Abstract
The developing kidney provides a useful model for study of the principles of organogenesis. In this report we use three independent platforms, Drop-Seq, Chromium 10x Genomics and Fluidigm C1, to carry out single cell RNA-Seq (scRNA-Seq) analysis of the E14.5 mouse kidney. Using the software AltAnalyze, in conjunction with the unsupervised approach ICGS, we were unable to identify and confirm the presence of 16 distinct cell populations during this stage of active nephrogenesis. Using a novel integrative supervised computational strategy, we were able to successfully harmonize and compare the cell profiles across all three technological platforms. Analysis of possible cross compartment receptor/ligand interactions identified the nephrogenic zone stroma as a source of GDNF. This was unexpected because the cap mesenchyme nephron progenitors had been thought to be the sole source of GDNF, which is a key driver of branching morphogenesis of the collecting duct system. The expression of Gdnf by stromal cells was validated in several ways, including Gdnf in situ hybridization combined with immunohistochemistry for SIX2, and marker of nephron progenitors, and MEIS1, a marker of stromal cells. Finally, the single cell gene expression profiles generated in this study confirmed and extended previous work showing the presence of multilineage priming during kidney development. Nephron progenitors showed stochastic expression of genes associated with multiple potential differentiation lineages.
Keywords: ICGS, kidney development, Gdnf, multilineage priming, scRNA-Seq
Introduction
Kidney development is a complex process that involves the coordinated differentiation of multiple cell types. The functional unit of the kidney is the nephron, which can be divided into segments including the glomerulus, proximal tubule, loop of Henle and distal tubule. The nephron connects to the collecting ducts and is surrounded by interstitial stromal cells. During kidney development the peripheral self renewing cap mesenchyme progenitor population is continually induced by the underlying branching ureteric bud to form nephrons through a series of intermediate structures including the renal vesicle, comma-shaped body, and S-shaped body (McMahon, 2016).
Gene expression profiling can be used to better define the gene expression programs driving the formation of the distinct differentiated kidney cell types. The Genito Urinary Development Molecular Anatomy Project (GUDMAP) consortium first used laser capture microdissection and microarrays to characterize gene expression of the different compartments of the developing kidney (Brunskill et al., 2008). Many compartments, however, include multiple cell types, confounding interpretation of results. It was not clear which cell type expressed which gene. GUDMAP also defined gene expression patterns of FACS purified specific cell types using transgenic mice with GFP reporter expression driven by Meis1 (stromal cells) (Brunskill and Potter, 2012a), Tie2 (endothelial cells) (Brunskill and Potter, 2010), Mafb (podocytes) (Brunskill et al., 2011), and Crym (cap mesenchyme) (Brunskill and Potter, 2012b). These results are more useful, but cannot serve to distinguish subtypes of cells. More recently we used Fluidigm C1 microfluidics/robotics to carry out scRNA-Seq analysis of selected cells during kidney development (Brunskill et al., 2014). Of particular interest, the cells of the renal vesicle showed multilineage priming, with stochastic expression of genes associated with many future potential differentiated cell types. Nevertheless, a limited number of cells were examined, and representing only the cap mesenchyme and renal vesicle compartments.
In this report we carry out scRNA-Seq analysis of the entire wild type E14.5 mouse kidney. This is an interesting stage of kidney development, with a few immature nephrons formed, and the process of nephrogenesis quite active. To provide a cross platform global validation of results we used three separate technologies, Drop-Seq (Macosko et al., 2015), the high throughput 800 cell IFC Fluidigm C1 (Fluidigm), and Chromium 10x Genomics InDrop (Chromium) (Klein et al., 2015). We developed a new computation strategy for multi-technology cell-classification in conjunction with a new version of the unsupervised cell-state prediction analysis tool Iterative Clustering and Guide-gene selection (ICGS) (Olsson et al., 2016), dividing the cells into 16 cell states. Consistent gene-level results and population frequencies were observed across all three technologies. The results provide an interactive single-cell resolution atlas of gene expression of the distinct kidney cell types during development, including progenitor populations, differentiated cells, and intermediates (http://altanalyze.org/ICGS.html). The process of multilineage priming was confirmed and extended to additional stages of nephrogenesis. We also examined the growth factors and receptors expressed by the different cell types and defined potential compartment crosstalk interactions. Unexpectedly, we show that a subset of nephrogenic zone stromal cells strongly express Gdnf, a key driver of ureteric bud branching morphogenesis, previously thought to be expressed only by cap mesenchyme progenitor cells.
