchling platform (https://benchling.com) maximizing both efficiency and specificity scores (Doench et al., 2014; Hsu et al., 2013) and cloned into the pL-CRISPR.EFS.GFP (Addgene #57818) or pL-CRISPR.EFS.tRFP (Addgene #57819) plasmids (Heckl et al., 2014) following published protocols (Ran et al., 2013). Lentiviral particles were prepared by transient transfection in 293T cells followed by concentration by ultracentrifugation. sgRNA sequences and targeted locations are provided in Table S4. phNPC lines generated and cultured as described in (Stein et al., 2014) were infected at 1 MOI after plating with control (spCas9-P2A-GFP and spCas9-P2A-tRFP lacking sgRNA) or a combination of two sgRNAs each co-expressing either spCas9-P2A-GFP or spCas9-P2A-tRFP. Next day, viral particles were removed by replacing with fresh medium and two days later phNPCs were induced to differentiate by replacing media with differentiation media (Stein et al., 2014). Following 2-3 weeks of differentiation to allow for maximal sgRNA expression and enhancer excision, dual GFP/tRFP positive cells were isolated by FACS and genomic DNA and RNA was extracted using DNeasy Blood & Tissue and miRNeasy Mini Kits (Qiagen), respectively. Genomic DNA amplification via PCR of the EOMES or FGFR2 locus to confirm enhancer excision was performed with Herculase II Fusion DNA Polymerase (Agilent Genomics) following manufacturers protocol using the following primers: EOMES; 5′-TGACATGGAGAATTCACAGGTGGA-3′ (Fwd), 5′-TCTTAGGGTCACACAGCTAGTT-3′ (Rev) (Figure 3H); FGFR2; 5′-CCGCCCCACCATCCCTTCTTTG-3′ (Fwd); 5′-TTGCCTTCTCCCCCACCACCTC-3′ (Rev) (Figure 6C). Quantification of enhancer target genes expression was assessed by qPCR using the SYBR Green I Master kit (Roche) on a Lightcycler 480 device using the following primers: EOMES; 5′- CACCGCCACCAAACTGAGAT -3′ (Fwd), 5′- CGAACACATTGTAGTGGGCAG-3′ (Rev) (Figure 3I); CMC1; 5′-CCAAGCGGCTACGTTCTTCT-3′ (Fwd), 5′- AGGAAGGGTAGACGAAGAGACA-3′ (Rev) (Figure S5H); AZI2; 5′- AAACCGGAAGCCCGT-3′ (Fwd), 5′- TGCAGTGACAAGAGCAAAATGG-3′ (Rev) (Figure S5H); FGFR2; 5′-TTTCGGCTGAGTCCAGCTCC-3′ (Fwd), 5′-CAATTCCCACTGCTTCCGCC-3′ (Rev) (Figure 6D); WDR11; 5′-CCGGGATGTTGCCCTACAC-3′ (Fwd), 5′-AGCAATTAAACCTTGCCAGCC-3′ (Rev) (Figure S6B). In parallel experiments, cultures infected with FGFR2 deletions displaying maximal FGFR2 reduction were differentiated for two weeks, fixed with 4% PFA, permeabilized with 0.1% TritonX-100/PBS, blocked with 10% normal goat serum (NGS) in 0.1% TritonX-100/PBS, and subjected to immunocytochemistry with antibodies to GFP (Abcam, ab13970,1:1000 dilution), tRFP (Evrogen, AB232, 1:500 dilution) and Tuj1 (Sigma, T8660, 1:500 dilution) to quantify cell fate of dual infected cells. All antibody incubations were performed in 0.02% Tween-20/PBS with 1% NGS for 1hr at room temperature (Figure 6E). For Tuj1 quantification (Figure 6E), p-values were calculated using a linear mixed-effect model using a random intercept for biological replicates to account for non-independence of the measurements (Aarts et al., 2014).
Quantification and Statistical Analysis
ATAC-seq pre-processing
Sequencing files from each sequencing lane were de-multiplexed and poor quality reads were excluded (PF=1 reads were retained). Reads were then mapped to the human genome including decoy sequences (hg19; 1000 Genomes Project Phase 2 reference assembly) using bwa mem (Li and Durbin, 2009) (v0.7.12). Optical and PCR duplicates were then removed using PicardTools (v1.128). De-duplicated BAM files from the same sample were merged from separate lanes and PicardTools was again used to remove duplicate reads. Only uniquely mapped reads mapping to chr 1-22 and X were kept (mitochondrial genome, Y chromosome, and unmapped contigs were removed). Insert size histograms were derived from PicardTools separately for chr1-22 and X (Figures S1C and S5B) as well as the mitochondrial genome (Figure S1D). All samples displayed the expected periodicity of DNA winding around nucleosomes in genomic (Buenrostro et al., 2013) (Figure S1C), but not mitochondrial DNA (Figure S1D). Peaks were called for each sample using MACS2 (Zhang et al., 2008) (v2.1.0.20140616). Peaks that overlapped with ENCODE blacklisted regions (http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeMapability/wgEncode DacMapabilityConsensusExcludable.bed.gz) were removed (Encode Project Consortium, 2012). Peak width in each sample, calculated in GenomicRanges (v1.16.4) (Lawrence et al., 2013), the number of peaks in each sample, and the number of peaks versus the length of each chromosome, derived from the BSgenome.Hsapiens.UCSC.hg19 (v1.32.0), were calculated (Figures S1E-G and S5C-E).
ATAC-seq differential accessibility analysis
R version 3.1.1 (2014-07-10) was used for all subsequent analyses. The DiffBind (Ross-Innes et al., 2012) package (v1.10.2) was used to find high confidence peaks present in at least 40% of samples. We used a 40% threshold for overlapping peaks across samples as a stringent threshold to define a peak, because each peak identified in this fashion must be present in all donors of a given condition (GZ or CP). This stringent threshold increases confidence that these peaks are reproducible, as each peak must be called multiple times in both technical and biological replicates. When we used lower thresholds ranging from 10-30% we found the expected decrease in correlation across replicates, further supporting the 40% threshold, which gave maximum confidence, while not requiring the peaks to be present in both GZ and CP, which would be the case if a higher threshold were used. Using our conservative thresholds we identified 62,005 peaks with an average merged peak width of 1188 bp (SD +/- 757 bp). The union across samples of those remaining peaks (called consensus peaksets in DiffBind, but hereafter simply referred to as peaks) was used for subsequent analyses. The number of reads within each peak was counted and then normalization factors for each peak across samples were calculated accounting for GC content, peak width, and total number of unique non-mitochondrial fragments sequenced using conditional quantile normalization (Hansen et al., 2012) from the cqn package (v1.10.0) (Hansen et al., 2012). Normalization by GC content within peaks was performed to offset observed biases of GC content on differential accessibility (Figures S1J-L). Differential accessibility across tissue type was calculated using a negative binomial regression with normalization based on the size factors from cqn and implemented in DEseq2 (v1.4.5) (Love et al., 2014) with default parameters (fitType=“parametric”,test=“Wald”). In order to find DA peaks across tissue type controlling for donor differences, the statistical model used included a regressor for tissue type (GZ or CP) and a factor regressor for the 3 donors included in the analysis. Heatmaps (Figures 1C and S5F) and coverage plots (Figures 1B, 2A, 3E, 6A, S3A, S5G and S6A) display normalized read counts - the number of reads within a peak divided by the normalization factor from cqn. Coverage plots were generated with Gviz (v1.8.4) (Hahne and Ivanek, 2016) with annotations from Gencode v19 (Harrow et al., 2012). In coverage plots, the normalization factor is averaged across all peaks for the sample.
Enrichment of peaks in genomic elements
Enrichment of DA peaks within annotated genic regions of the genome or epigenetically annotated regions of the genome was calculated using the ratio between the (#bases in state AND overlap feature)/(#bases in genome) and the [(#bases overlap feature)/(#bases in genome) × (#bases in state)/(#bases in genome)] as described previously by the Roadmap Epigenomics Consortium (Roadmap Epigenomics et al., 2015) (see Figure 1E-F). The significance of this enrichment was calculated using a binomial test as in the GREAT algorithm (McLean et al., 2010) (Figures S3B, S3D and 5B). Gene based annotations of the genome were derived from Gencode v19. For these analyses, transcription start site (TSS) was defined as 1kb upstream and 1kb downstream from the true TSS, promoter was defined as 2kb upstream and 1kb downstream from the TSS, and transcription end site (TES) was defined as 1kb upstream and 1kb downstream from the true TES. Validated human forebrain enhancers and non-forebrain enhancers were downloaded from the VISTA Enhancer browser (Visel et al., 2007) (http://enhancer.lbl.gov/) (Figure 1E-F). Human-gained enhancers were downloaded from GEO (GSE63648) (Reilly et al., 2015) (Figure 5B) using data from 12 PCW from either frontal or occipital cortex marked by H3K27ac or H3K4me2. Chromatin state definitions from an imputed 25-state model were derived from fetal brain tissue (E081) by the Roadmap Epigenomics project (Ernst and Kellis, 2015; Roadmap Epigenomics et al., 2015) and acquired from (http://www.broadinstitute.org/∼jernst/MODEL_IMPUTED12MARKS/). Chromatin state annotations are as follows:
| TssA | Active TSS |
| PromU | Promoter Upstream TSS |
| PromD1 | Promoter Downstream TSS with DNase |
| PromD2 | Promoter Downstream TSS |
| Tx5' | Transcription 5' |
| Tx | Transcription |
| Tx3' | Transcription 3' |
| TxWk | Weak transcription |
| TxReg | Transcription Regulatory |
| TxEnh5' | Transcription 5' Enhancer |
| TxEnh3' | Transcription 3' Enhancer |
| TxEnhW | Transcription Weak Enhancer |
| EnhA1 | Active Enhancer 1 |
| EnhA2 | Active Enhancer 2 |
| EnhAF | Active Enhancer Flank |
| EnhW1 | Weak Enhancer 1 |
| EnhW2 | Weak Enhancer 2 |
| EnhAc | Enhancer Acetylation Only |
| DNase | DNase only |
| ZNF/Rpts | ZNF genes & repeats |
| Het | Heterochromatin |
| PromP | Poised Promoter |
| PromBiv | Bivalent Promoter |
| ReprPC | Repressed PolyComb |
| Quies | Quiescent |
GO analysis at the TSS for DA peaks was completed by first overlapping DA peaks with a region 2kb upstream and 1kb downstream of the TSS of genes defined by Gencode v19. Protein-coding genes at DA peaks were input into GO-Elite (Zambon et al., 2012) (v1.2.5) using ontologies from EnsMart77Plus with a background list of all protein-coding genes (Figure 2D). The number of DA sites was calculated for GZ>CP and CP>GZ sites at the Gencode v19 defined TSS for multiple gene biotypes acquired from ENSEMBL through biomaRt (v2.20.0) (Durinck et al., 2009) (Figure S3C).
Correlation of peaks with DNAse-I HS data
The number of reads mapping to each peak was correlated between ATAC-seq replicates, between biological types within ATAC-seq, between ATAC-seq and DNAse-I Hypersensitivity from human fetal brain from the Roadmap Epigenomics Consortium (GSM1027328), and between ATAC-seq and DNAse-I Hypersensitivity from left human fetal lung from the Roadmap Epigenomics Consortium (GSM1027345) (Figure S2C-D). Reads were counted within peaks defined in the ATAC-seq data as described above. Data were plotted using smoothScatter() in R (Figure S2). Roadmap Epigenomics data was downloaded from SRA, mapped to the same genome build as described above (hg19) using the same software (bwa mem), duplicates were removed with PicardTools, and only uniquely mapped reads were retained.
RNA-seq differential expression analysis
RNA sequencing files were mapped to the genome (hg19; Homo_sapiens.GRCh37.75.dna.primary_assembly.fa; ftp://ftp.ensembl.org/pub/release-75/fasta/homo_sapiens/dna/) using RNA-STAR (Dobin et al., 2013) (v2.4.0h1). Gencode v19 annotations were used from ENSEMBL. ERCC control sequences were added to both the genome sequence and annotation files. Duplicates were marked and samples merged across lanes using PicardTools (v1.128). The number of fragments mapping to each exon was counted using the summarizeOverlaps() function from the GenomicAlignments package (v1.0.6) (Lawrence et al., 2013). Exons were retained in the analysis if found in at least 40% of samples with at least 10 fragments mapping. Normalization factors for each exon across samples were calculated accounting for GC content, exon width, and total number uniquely aligned fragments using conditional quantile normalization (Hansen et al., 2012) from the cqn package (v1.10.0). Normalized FPKM values were used as a measure of gene expression. A heatmap of RNA expression was produced using the correlation of normalized gene-level FPKM values across samples with the heatmap.2() function in gplots package (v2.17.0) (Figure 1D). In order to find DE genes across tissue type controlling for donor differences and RIN, a linear regression was used on normalized gene-level FPKM values with a statistical model including a regressor for tissue type (GZ or CP), a factor regressor for the 3 donors included in the analysis, and a numeric regressor for RIN. Gene level differential expression statistics were compared to data from laser-capture microdissection of the human fetal cortical wall (Miller et al., 2014) through transition mapping (Stein et al., 2014) (Figure S1A) and correlation (Figure S1B). DE protein-coding genes were input into GO-Elite (Zambon et al., 2012) (v1.2.5) using ontologies from EnsMart77Plus with a background list of all protein-coding genes (Figure S4A-B). In order to find DE exons across tissue type controlling for donor differences and RIN, a linear regression was used on normalized exon-level FPKM values with an identical statistical model described above. Adjusting significance for multiple comparisons across DE exons was evaluated with FDR (Benjamini and Hochberg, 1995).
Accessibility and gene expression correlation
The correlation between gene expression and chromatin accessibility was assessed by first mapping each peak overlapping with a promoter region (2kb upstream, 1kb downstream of TSS) to that gene. Promoter peaks marking two separate genes were removed (for example one promoter peak marking two genes on opposite strands). The differential accessibility of these promoters as measured by log2 (fold change) was plotted against the differential expression of the exon closest to the promoter. Only those peak-exon pairs with both significantly different gene expression and chromatin accessibility (FDR adjusted P < 0.05) were retained in the analysis (Figure 1G).
Mapping regulatory elements to cognate gene
The correlation between ATAC-seq peaks and the promoter was assessed by first identifying a peak as belonging to the promoter of a given gene if it fell within 2kb upstream of the TSS. Then, the correlation between the promoter peak and all peaks ±1 Mb from the promoter was assessed using the log2 transformation of normalized counts across samples. A 1Mb window (1Mb upstream to 1Mb downstream) was chosen, as most regulatory variation has been identified within this window (Jin et al., 2013). The Pearson's correlation and its significance were assessed using a one-sided test implemented with cor.test() in R. Multiple comparisons correction was performed on the p-values using FDR correction (Benjamini and Hochberg, 1995). A significant correlation between a peak and a promoter (adjusted P < 0.05) was used as evidence (chromatin accessibility correlation) that the peak was a DRE of the gene.