Materials and methods
Single-cell RNA-Seq
Wild type E14.5 kidneys were sliced into quarters, placed into 200uL TrypLE Select 10X (Invitrogen) and incubated 5 min at 37° C with trituration. Then 1mL of ice cold DMEM with 10% fetal bovine serum was added, samples were filtered through a 20μ filter, and centrifuged 5 min at 1,600 g at 4° C. Supernatant was removed, and cells were re-suspended in 10% DMSO, 25% fetal bovine serum, 65% DMEM at 200 cells/μl. Cells underwent a slow freeze in a styrofoam container at −80° C, and were then transferred to liquid nitrogen. For scRNA-Seq the cells were rapidly thawed in a 37° C water bath, an equal volume of DMEM was added, cells were pelleted for 5 min at 1,600 g at 4° C and rinsed twice with PBS. Drop-Seq was carried out as previously described (Macosko et al., 2015), and Fluidigm C1 HT IFC, and Chromium 10x Genomics as per the manufacturer’s recommendations. Two Fluidigm C1 800 cell IFCs were used. With the Fluidigm C1 system, cell capture points were imaged and chambers with zero or multiple cells were removed. A total of 705 chambers were removed based on these criteria. We compared fresh and frozen wild type E14.5 kidney scRNA-Seq data and found no detectable differences (data not shown).
Data analysis
To obtain read counts for all known murine genes, paired-end reads were evaluated using standard sequence alignment methods following barcode and unique molecular index (UMI) deconvolution as previously described (Brunskill and Potter, 2010; Klein et al., 2015; Macosko et al., 2015). All FASTQ sequence files were aligned with the software STAR to mouse genome build mm10. Using this method Fluidigm captured cells had ~190,000 average reads per cell and Drop-Seq cells had 26,181 average reads per cell. For Chromium analyses, the CellRanger workflow was applied to bcl2 sequence data to obtain FASTQ files for subsequent alignment. 2,295 cells with ~7,000 UMI counts per cell on average (42,000 reads on average) were selected by CellRanger for further analysis (Klein et al., 2015). For all datasets, gene-level unique molecular indexes (UMIs) were normalized (divided by) the total UMIs for each barcode, multiplied by 10,000 to derive counts per ten thousand reads by custom scripts for the Drop-Seq and Fluidigm data or by the software AltAnalyze version 2.1.1 from the CellRanger Chromium sparse matrix file. From the associated input files from each scRNA-Seq platform the algorithm Iterative Clustering and Guide-gene Selection (ICGS) was used to predict de novo cell populations as previously described (Olsson et al., 2016). AltAnalyze only considered cells with at least 1,000 genes expressed (exclude outlier option). For Drop-Seq, 4,738 cells with ~6,000 UMI counts per cell were obtained on average (~26,000 reads on average). The options applied for ICGS were a Pearson correlation threshold of 0.3, at least 4 cells with a 4-fold difference in expression using conservative Cell Cycle effects exclusion to identify the major population variation for all technological platforms, according to the software defaults for 3′ transcript biased scRNA-Seq platforms. For the Drop-Seq analyses, following the initial identification of 11 cell-states, ICGS was re-run with the same options on each of the major identified epithelial and stromal cell states (ICGS BioMarker GO-Elite predicted) to identify additional subpopulation heterogeneity, resulting in 16 final combined cell states. From all ICGS analyses, cell-state restricted genes were identified using automatically produced MarkerFinder results (Sigdel et al., 2014). Gene set enrichment analysis of ToppGene defined GUDMAP genes was performed using the GO-Elite option in the hierarchical clustering and heatmap viewer user-interface in AltAnalyze (Chen et al., 2009; Zambon et al., 2012). All t-SNE and heatmap clustering results were additionally generated through AltAnalyze. These data are deposited in the Gene Expression Omnibus (Accession GSE104396). All processed datasets, ICGS results and classification outputs have been further deposited in the online Synapse data portal (https://www.synapse.org/#!Synapse:syn11001759) or through an interactive web browser (http://altanalyze.org/ICGS/Kidney.php).