Hi-C data available through GEO and dbGaP under the accession number GSE77565 and phs001190.v1.p1, respectively were used to integrate with chromatin accessibility correlation profiles. Hi-C and ATAC-seq were performed on different donor tissues dissected following the same protocol and in the same laboratory. As the highest resolution available for the current Hi-C data was 10kb, we assigned these ATAC-seq peaks to 10kb bins and obtained the Hi-C interaction profile for 1Mb flanking region of each bin. Notably, the bin immediately adjacent to the seed region was not assessed for interaction. We made a background Hi-C interaction profile by pooling (1) 255,698 H3K27ac sites from frontal and occipital cortex at 12 PCW for human-gained enhancers (Reilly et al., 2015) and (2) 93,534 random regions of the genome with matched GC-content (within 5%) and length as Gencode v19 promoters. To avoid significant Hi-C interactions affecting the distribution fitting as well as parameter estimation, we used the lowest 95 percentiles of Hi-C contacts and removed zero contact values. Using these background Hi-C interaction profiles, we fit the distribution of Hi-C contacts at each distance for each chromosome using fitdistrplus package (Delignette-Muller and Dutang, 2015). Significance for a given Hi-C contact was calculated as the probability of observing a stronger contact under the fitted Weibull distribution matched by chromosome and distance. Hi-C contacts with FDR<0.01 were selected as significant interactions. We considered evidence for chromatin interaction when overlap of an ATAC-seq peak-containing bin showed significant contact with another peak-containing bin along with a flanking region of ±5 kb to account for edge effects of the bin. We used Hi-C data from the cognate compartment GZ or CP to overlap with ATAC-Seq as GZ>CP peaks showed higher peak correlation when supported by GZ Hi-C as compared to CP Hi-C (P=0.0155). Similarly, CP>GZ peaks show higher peak correlation when supported by CP Hi-C as compared to GZ Hi-C (P= 5.71×10-5) (Figure 3B). Evidence of both chromatin interaction and chromatin accessibility correlation was used to map DREs to their cognate genes (Figure 3A and Table S2). Cognate genes for HGEs distal to the TSS were mapped in the same way. As described above, the resolution of Hi-C is limited to 10kb bins and the bins immediately adjacent or containing the HGE were not evaluated for chromatin interaction, therefore we instead used chromatin accessibility correlation alone (without chromatin interaction) to map the targets of those regulatory elements when close to the TSS (Figure 5A). The chromatin accessibility correlation between distal peaks and the TSS peak was separated into categories based on the directionality of the chromatin accessibility change and whether chromatin interaction was present. The statistical significance of mean differences between chromatin accessibility correlation values in the different categories was calculated by transforming the correlations to Z-scores using Fisher z-transformation and then fitting a linear model (Figure 3B). Correlation between gene expression and distal enhancer chromatin accessibility was performed as above but using DREs supported by chromatin accessibility alone or chromatin accessibility and chromatin interaction (Figure 3C). GO of distal enhancers cognate genes or HGE targets was evaluated using mapped protein-coding genes input into GO-Elite (Zambon et al., 2012) (v1.2.5) using ontologies from EnsMart77Plus with a background list of all protein-coding genes within +/- 1 Mb of all DREs (Figure 3D) or all protein-coding genes within +/- 1 Mb of all HGEs (Figure 5C).
Differential TF binding analysis
Potential transcription factor binding sites were called in the human genome using TFBSTools (v1.4.0) (Tan and Lenhard, 2016) with a minimum score threshold of 80% based on position weight matrices from the JASPAR2016 (Mathelier et al., 2016) core database, selecting vertebrates as the taxonomic group. Only the most recent version of the PWM for a given TF was used. To select regions of the genome that are highly conserved amongst vertebrates, and likely functional, regions of the genome with 100-way phastCons (Pollard et al., 2010) scores > 0.4 in regions ≥ 20 bp were saved (downloaded from UCSC genome browser). Only called TFBS sites within conserved regions were retained for further analyses. Differential motif enrichment analysis was performed using a logistic regression model to identify motifs present more often in GZ>CP peaks as compared to CP>GZ peaks, or vice versa. Logistic regression explicitly controlled for differences in peak width and peak conservation between GZ>CP and CP>GZ DA peaks. The analysis was implemented in R as: glm(TFBS ∼ GZCP + peakwidth + conservedbppercent, family = “binomial”). The dependent variable (TFBS) was a binary representation of whether each DA peak contained a motif of a TF or not. The independent variable of interest marked whether a peak was GZ>CP (GZCP=1) or CP>GZ (GZCP=0). Other covariates included peak width (peakwidth) and the percentage of the peak with conservation (conservedbppercent) as defined above. Significant differential motif enrichment was determined by a FDR adjusted P < 0.05 threshold of the GZCP covariate as well as evidence of expression within the RNA-seq data. gzTFs were defined as significant differentially enriched motifs present more often in GZ>CP as compared to CP>GZ peaks, whereas cpTFs were defined as significant differentially enriched motifs present more often in CP>GZ as compared to GZ>CP peaks (Figures 4A and 4C, Table S3). The class of each TF was acquired through JASPAR2016 and a Fisher's exact test was used to calculate if a significant enrichment or depletion of that class was present in significant gzTFs or cpTFs as compared to all TFs found in JASPAR2016. Those classes with ≥ 5 significant TFs are shown in Figures 4B and 4D. The same analysis was implemented on hESC and Fibroblast DNAse-seq data (Figure S4C) that were downloaded from www.encodeproject.org (Experiment IDs for hESC: ENCSR915BSC, ENCFF251WIB, ENCSR000EMU, ENCSR000EMZ, ENCSR000EMZ, ENCSR820WLP, ENCSR820WLP, ENCSR678ILN, ENCSR678ILN; Experiment IDs for Fibroblast: ENCSR067XUN, ENCSR360LBD, ENCSR670VQZ, ENCSR587STM, ENCSR562ACY, ENCSR648GDP, ENCSR835BNW, ENCSR555TFE, ENCSR371KRY).
Expression trajectories of HGE targets
Expression trajectories of HGE targets in prenatal cortex (PCW 6-37) were plotted using the human brain transcriptomic atlas dataset (Kang et al., 2011). Gene expression values for each HGE target set (GZ or CP) were log2 transformed and centered to the mean of all expressed genes using the R function scale(x, center = T, scale =F) +1 (Figure 5D).
Laminar and single-cell gene enrichment
To assess the enrichment of human-gained enhancer target genes within specific fetal cortical lamina or cell types, we used logistic regression (Figure 5E-F). Genes enriched within specific cortical lamina were determined by calculating differential gene expression between all genes in a given lamina against all other lamina (FDR adjusted P < 0.05) as described (Parikshak et al., 2013) using data from laser-capture microdissected cortex from PCW 15, 16 and 21 (Miller et al., 2014). Cell-specific gene lists for major cell types within human fetal cortex were generated by taking the Pearson correlation of each gene to an idealized uniformly expressed cluster marker as provided in (Pollen et al., 2015) and converting them to p-values using the function corPvalueStudent(), where n=393 (number of cells profiled). “Enriched genes” are those with an FDR adjusted P < 0.05 for its respective cell type and “unique genes” are those fulfilling the same criteria, but not enriched in any other cell type.
Partitioned heritability analysis
Partitioned heritability was assessed using LD score regression (v1.0.0) (Finucane et al., 2015). Heritability is calculated by comparing the association statistics for common genetic variants falling within a given annotation, in this case DA peaks, with the LD-score, a measure of the extent of the LD block (Figure 7A). First, an annotation file was created which marked all HapMap3 SNPs that fell within GZ>CP or CP>GZ DA regions (Figures 7B and 7C). To control for the total number of peaks, we randomly subsampled the peaks GZ>CP peaks to have the same number as the CP>GZ peaks (Figure S7A), demonstrating that GZ>CP enrichments are not due to the greater number of peaks identified in GZ>CP analysis. In addition, separate annotation files were created by finding peaks in ATAC-seq data from 182 adult neocortex samples from the PsychENCODE project where peaks were called with MACS2 (downloaded from www.synapse.org with synapse ID syn5383038) (Figure S7B). Adult neocortical consensus peaksets were derived using the DiffBind package with a minimum overlap of 2 samples to call a consensus peakset, resulting in 106,983 peaks. LD-scores were calculated for these SNPs within 1 cM windows using the 1000 Genomes EUR data. These LD-scores were included simultaneously with the baseline distributed annotation file from Finucane et al., 2015 (Finucane et al., 2015). Subsequently, the heritability explained by these annotated regions of the genome was assessed from phenotypes for 17 genome-wide association studies (references given below). The enrichment was calculated as the heritability explained for each phenotype within a given annotation divided by the proportion of SNPs in the genome and FDR correction within each GWAS was used to correct for multiple comparisons.
Data and Software Availability
The ATAC-seq peaks from this study have been deposited in the GEO and dbGaP databases under ID codes GSE95023 and phs001438, respectively. The RNA-seq data from this study have been deposited in the dbGaP database under ID code phs001438.
Supplementary Material
Figure S1. Related to Figure 1: Tissue dissection and ATAC-seq data quality measures.
(A) Transition mapping (Stein et al., 2014) was used to display the similarity between the differential expression profiles from laser capture micro-dissected data (Miller et al., 2014) from each of the lamina listed as compared to RNA-seq data acquired in this study. As expected, RNA-seq data acquired in this study are most similar to the transition between the proliferative ventricular zone (VZ) and subventricular zone (SVZ) as compared to post-mitotic inner and outer cortical plate (CPi/CPo) and subplate (SP).
(B) The correlation between differential expression fold changes in RNA-seq data acquired in this study and fold changes from laser capture microdissection data from the SVZ as compared to the CPi is very high indicating strong correspondence with previous transcriptomic measurements in lamina of fetal brains.
(C) Insert size histogram shows the expected pattern of transposase insertion between nucleosomes for both GZ and CP samples.
(D) The insert size histogram for reads mapping to the mitochondrial chromosome shows no clear periodicity as mitochondrial DNA does not have a precisely ordered nucleosome structure.
(E) Peak width distribution for individual samples. Note that consensus peaksets, used in all subsequent analyses, represent the union of multiple peaks and therefore are wider.
(F) The number of peaks called in each sample by MACS2. Note that more peaks are called in samples 13-19 because more reads were acquired for those samples.
(G) As expected, the percentage of peaks within a given chromosome scales with the size of that chromosome for each sample.
(H) Overlap of identified peaks using all donors compared to excluding samples 13-19 (PCW 17 donor). The majority of peaks are shared when including or excluding samples 13-19.
(I) The effect size of peak differential accessibility is well preserved across donors.
(J) An MA-plot prior to conditional quantile normalization (CQN) for GC content within peaks shows a clear bias of high-GC content peaks within GZ>CP peaks (fold change > 0) and low-GC content peaks in CP>GZ peaks (fold change < 0), similar to previous work in differential expression from RNA-seq (Hansen et al., 2012).
(K) After CQN correction, less bias is observed.
(L) In order to demonstrate that CQN normalization is not removing important biological variance, the mean of log2(counts) of ATAC-seq reads prior to CQN normalization was graphed within GC content deciles. Similar patterns are observed for both GZ and CP samples. This indicates that normalizing across GC content is not removing major biological variance.
Figure S2. Related to Figure 1: Reproducibility of chromatin accessibility across technical, biological, and methodological replicates.
(A) Technical replicates (same region, same donor, different ATAC-seq library preparation) show extremely high correlation of normalized reads within peaks.
(B) Biological replicates (same region, different donor, different ATAC-seq library preparation) also show high correlation but somewhat lower than technical replicates.
(C) Methodological replicates (fetal brain tissue processed with DNAse-seq Roadmap Epigenomics et al., 2015) compared with GZ or CP dissected fetal brain tissue processed with ATAC-seq) show a lower but still strong relationship with ATAC-seq from fetal brain indicating reproducibility across methods for assaying chromatin accessibility. Note that peaks were defined across the ATAC-seq sample and reads were counted in those defined genomic locations from the DNAse-seq data.
(D) As expected, ATAC-seq from fetal brain showed a weaker relationship with DNAse-seq from fetal lung, indicating the expected tissue specificity of accessible chromatin. Correlations between samples not shown were not included for space limitations and were not substantially different than those displayed.
Figure S3. Related to Figure 1: Enrichment of DA chromatin within defined genomic elements.
(A) DA peaks (highlighted in sky blue) above coverage maps of normalized reads from ATAC-seq are shown near the PAX6 gene on chromosome 11. Individual sample coverage maps are displayed to indicate reproducibility across replicates and donors.
(B) The significance calculation for the enrichment of DA peaks within annotated genomic regions (Roadmap Epigenomics et al., 2015) using a binomial test similar to implementation by the GREAT tool (McLean et al., 2010) corresponding to the fold enrichments shown in Figure 1E.
(C) The number of DA peaks at the transcription start site (TSS) categorized by the transcript biotype. Numbers are roughly similar between lamina.
(D) The significance calculation for the enrichment of DA peaks within chromatin states defined in fetal tissue using a binomial test (McLean et al., 2010) corresponding to the fold enrichments shown in Figure 1F.
Figure S4. Related to Figure 2 and Figure 4: GO and pathway analysis of DE genes and DA promoters.
(A) DE genes with higher expression in GZ than CP show the expected enrichment in cell cycle and mitotic processes. Conversely, DE genes with higher expression in CP than GZ show the expected enrichment in neuronal and synaptic pathways.
(B) KEGG pathway analysis of GZ>CP DA promoters show enrichment in axon guidance, Notch, and Wnt signaling pathways, whereas CP>GZ DA promoters show enrichment in synaptic pathways.
(C) TFs with significant differentially enriched conserved motifs present more often in hESC > fibroblast DA peaks as compared to fibroblast > hESC peaks. Red arrows indicate known pluripotency-associated TFs identified with this approach, including OCT4 (POU5F1), SOX2, KLF4, ESRRB, TCF3 and ZIC3.
Figure S5. Related to Figure 3: Enrichment of distal enhancers in cortical lamina, quality control measures of ATAC-seq data in phNPCs and negative controls for EOMES qPCR.
(A) Enrichment of genes regulated by GZ/CP differential distal enhancers within specific human fetal cortical laminae (Miller et al., 2014) (VZ: ventricular zone; i/oSVZ: inner/outer subventricular zone; IZ: intermediate zone; CPi/o: inner/outer cortical plate; MZ: marginal zone). Enrichment of GZ>CP distal enhancer targets is observed in the progenitor-enriched VZ, iSVZ and oSVZ, whereas CP>GZ distal enhancer targets show enrichment in the neuron-enriched cortical plate (CPi/o) and in the MZ.
(B) Insert size histograms showing the expected pattern of transposase insertion between nucleosomes in phNPCs at various time points post-differentiation.
(C) Peak width distribution for individual samples. Note that consensus peaksets, used in all subsequent analyses, represent the union of multiple peaks and therefore are wider.
(D) The number of peaks called in each sample by MACS2. Note that more peaks are called in samples 41-50 because more reads were acquired for those samples.
(E) As expected, the percentage of peaks within a given chromosome scales with the size of that chromosome for each sample.
(F) Hierarchical clustering based on ATAC-seq genome-wide normalized reads within peaks shows high technical reproducibility with clustering driven by phNPC differentiation stage.
(G) Coverage maps of normalized ATAC-seq reads from in vivo fetal cortex or in vitro differentiated phNPCs covering the EOMES locus and predicted enhancers (highlighted in orange). The EOMES enhancer peak targeted for functional validation (dashed red rectangle) is preserved in phNPCs. The location of a chromosomal breakpoint for a balanced chromosomal translocation that leads to complete absence of EOMES expression is indicated by a dashed vertical black line.
(H) Excision of the EOMES enhancer in human neural progenitors does not change the expression of the nearest genes CMC1 and AZI2, which are not predicted targets of this enhancer (ANOVA, with Bonferroni post-hoc test, n=4, mean ± SEM shown).
Figure S6. Related to Figure 6: Preservation of FGFR2 enhancer in phNPCs.
(A) Coverage maps of normalized ATAC-seq reads from in vivo fetal cortex or in vitro differentiated phNPCs covering the FGFR2 locus and predicted enhancer (highlighted in orange). The FGFR2 enhancer peak targeted for functional validation is preserved in phNPCs.
(B) Excision of the FGFR2 HGE in phNPCs does not change the expression of the nearest gene WDR11, which is not a predicted target of this enhancer (ANOVA, with Bonferroni post-hoc test, n=4, mean ± SEM shown).
Figure S7. Related to Figure 7: Partitioned heritability sub-sampling to control for potential biases related to peak number and enrichment of chromatin accessibility peaks in adult or fetal brain.
(A) FDR-corrected significance of partitioned heritability enrichment across brain traits controlling for differences in peak number through random sub-sampling of GZ peaks to equal the number of CP peaks. Significant enrichment in brain traits and not in IBD or finger whorl within GZ peaks is not driven by differences in peak number. References for each GWAS is found in the STAR*Methods.
(B) FDR-corrected significance (left) of partitioned heritability enrichment across brain traits in fetal or adult ATAC-seq peaks (from PsychENCODE consortium; www.synapse.org; Synapse ID: syn5383038) and enrichment values (right). An enrichment in heritability explained was found in both fetal and adult chromatin accessibility peaks for schizophrenia and educational attainment. No enrichment was found for adult chromatin accessibility peaks for depressive symptoms, ADHD, neuroticism, or ICV. Error bars represent standard error.
Table S1. Related to Figures 1 and 2: Statistics for differential accessibility at each peak. Chr, peakstart, peakend are the genomic locations of the peaks on hg19; baseMean is the mean of normalized counts for all samples in the analysis; log2FoldChange is a measure of effect size between GZ and CP with GZ>CP corresponding to log2(Fold Change)>0; lfcse is the standard error of log2FoldChange; stat is the Wald statistic; pvalue is the nominal p-value of differential accessibility; padj is the Benjamini-Hochberg FDR adjusted p-value; GC content is the percentage of guanines and cytosines within the peak. ENSGIDatTSS, HGNCatTSS and biotypeatTSS denote gene identifiers and biotype for peaks falling within a TSS.