Immunofluorescence
E14.5 kidney samples were fixed in RNAse free 4% PFA in PBS overnight at 4°C. Tissue was then dehydrated in a graded ethanol to xylene series followed by paraffin embedding. Sections were taken at a 7μm thickness. Slides for immunofluorescence were processed as previously described (Raines et al., 2015). Primary antibodies against SIX2 (1:200 Proteintech), CRABP1 (1:200 Cell Signaling) and secondary antibodies Alexa Fluor anti-rabbit and anti-mouse were used. Slides were imaged using a Nikon wide field imaging system.
Dual In Situ Hybridization and Immunohistochemistry
Samples were initially collected and processed as above. In situ hybridization protocol used was previously described (Mugford et al., 2009). Seven-micron thick paraffin sections were de-waxed and rehydrated through RNase free Xylene and Ethanol washes. Samples were developed using BM-purple (Sigma-Aldrich). Previously described Gdnf probe (a generous gift from Dr. Rulang Jiang) was used for in situ hybridization (James et al., 2006). After BM-purple development of sections, slides were washed in PBS and processed for immunohistochemistry (IHC) as previously described (Dave et al., 2008). Primary antibodies against SIX2 (1:2,000 Proteintech) and MEIS1 (1:8,000 Abcam) and secondary biotinylated anti-rabbit antibody (1:200 Vector labs) were used.
Cell-Cell Protein Interaction analysis
A previous study reported a comprehensive list 1179 known ligand-receptors pairs (Ramilowski et al., 2015). Matching to this list, we identified the markers in each cell type that interacted with other markers of the same or different cell type. Circos (Krzywinski et al., 2009) was used to visualize these interactions between MarkerFinder Drop-Seq marker genes identified across the 16 cell types (Pearson correlation coefficient >0.3).
Results and discussion
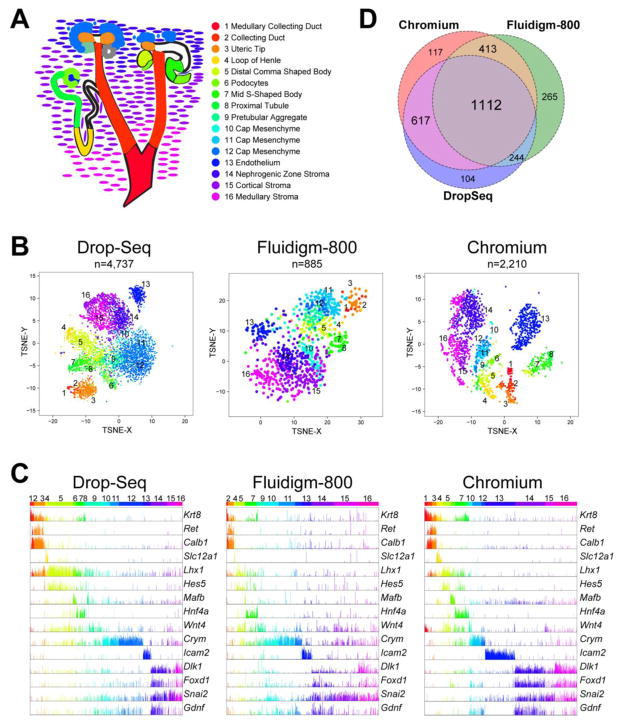
To create a single cell resolution atlas of gene expression in the E14.5 developing murine kidney we used three independent cross validating scRNA-Seq technologies, Drop-Seq, 800 cell IFC Fluidigm C1 (Fluidigm) and Chromium 10x Genomics (Chromium). The E14.5 kidney is undergoing active nephrogenesis, with a limited number of nephrons already formed, and all intermediate stages of nephron formation represented. Entire E14.5 kidneys were dissociated and used for scRNA-Seq analysis. In total we generated and examined gene expression profiles of over 8,000 E14.5 kidney single cells. We developed a novel integrative analysis workflow to identify initial heterogeneity within these datasets, harmonize the data using Drop-Seq as a reference and evaluate compartmental specific gene expression patterns (Fig. 1). For all three platforms we adapted and applied the Iterative Clustering and Guide-gene Selection (ICGS) algorithm from the open-source software AltAnalyze (http://www.altanalyze.org/) to identify distinct populations and compare those populations across profiling technologies (Fig. 2A). Application of ICGS to the top 2,000 Drop-Seq cells (barcodes) with the highest overall sequencing depth identified 16 cell populations, following two rounds of successive analysis (11 cell states in round 1). Only blood cells and macrophages were excluded from these 16 cell states (preliminary ICGS round). Generally similar states were captured by the Fluidigm and Chromium platforms as determined by t-SNE visualization, gene set enrichment analysis against the 16 Drop-Seq identified population markers and expression of previously defined embryonic kidney cell-type/compartmental specific marker genes (Fig. 2B–C). Importantly, no new additional populations could be identified using the software Seurat (Macosko et al., 2015) applied to the Drop-Seq, Chromium or Fluidigm data (data not shown).