Table S2. Related to Figure 3: DREs mapped to genes through both chromatin accessibility correlation and chromatin interaction assays for genes with detectable expression and without overlapping promoters with other genes. Note this list contains data where the DRE is supported by either GZ or CP (or both) chromatin interaction assays. enhancerchr, enhancerstart, enhancerend are the genomic location of the DRE peak on hg19; cor, cor.p.value, cor.p.fdr are the correlation, significance, and FDR adjusted significance of the ATAC-seq peak correlation between the DRE peak and the promoter peak. Only peaks with cor.p.fdr < 0.05 are shown. ENSGID, hgnc_symbol, gene_biotype are the Ensembl gene ID, HGNC symbol, and type of gene at the promoter. promoterpeakchr, promoterpeakstart, promoterpeakend are the genomic location of the ATAC-seq peak at the promoter. expr.beta, expr.pvalue, expr.padj are the differential expression values (expr.beta is log2(Fold Change) with GZ>CP corresponding to log2(Fold Change) > 0 corresponding to the significance and corrected significance values of expr.pvalue and expr.padj) for the exon next to the promoter. exonseqnames, exonstart, and exonend represent the genomic location of the exon. promchr, promstart, promend, promstrand are the genomic location of the reduced promoter (2kb upstream and 1kb downstream from the TSS). Note that if multiple isoforms exist with TSSs close to each other, the promoter region is expanded to include the promoters of all connected isoforms. baseMean, log2FoldChange, lfcSE, stat, pvalue.diffacc, padj.diffacc, GCcontent are the differential accessibility statistics for the DRE as described in Table S1.
Table S3. Related to Figure 4: Differentially enriched TFs in DA peaks. TFname, TFID are the names of the TF from JASPAR2016. pval, estimate, and Padj are outputs from the differential motif enrichment analysis logistic regression evaluating whether motifs are present more often in GZ>CP as compared to CP>GZ peaks (estimate > 0; gzTF) or CP>GZ as compared to GZ>CP peaks (estimate < 0; cpTF). p-value adjustments for multiple comparisons are via FDR. Type represents a gzTF or cpTF.
Table S4. Related to Figures 3 and 6, and STAR*Methods: First tab: Sequences, target genomic coordinates, and specificity and efficiency scores for sgRNAs used in this study. All output metrics obtained from Benchling using published algorithms (Doench et al., 2014; Hsu et al., 2013). Second Tab: Primers used in this study.
Highlights.
We define non-coding regions regulating gene expression in developing human cortex
Human-gained enhancers preferentially regulate genes expressed in outer radial glia
A distal human-gained enhancer regulates FGFR2 during neocortical development
Genetic variation influencing cognition and brain size act during neurogenesis
Acknowledgments
This work was supported by NIH grants to D.H.G. (5R01MH060233; 5R01MH100027; 3U01MH103339; 1R01MH110927; 1R01MH094714), J.L.S. (R00MH102357), and H.W. (K99MH113823), and the California Institute for Regenerative Medicine (CIRM)-BSCRC Training Grant (TG2-01169) to L.T.U. ATAC-seq libraries were sequenced by the UCLA BSCRC, RNA-seq libraries were prepared and sequenced by the UCLA Neuroscience Genomics Core, and fetal tissue was collected from the UCLA CFAR (5P30 AI028697). Adult ATAC-seq data were generated as part of the PsychENCODE Consortium, supported by: U01MH103339, U01MH103365, U01MH103392, U01MH103340, U01MH103346, R01MH105472, R01MH094714, R01MH105898, R21MH102791, R21MH105881, R21MH103877, and P50MH106934 awarded to: S. Akbarian (ISMMS), G.Crawford (Duke), S. Dracheva (ISMMS), P. Farnham (USC), M. Gerstein (Yale), D. Geschwind (UCLA), T. M. Hyde (LIBD), A. Jaffe (LIBD), J. A. Knowles (USC), C. Liu (UIC), D. Pinto (ISMMS), N. Sestan (Yale), P. Sklar (ISMMS), M. State (UCSF), P. Sullivan (UNC), F. Vaccarino (Yale), S. Weissman (Yale), K. White (UChicago) and P. Zandi (JHU). pL-CRISPR.EFS.GFP and pL-CRISPR.EFS.tRFP plasmids were a gift from Benjamin Ebert. Finger whorl GWAS data were kindly provided by Sarah Medland. We thank M. Gandal, E. Ruzzo, and members of the Geschwind lab for helpful discussions and critical reading of the manuscript
Footnotes
Author contributions: L.T.U. and J.L.S. designed the experiments, implemented the analysis pipeline, interpreted results of and performed integration with other expression and epigenetic data, and co-wrote the manuscript. L.T.U., C.K.O., and D. Lu performed sample collection, brain dissections, and genome editing experiments. L.T.U. performed the ATAC-seq library preparation. H.W. performed the chromatin interaction analyses and contributed to the integrative statistical analysis. J.L.S. and D. Liang implemented the differential motif enrichment analysis. D.H.G. conceived and supervised experimental design and analysis, interpreted results, provided funding, and co-wrote the manuscript.
The authors declare no financial conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References Cited
- Aarts E, Verhage M, Veenvliet JV, Dolan CV, van der Sluis S. A solution to dependency: using multilevel analysis to accommodate nested data. Nat Neurosci. 2014;17:491–496. doi: 10.1038/nn.3648. [DOI] [PubMed] [Google Scholar]
- Adams HH, Hibar DP, Chouraki V, Stein JL, Nyquist PA, Renteria ME, Trompet S, Arias-Vasquez A, Seshadri S, Desrivieres S, et al. Novel genetic loci underlying human intracranial volume identified through genome-wide association. Nat Neurosci. 2016;19:1569–1582. doi: 10.1038/nn.4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano T, Sagai T, Tanabe H, Mizushina Y, Nakazawa H, Shiroishi T. Chromosomal dynamics at the Shh locus: limb bud-specific differential regulation of competence and active transcription. Developmental cell. 2009;16:47–57. doi: 10.1016/j.devcel.2008.11.011. [DOI] [PubMed] [Google Scholar]
- Arnold SJ, Huang GJ, Cheung AF, Era T, Nishikawa S, Bikoff EK, Molnar Z, Robertson EJ, Groszer M. The T-box transcription factor Eomes/Tbr2 regulates neurogenesis in the cortical subventricular zone. Genes Dev. 2008;22:2479–2484. doi: 10.1101/gad.475408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baala L, Briault S, Etchevers HC, Laumonnier F, Natiq A, Amiel J, Boddaert N, Picard C, Sbiti A, Asermouh A, et al. Homozygous silencing of T-box transcription factor EOMES leads to microcephaly with polymicrogyria and corpus callosum agenesis. Nat Genet. 2007;39:454–456. doi: 10.1038/ng1993. [DOI] [PubMed] [Google Scholar]
- Bae BI, Jayaraman D, Walsh CA. Genetic Changes Shaping the Human Brain. Dev Cell. 2015;32:423–434. doi: 10.1016/j.devcel.2015.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakken TE, Miller JA, Ding SL, Sunkin SM, Smith KA, Ng L, Szafer A, Dalley RA, Royall JJ, Lemon T, et al. A comprehensive transcriptional map of primate brain development. Nature. 2016;535:367–375. doi: 10.1038/nature18637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- Briggs JA, Wolvetang EJ, Mattick JS, Rinn JL, Barry G. Mechanisms of Long Non-coding RNAs in Mammalian Nervous System Development, Plasticity, Disease, and Evolution. Neuron. 2015;88:861–877. doi: 10.1016/j.neuron.2015.09.045. [DOI] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y, Faddah DA, Jaenisch R. Mechanisms and models of somatic cell reprogramming. Nat Rev Genet. 2013;14:427–439. doi: 10.1038/nrg3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystron I, Blakemore C, Rakic P. Development of the human cerebral cortex: Boulder Committee revisited. Nat Rev Neurosci. 2008;9:110–122. doi: 10.1038/nrn2252. [DOI] [PubMed] [Google Scholar]
- Cohen MM, Jr, Kreiborg S. The central nervous system in the Apert syndrome. American journal of medical genetics. 1990;35:36–45. doi: 10.1002/ajmg.1320350108. [DOI] [PubMed] [Google Scholar]
- de la Torre-Ubieta L, Won H, Stein JL, Geschwind DH. Advancing the understanding of autism disease mechanisms through genetics. Nat Med. 2016;22:345–361. doi: 10.1038/nm.4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, Kou Y, Liu L, Fromer M, Walker S, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delignette-Muller ML, Dutang C. fitdistrplus: An R Package for Fitting Distributions. 2015;64:34. 2015. [Google Scholar]
- Demontis D, Walters RK, Martin J, Mattheisen M, Als TD, Agerbo E, Belliveau R, Bybjerg-Grauholm J, Bækved-Hansen M, Cerrato F, et al. Discovery Of The First Genome-Wide Significant Risk Loci For ADHD. bioRxiv 2017 [Google Scholar]