Figure 1. Integrative workflow for multi-platform single-cell analysis.
Experimental design included dissociation of E14.5 mouse kidney cells, scRNA-Seq data generation with Chromium 10X Genomics, Drop-Seq and Fluidigm HT 800 cell IFC platforms, unsupervised bioinformatics classification of the resulting scRNA-Seq profiles, supervised harmonization of the three datasets and downstream cell-population level analyses.
Figure 2. Unsupervised cell-population analysis for Drop-Seq, Fluidigm and Chromium E14.5 kidney scRNA-Seq data.
A) De novo identified cell populations from the software ICGS are shown for each scRNA-Seq platform. The displayed heatmaps were produced by the MarkerFinder algorithm, downstream of the ICGS population predictions, with yellow indicating high relative gene expression and blue or black, low or no gene expression in the associated genes (rows). Prior established embryonic kidney marker genes corresponding to compartments are shown in panel C. Text to the left of each heatmap indicates the statistical enrichment of genes from the Drop-Seq ICGS analysis for the 16 identified populations (MarkerFinder) using the embedded gene-set enrichment analysis tool GO-Elite in AltAnalyze. B–C) t-SNE plot derived from the ICGS heatmaps in panel A, where each dot represents individual cells colored according to its B) ICGS cluster annotation or C) prior established population specific genes. CD: Collecting duct, UT: Ureteric Tip, LOH: Loop of Henle, RV: Renal vesicle, DCSB: Distal comma shaped body, Pod: podocyte, PT: Proximal Tubule, PA: Pre-tubular aggregate, CM: Cap mesenchyme, Endo: Endothelium, NZS: Nephrogenic Stroma, CS: Cortical Stroma.
While several transcriptionally distinct cell populations were evident from these unsupervised analyses, such as Endothelial (Icam2 positive), Cap mesenchyme (Crym positive) and Uteric tip (Ret positive), a precise 1-to-1 mapping of distinct cell type predictions was not obtained. To address this challenge we attempted to devise a strategy to directly compare the results between platforms using applicable supervised multiclass classification algorithms. Such analyses have not previously been described to our knowledge. To leverage the large number of cells captured by the Drop-Seq platform (>4,700), we choose to consider this platform as a reference for the other technologies. Although the Fluidigm cell-libraries were sequenced at a greater depth, we surmised that using results with shallower sequencing depth would improve our likelihood of consistently detecting the same assigned population-specific markers. Ultimately, two algorithms were tested on all datasets: 1) a k-nearest neighbor (knn, k=1) centroid classification method we previously developed to classify blood samples from renal allograft transplant patients (Roedder et al., 2014) and 2) the algorithm MarkerFinder applied to cell profiles rather than gene-profiles (transposed expression matrix of Drop-Seq marker genes). The latter algorithm was selected as a potential platform agnostic approach, as it will attempt to identify cells with genes that have expression largely restricted to a single cell-state, rather than evaluating similarity to a reference cell population centroid. In both cases, the classification analyses were restricted to the informative marker genes identified from the Drop-Seq ICGS analysis. The software MarkerFinder in AltAnalyze identifies genes specific to each ICGS identified cell population by identifying genes with expression most similar to an idealized cell-state specific pattern (expression = 1 in a single-cell state and expression = 0 in all other cells). Using this same approach, we transposed the expression matrix of the cells from which to classify from the alternative single-cell platforms to identify cells with an idealized expression profile matching these prior defined 16 populations (genes with expression only in a single cell state-profile), where each cell is assigned to a single-cell state (best match).
Application of the two test algorithms to the Fluidigm and Chromium single-cell datasets classified cells from each platform into cell-state bins largely similar those observed from Drop-Seq (Fig. 3A–C). Importantly, all major cell populations were observed across all three technologies with some differences in the relative frequency of the cell populations. For example, cell populations identified by Drop-Seq, such as cluster 5 – Distal Comma Shaped Body marker gene predicted, were present but reduced in frequency by both Fludigim and Chromium analyses. However, cluster 13 (Endothelial) was increased by both classification algorithms. Hence, these cell frequency differences are most likely due to variation in sample dissociation rather than classification accuracy. While generally both classification approaches identified cells from all 16 Drop-Seq populations, notable differences in discrete population assignments were found. To determine which algorithm is likely to provide the most accurate cross-platform classifications, we examined genes with the highest predicted population specificity in all three scRNA-Seq platforms (MarkerFinder-gene algorithm) (Fig. 3B, C, lower panels and Table S1). From both the knn and MarkerFinder-cell analysis, we identified ~1,100 genes-specific to a single-cell population consistently across the three scRNA-Seq platforms. 855 of the genes were consistently identified between both classification algorithms suggesting overall high concordance in the cell state assignments (Fig. 3D). However, comparison of genes with consistent population assignments between the scRNA-Seq platforms indicates an improvement in the detection of relatively infrequent cell types (e.g., cluster 2 - Collecting Duct, cluster 15 - Cortical Stroma) by the knn approach (Fig. 3E). Hence, we applied the results from the knn algorithm for all downstream comparison analyses. Interactive navigation of the results from both classification algorithms can be queried online through a web portal at http://altanalyze.org/ICGS/Kidney.php.
Figure 3. Classification and comparison of kidney cellular heterogeneity between scRNA-Seq platforms.
A) ICGS delineated cellular heterogeneity in the top ~2,000 DropSeq captured barcodes, segregated into 16 populations. B–C) Supervised classification of Fluidigm-800 chip captured libraries and 10X Genomics Chromium In-Drop barcodes using B) K-nearest neighbor (knn) classification against the Drop-Seq population centroids or C) MarkerFinder classified cells (rather than genes) for the top 862 population-specific genes from the Drop-Seq analysis. The upper panel displays the 862 Drop-Seq population specific genes and lower panel the top de novo MarkerFinder-gene results obtained following cell-population assignment. D) Comparison of population-specific genes jointly identified by all three scRNA-Seq technological platforms from the knn or MarkerFinder analysis. E) Number of genes jointly identified population-specific genes for each individual population for the two classification approaches. DCSB: Distal comma shaped body.
Cell-type annotation and specificity of ICGS kidney cell states
To more accurately define the likely cell types associated with the 16 cell clusters identified from our unsupervised and supervised analyses, we performed gene set enrichment analyses for each marker gene cluster against the GUDMAP ToppGene gene-set database (http://www.gudmap.org, https://toppgene.cchmc.org) and rigorously examined prior defined marker genes from this database (Fig. 4A). From this analysis, we were able to assign cells from the medullary collecting duct (C1), cortical collecting duct (C2), ureteric tip (C3), loop of Henle (C4), distal comma shaped body (C5), podocytes (C6), mid S-shaped body (C7), early proximal tubule (C8), pre-tubular aggregate (C9), three cap mesenchyme groups (C10, C11, and C12), endothelium (C13), nephrogenic zone stroma (C14), cortical stroma (C15), and medullary stroma (C16) (Fig. 4A). Table S2 summarizes some of the previously defined marker genes used for cell type identification, as well as additional genes with compartment enriched expression that emerged. Many of the early differentiating nephron cells, for example renal vesicle and comma shaped body, show strongly overlapping gene expression patterns and therefore some of these classifications are less definitive.