- Dimidschstein J, Chen Q, Tremblay R, Rogers SL, Saldi GA, Guo L, Xu Q, Liu R, Lu C, Chu J, et al. A viral strategy for targeting and manipulating interneurons across vertebrate species. Nat Neurosci. 2016;19:1743–1749. doi: 10.1038/nn.4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan RN, Bae BI, Cubelos B, Chang C, Hossain AA, Al-Saad S, Mukaddes NM, Oner O, Al-Saffar M, Balkhy S, et al. Mutations in Human Accelerated Regions Disrupt Cognition and Social Behavior. Cell. 2016;167:341–354 e312. doi: 10.1016/j.cell.2016.08.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Hartenian E, Graham DB, Tothova Z, Hegde M, Smith I, Sullender M, Ebert BL, Xavier RJ, Root DE. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol. 2014;32:1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc. 2009;4:1184–1191. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encode Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kellis M. Large-scale imputation of epigenomic datasets for systematic annotation of diverse human tissues. Nat Biotechnol. 2015;33:364–376. doi: 10.1038/nbt.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estivill-Torrus G, Pearson H, van Heyningen V, Price DJ, Rashbass P. Pax6 is required to regulate the cell cycle and the rate of progression from symmetrical to asymmetrical division in mammalian cortical progenitors. Development. 2002;129:455–466. doi: 10.1242/dev.129.2.455. [DOI] [PubMed] [Google Scholar]
- Finucane HK, Bulik-Sullivan B, Gusev A, Trynka G, Reshef Y, Loh PR, Anttila V, Xu H, Zang C, Farh K, et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. 2015;47:1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank CL, Liu F, Wijayatunge R, Song L, Biegler MT, Yang MG, Vockley CM, Safi A, Gersbach CA, Crawford GE, et al. Regulation of chromatin accessibility and Zic binding at enhancers in the developing cerebellum. Nat Neurosci. 2015;18:647–656. doi: 10.1038/nn.3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind DH, Rakic P. Cortical evolution: judge the brain by its cover. Neuron. 2013;80:633–647. doi: 10.1016/j.neuron.2013.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham V, Khudyakov J, Ellis P, Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39:749–765. doi: 10.1016/s0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- Gray LT, Yao Z, Nguyen TN, Kim TK, Zeng H, Tasic B. Layer-specific chromatin accessibility landscapes reveal regulatory networks in adult mouse visual cortex. eLife. 2017;6 doi: 10.7554/eLife.21883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx C. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemot F. Spatial and temporal specification of neural fates by transcription factor codes. Development. 2007;134:3771–3780. doi: 10.1242/dev.006379. [DOI] [PubMed] [Google Scholar]
- Hahne F, Ivanek R. Visualizing Genomic Data Using Gviz and Bioconductor. Methods Mol Biol. 2016;1418:335–351. doi: 10.1007/978-1-4939-3578-9_16. [DOI] [PubMed] [Google Scholar]
- Hansen KD, Irizarry RA, Wu Z. Removing technical variability in RNA-seq data using conditional quantile normalization. Biostatistics. 2012;13:204–216. doi: 10.1093/biostatistics/kxr054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckl D, Kowalczyk MS, Yudovich D, Belizaire R, Puram RV, McConkey ME, Thielke A, Aster JC, Regev A, Ebert BL. Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat Biotechnol. 2014;32:941–946. doi: 10.1038/nbt.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Ren B. The gateway to transcription: identifying, characterizing and understanding promoters in the eukaryotic genome. Cell Mol Life Sci. 2007;64:386–400. doi: 10.1007/s00018-006-6295-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho YY, Evans DM, Montgomery GW, Henders AK, Kemp JP, Timpson NJ, St Pourcain B, Heath AC, Madden PA, Loesch DZ, et al. Common Genetic Variants Influence Whorls in Fingerprint Patterns. J Invest Dermatol. 2016;136:859–862. doi: 10.1016/j.jid.2015.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, O'Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, Stessman HA, Witherspoon KT, Vives L, Patterson KE, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamsheer A, Sowinska A, Trzeciak T, Jamsheer-Bratkowska M, Geppert A, Latos-Bielenska A. Expanded mutational spectrum of the GLI3 gene substantiates genotype-phenotype correlations. Journal of applied genetics. 2012;53:415–422. doi: 10.1007/s13353-012-0109-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F, Li Y, Dixon JR, Selvaraj S, Ye Z, Lee AY, Yen CA, Schmitt AD, Espinoza CA, Ren B. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AM, Pletikos M, Meyer KA, Sedmak G, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang W, Wong LC, Shi SH, Hebert JM. The transition from radial glial to intermediate progenitor cell is inhibited by FGF signaling during corticogenesis. J Neurosci. 2009;29:14571–14580. doi: 10.1523/JNEUROSCI.3844-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasnegor NA, Lyon GR, Goldman-Rakic PS. Development of the prefrontal cortex : evolution, neurobiology, and behavior. Baltimore: P.H. Brookes Pub. Co.; 1997. [Google Scholar]
- Lawrence M, Huber W, Pages H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ. Software for computing and annotating genomic ranges. PLoS Comput Biol. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Xiao G, Yin H, Hippenmeyer S, Horowitz JM, Ghashghaei HT. Neural development is dependent on the function of specificity protein 2 in cell cycle progression. Development. 2013;140:552–561. doi: 10.1242/dev.085621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathelier A, Fornes O, Arenillas DJ, Chen CY, Denay G, Lee J, Shi W, Shyr C, Tan G, Worsley-Hunt R, et al. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2016;44:D110–115. doi: 10.1093/nar/gkv1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JA, Ding SL, Sunkin SM, Smith KA, Ng L, Szafer A, Ebbert A, Riley ZL, Royall JJ, Aiona K, et al. Transcriptional landscape of the prenatal human brain. Nature. 2014;508:199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan A, Yardimci GG, Sheffield NC, Crawford GE, Ohler U. Predicting cell-type-specific gene expression from regions of open chromatin. Genome Res. 2012;22:1711–1722. doi: 10.1101/gr.135129.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nord AS, Pattabiraman K, Visel A, Rubenstein JL. Genomic Perspectives of Transcriptional Regulation in Forebrain Development. Neuron. 2015;85:27–47. doi: 10.1016/j.neuron.2014.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okbay A, Baselmans BM, De Neve JE, Turley P, Nivard MG, Fontana MA, Meddens SF, Linner RK, Rietveld CA, Derringer J, et al. Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nat Genet. 2016a;48:624–633. doi: 10.1038/ng.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okbay A, Beauchamp JP, Fontana MA, Lee JJ, Pers TH, Rietveld CA, Turley P, Chen GB, Emilsson V, Meddens SF, et al. Genome-wide association study identifies 74 loci associated with educational attainment. Nature. 2016b;533:539–542. doi: 10.1038/nature17671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikshak NN, Gandal MJ, Geschwind DH. Systems biology and gene networks in neurodevelopmental and neurodegenerative disorders. Nat Rev Genet. 2015;16:441–458. doi: 10.1038/nrg3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikshak NN, Luo R, Zhang A, Won H, Lowe JK, Chandran V, Horvath S, Geschwind DH. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell. 2013;155:1008–1021. doi: 10.1016/j.cell.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollen AA, Nowakowski TJ, Chen J, Retallack H, Sandoval-Espinosa C, Nicholas CR, Shuga J, Liu SJ, Oldham MC, Diaz A, et al. Molecular Identity of Human Outer Radial Glia during Cortical Development. Cell. 2015;163:55–67. doi: 10.1016/j.cell.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin S, Zhang CL. Role of Kruppel-like factor 4 in neurogenesis and radial neuronal migration in the developing cerebral cortex. Mol Cell Biol. 2012;32:4297–4305. doi: 10.1128/MCB.00838-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raballo R, Rhee J, Lyn-Cook R, Leckman JF, Schwartz ML, Vaccarino FM. Basic fibroblast growth factor (Fgf2) is necessary for cell proliferation and neurogenesis in the developing cerebral cortex. J Neurosci. 2000;20:5012–5023. doi: 10.1523/JNEUROSCI.20-13-05012.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rackham OJ, Firas J, Fang H, Oates ME, Holmes ML, Knaupp AS, Consortium F, Suzuki H, Nefzger CM, Daub CO, et al. A predictive computational framework for direct reprogramming between human cell types. Nat Genet. 2016;48:331–335. doi: 10.1038/ng.3487. [DOI] [PubMed] [Google Scholar]
- Rakic P. Evolution of the neocortex: a perspective from developmental biology. Nat Rev Neurosci. 2009;10:724–735. doi: 10.1038/nrn2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly SK, Yin J, Ayoub AE, Emera D, Leng J, Cotney J, Sarro R, Rakic P, Noonan JP. Evolutionary genomics. Evolutionary changes in promoter and enhancer activity during human corticogenesis. Science. 2015;347:1155–1159. doi: 10.1126/science.1260943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics C. Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson EB, St Pourcain B, Anttila V, Kosmicki JA, Bulik-Sullivan B, Grove J, Maller J, Samocha KE, Sanders SJ, Ripke S, et al. Genetic risk for autism spectrum disorders and neuropsychiatric variation in the general population. Nat Genet. 2016 doi: 10.1038/ng.3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronan JL, Wu W, Crabtree GR. From neural development to cognition: unexpected roles for chromatin. Nat Rev Genet. 2013;14:347–359. doi: 10.1038/nrg3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, Brown GD, Gojis O, Ellis IO, Green AR, et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481:389–393. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein JLR, Rakic P. Comprehensive developmental neuroscience : patterning and cell type specification in the developing CNS and PNS. First. Amsterdam: Elsevier/Academic Press; 2013. [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics, C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa A, Mao CA, Hadjantonakis AK, Klein WH, Broccoli V. Tbr2 directs conversion of radial glia into basal precursors and guides neuronal amplification by indirect neurogenesis in the developing neocortex. Neuron. 2008;60:56–69. doi: 10.1016/j.neuron.2008.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein JL, de la Torre-Ubieta L, Tian Y, Parikshak NN, Hernandez IA, Marchetto MC, Baker DK, Lu D, Hinman CR, Lowe JK, et al. A quantitative framework to evaluate modeling of cortical development by neural stem cells. Neuron. 2014;83:69–86. doi: 10.1016/j.neuron.2014.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens HE, Smith KM, Maragnoli ME, Fagel D, Borok E, Shanabrough M, Horvath TL, Vaccarino FM. Fgfr2 is required for the development of the medial prefrontal cortex and its connections with limbic circuits. J Neurosci. 2010;30:5590–5602. doi: 10.1523/JNEUROSCI.5837-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Hevner RF. Growth and folding of the mammalian cerebral cortex: from molecules to malformations. Nat Rev Neurosci. 2014;15:217–232. doi: 10.1038/nrn3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan G, Lenhard B. TFBSTools: an R/bioconductor package for transcription factor binding site analysis. Bioinformatics. 2016;32:1555–1556. doi: 10.1093/bioinformatics/btw024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna E, Gotz M, Huttner WB. The cell biology of neurogenesis: toward an understanding of the development and evolution of the neocortex. Annu Rev Cell Dev Biol. 2014;30:465–502. doi: 10.1146/annurev-cellbio-101011-155801. [DOI] [PubMed] [Google Scholar]
- Varki A, Geschwind DH, Eichler EE. Explaining human uniqueness: genome interactions with environment, behaviour and culture. Nat Rev Genet. 2008;9:749–763. doi: 10.1038/nrg2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA Enhancer Browser--a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35:D88–92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Taher L, Girgis H, May D, Golonzhka O, Hoch RV, McKinsey GL, Pattabiraman K, Silberberg SN, Blow MJ, et al. A high-resolution enhancer atlas of the developing telencephalon. Cell. 2013;152:895–908. doi: 10.1016/j.cell.2012.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward LD, Kellis M. Interpreting noncoding genetic variation in complex traits and human disease. Nat Biotechnol. 2012;30:1095–1106. doi: 10.1038/nbt.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won H, de la Torre-Ubieta L, Stein JL, Parikshak NN, Huang J, Opland CK, Gandal MJ, Sutton GJ, Hormozdiari F, Lu D, et al. Chromosome conformation elucidates regulatory relationships in developing human brain. Nature. 2016;538:523–527. doi: 10.1038/nature19847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambon AC, Gaj S, Ho I, Hanspers K, Vranizan K, Evelo CT, Conklin BR, Pico AR, Salomonis N. GO-Elite: a flexible solution for pathway and ontology over-representation. Bioinformatics. 2012;28:2209–2210. doi: 10.1093/bioinformatics/bts366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Related to Figure 1: Tissue dissection and ATAC-seq data quality measures.
(A) Transition mapping (Stein et al., 2014) was used to display the similarity between the differential expression profiles from laser capture micro-dissected data (Miller et al., 2014) from each of the lamina listed as compared to RNA-seq data acquired in this study. As expected, RNA-seq data acquired in this study are most similar to the transition between the proliferative ventricular zone (VZ) and subventricular zone (SVZ) as compared to post-mitotic inner and outer cortical plate (CPi/CPo) and subplate (SP).
(B) The correlation between differential expression fold changes in RNA-seq data acquired in this study and fold changes from laser capture microdissection data from the SVZ as compared to the CPi is very high indicating strong correspondence with previous transcriptomic measurements in lamina of fetal brains.
(C) Insert size histogram shows the expected pattern of transposase insertion between nucleosomes for both GZ and CP samples.
(D) The insert size histogram for reads mapping to the mitochondrial chromosome shows no clear periodicity as mitochondrial DNA does not have a precisely ordered nucleosome structure.
(E) Peak width distribution for individual samples. Note that consensus peaksets, used in all subsequent analyses, represent the union of multiple peaks and therefore are wider.
(F) The number of peaks called in each sample by MACS2. Note that more peaks are called in samples 13-19 because more reads were acquired for those samples.
(G) As expected, the percentage of peaks within a given chromosome scales with the size of that chromosome for each sample.
(H) Overlap of identified peaks using all donors compared to excluding samples 13-19 (PCW 17 donor). The majority of peaks are shared when including or excluding samples 13-19.
(I) The effect size of peak differential accessibility is well preserved across donors.
(J) An MA-plot prior to conditional quantile normalization (CQN) for GC content within peaks shows a clear bias of high-GC content peaks within GZ>CP peaks (fold change > 0) and low-GC content peaks in CP>GZ peaks (fold change < 0), similar to previous work in differential expression from RNA-seq (Hansen et al., 2012).
(K) After CQN correction, less bias is observed.
(L) In order to demonstrate that CQN normalization is not removing important biological variance, the mean of log2(counts) of ATAC-seq reads prior to CQN normalization was graphed within GC content deciles. Similar patterns are observed for both GZ and CP samples. This indicates that normalizing across GC content is not removing major biological variance.
Figure S2. Related to Figure 1: Reproducibility of chromatin accessibility across technical, biological, and methodological replicates.
(A) Technical replicates (same region, same donor, different ATAC-seq library preparation) show extremely high correlation of normalized reads within peaks.
(B) Biological replicates (same region, different donor, different ATAC-seq library preparation) also show high correlation but somewhat lower than technical replicates.
(C) Methodological replicates (fetal brain tissue processed with DNAse-seq Roadmap Epigenomics et al., 2015) compared with GZ or CP dissected fetal brain tissue processed with ATAC-seq) show a lower but still strong relationship with ATAC-seq from fetal brain indicating reproducibility across methods for assaying chromatin accessibility. Note that peaks were defined across the ATAC-seq sample and reads were counted in those defined genomic locations from the DNAse-seq data.
(D) As expected, ATAC-seq from fetal brain showed a weaker relationship with DNAse-seq from fetal lung, indicating the expected tissue specificity of accessible chromatin. Correlations between samples not shown were not included for space limitations and were not substantially different than those displayed.
Figure S3. Related to Figure 1: Enrichment of DA chromatin within defined genomic elements.
(A) DA peaks (highlighted in sky blue) above coverage maps of normalized reads from ATAC-seq are shown near the PAX6 gene on chromosome 11. Individual sample coverage maps are displayed to indicate reproducibility across replicates and donors.
(B) The significance calculation for the enrichment of DA peaks within annotated genomic regions (Roadmap Epigenomics et al., 2015) using a binomial test similar to implementation by the GREAT tool (McLean et al., 2010) corresponding to the fold enrichments shown in Figure 1E.
(C) The number of DA peaks at the transcription start site (TSS) categorized by the transcript biotype. Numbers are roughly similar between lamina.
(D) The significance calculation for the enrichment of DA peaks within chromatin states defined in fetal tissue using a binomial test (McLean et al., 2010) corresponding to the fold enrichments shown in Figure 1F.
Figure S4. Related to Figure 2 and Figure 4: GO and pathway analysis of DE genes and DA promoters.
(A) DE genes with higher expression in GZ than CP show the expected enrichment in cell cycle and mitotic processes. Conversely, DE genes with higher expression in CP than GZ show the expected enrichment in neuronal and synaptic pathways.
(B) KEGG pathway analysis of GZ>CP DA promoters show enrichment in axon guidance, Notch, and Wnt signaling pathways, whereas CP>GZ DA promoters show enrichment in synaptic pathways.
(C) TFs with significant differentially enriched conserved motifs present more often in hESC > fibroblast DA peaks as compared to fibroblast > hESC peaks. Red arrows indicate known pluripotency-associated TFs identified with this approach, including OCT4 (POU5F1), SOX2, KLF4, ESRRB, TCF3 and ZIC3.
Figure S5. Related to Figure 3: Enrichment of distal enhancers in cortical lamina, quality control measures of ATAC-seq data in phNPCs and negative controls for EOMES qPCR.
(A) Enrichment of genes regulated by GZ/CP differential distal enhancers within specific human fetal cortical laminae (Miller et al., 2014) (VZ: ventricular zone; i/oSVZ: inner/outer subventricular zone; IZ: intermediate zone; CPi/o: inner/outer cortical plate; MZ: marginal zone). Enrichment of GZ>CP distal enhancer targets is observed in the progenitor-enriched VZ, iSVZ and oSVZ, whereas CP>GZ distal enhancer targets show enrichment in the neuron-enriched cortical plate (CPi/o) and in the MZ.
(B) Insert size histograms showing the expected pattern of transposase insertion between nucleosomes in phNPCs at various time points post-differentiation.
(C) Peak width distribution for individual samples. Note that consensus peaksets, used in all subsequent analyses, represent the union of multiple peaks and therefore are wider.
(D) The number of peaks called in each sample by MACS2. Note that more peaks are called in samples 41-50 because more reads were acquired for those samples.
(E) As expected, the percentage of peaks within a given chromosome scales with the size of that chromosome for each sample.
(F) Hierarchical clustering based on ATAC-seq genome-wide normalized reads within peaks shows high technical reproducibility with clustering driven by phNPC differentiation stage.
(G) Coverage maps of normalized ATAC-seq reads from in vivo fetal cortex or in vitro differentiated phNPCs covering the EOMES locus and predicted enhancers (highlighted in orange). The EOMES enhancer peak targeted for functional validation (dashed red rectangle) is preserved in phNPCs. The location of a chromosomal breakpoint for a balanced chromosomal translocation that leads to complete absence of EOMES expression is indicated by a dashed vertical black line.
(H) Excision of the EOMES enhancer in human neural progenitors does not change the expression of the nearest genes CMC1 and AZI2, which are not predicted targets of this enhancer (ANOVA, with Bonferroni post-hoc test, n=4, mean ± SEM shown).
Figure S6. Related to Figure 6: Preservation of FGFR2 enhancer in phNPCs.
(A) Coverage maps of normalized ATAC-seq reads from in vivo fetal cortex or in vitro differentiated phNPCs covering the FGFR2 locus and predicted enhancer (highlighted in orange). The FGFR2 enhancer peak targeted for functional validation is preserved in phNPCs.
(B) Excision of the FGFR2 HGE in phNPCs does not change the expression of the nearest gene WDR11, which is not a predicted target of this enhancer (ANOVA, with Bonferroni post-hoc test, n=4, mean ± SEM shown).
Figure S7. Related to Figure 7: Partitioned heritability sub-sampling to control for potential biases related to peak number and enrichment of chromatin accessibility peaks in adult or fetal brain.
(A) FDR-corrected significance of partitioned heritability enrichment across brain traits controlling for differences in peak number through random sub-sampling of GZ peaks to equal the number of CP peaks. Significant enrichment in brain traits and not in IBD or finger whorl within GZ peaks is not driven by differences in peak number. References for each GWAS is found in the STAR*Methods.
(B) FDR-corrected significance (left) of partitioned heritability enrichment across brain traits in fetal or adult ATAC-seq peaks (from PsychENCODE consortium; www.synapse.org; Synapse ID: syn5383038) and enrichment values (right). An enrichment in heritability explained was found in both fetal and adult chromatin accessibility peaks for schizophrenia and educational attainment. No enrichment was found for adult chromatin accessibility peaks for depressive symptoms, ADHD, neuroticism, or ICV. Error bars represent standard error.
Table S1. Related to Figures 1 and 2: Statistics for differential accessibility at each peak. Chr, peakstart, peakend are the genomic locations of the peaks on hg19; baseMean is the mean of normalized counts for all samples in the analysis; log2FoldChange is a measure of effect size between GZ and CP with GZ>CP corresponding to log2(Fold Change)>0; lfcse is the standard error of log2FoldChange; stat is the Wald statistic; pvalue is the nominal p-value of differential accessibility; padj is the Benjamini-Hochberg FDR adjusted p-value; GC content is the percentage of guanines and cytosines within the peak. ENSGIDatTSS, HGNCatTSS and biotypeatTSS denote gene identifiers and biotype for peaks falling within a TSS.
Table S2. Related to Figure 3: DREs mapped to genes through both chromatin accessibility correlation and chromatin interaction assays for genes with detectable expression and without overlapping promoters with other genes. Note this list contains data where the DRE is supported by either GZ or CP (or both) chromatin interaction assays. enhancerchr, enhancerstart, enhancerend are the genomic location of the DRE peak on hg19; cor, cor.p.value, cor.p.fdr are the correlation, significance, and FDR adjusted significance of the ATAC-seq peak correlation between the DRE peak and the promoter peak. Only peaks with cor.p.fdr < 0.05 are shown. ENSGID, hgnc_symbol, gene_biotype are the Ensembl gene ID, HGNC symbol, and type of gene at the promoter. promoterpeakchr, promoterpeakstart, promoterpeakend are the genomic location of the ATAC-seq peak at the promoter. expr.beta, expr.pvalue, expr.padj are the differential expression values (expr.beta is log2(Fold Change) with GZ>CP corresponding to log2(Fold Change) > 0 corresponding to the significance and corrected significance values of expr.pvalue and expr.padj) for the exon next to the promoter. exonseqnames, exonstart, and exonend represent the genomic location of the exon. promchr, promstart, promend, promstrand are the genomic location of the reduced promoter (2kb upstream and 1kb downstream from the TSS). Note that if multiple isoforms exist with TSSs close to each other, the promoter region is expanded to include the promoters of all connected isoforms. baseMean, log2FoldChange, lfcSE, stat, pvalue.diffacc, padj.diffacc, GCcontent are the differential accessibility statistics for the DRE as described in Table S1.
Table S3. Related to Figure 4: Differentially enriched TFs in DA peaks. TFname, TFID are the names of the TF from JASPAR2016. pval, estimate, and Padj are outputs from the differential motif enrichment analysis logistic regression evaluating whether motifs are present more often in GZ>CP as compared to CP>GZ peaks (estimate > 0; gzTF) or CP>GZ as compared to GZ>CP peaks (estimate < 0; cpTF). p-value adjustments for multiple comparisons are via FDR. Type represents a gzTF or cpTF.
Table S4. Related to Figures 3 and 6, and STAR*Methods: First tab: Sequences, target genomic coordinates, and specificity and efficiency scores for sgRNAs used in this study. All output metrics obtained from Benchling using published algorithms (Doench et al., 2014; Hsu et al., 2013). Second Tab: Primers used in this study.