Figure 4. Orthogonal scRNA-Seq platforms identify equivalent kidney cell-populations and population-specific genes.
A) Cell-population annotation predictions assigned from literature and GUDMAP gene-set annotations. Corresponding kidney compartments are colored according to the ICGS cell cluster colors. B) t-SNE analysis of the harmonized knn classified cell states using the de novo identified MarkerFinder genes for each scRNA-Seq platform. The number of cells present in the plot are indicated. C) Gene expression bar chart (log2) for prior annotated kidney developmental marker genes from the knn classified datasets. D) Comparison of population-specific genes consistently identified by two or more scRNA-Seq platforms or that are specific to a single platform by MarkerFinder-gene analysis.
Analysis of the marker genes obtained following cell classification (Fig. 3B) by t-SNE dimensionality reduction, suggests qualitative differences in the global population-specific expression profiles observed from the three analyzed platforms (Fig. 4B). Specifically, while distinct “clusters” of cells associated with each cell state are observed with each of the platforms, in agreement with the ICGS classification results, the Chromium cell populations appear to provide more discrete cell populations, with fewer intermediate cells/cell-states. The Fluidigm system failed to generate data for podocytes, likely due to their large size and/or unusual structure. To determine if one platform provides improved lineage markers based on prior knowledge, we visualized the expression of known compartmental specific markers (Fig. 4C). The three different scRNA-Seq platforms displayed extremely similar results for known marker genes.
Careful evaluation of these prior defined markers suggests some ICGS populations reflect subtle heterogeneity while others are highly specific to a single cell states (Fig. 4B,C). For example, in the Drop-Seq t-SNE identified endothelial cells (cluster 13) form a well separated cluster, as do the collecting duct cells (clusters 1,2,3), and to a lesser extent the stromal cells (clusters 14, 15, 16). The cap mesenchyme progenitors form a rather diffuse group (clusters 10, 11, 12), next to the pretubular aggregate (cluster 9), with more differentiating cells extending out in three prongs, to form the loop of Henle (cluster 4), proximal tubules (cluster 8) and podocytes (cluster 6). We find a similar overlap in gene expression within the differentiating nephron progenitors, including the presumptive Distal Comma Shaped body and S-shaped bodies. At this stage of development cells with distal tubule differentiation markers are not yet present.
Consistent Gene Expression Predictions Between Single-Cell Platforms
To assess the relative agreement of gene-to-cell population predictions between the three evaluated technology platforms, we directly compared the MarkerFinder gene predictions from each dataset (Fig. 4D, Table S1). This analysis found that the majority of genes called by MarkerFinder were consistently associated with same cell populations for each technology (n=1112), with Fluidigm finding the greatest number of unique genes (265), followed by Chromium (117), and Drop-Seq (104). While Fluidigm had the largest number of unique genes detected on its own, Chromium was found to have the highest overall agreement with the other platforms. We asked if any of the platform specific marker genes were previously validated. Although we were unable to identify marker genes obtained by a single-platform that have well-documented cell specificity in the literature, many of the population specific genes identified were predicted to correspond to assigned population labels by ToppGene analysis (Table S3).
Validation of Novel Population-Specific Markers
To evaluate the specificity of the scRNA-Seq predictions, we validated the expression patterns of multiple predicted markers by using immunofluorescence, the Allen Brain Atlas and GUDMAP public resources. These analyses validate the specificity of Rprm expression in the ureteric tip, Pcp4 in podocytes, and Spock2 in the cap mesenchyme (Fig. 5 A–C). The c14 compartment contains the nephrogenic zone stroma cells, although genes associated with these cells show some variation in expression domains. While CRABP1 is restricted to the nephrogenic zone stroma, Gpc3 was expressed at the highest level within the nephrogenic zone stroma but also at a lower level in the underlying cortical stroma region (Fig. 5 D,E). Penk and Alx1 showed strongest expression in the stromal cells flanking the collecting ducts, while Col6a1 showed a somewhat complementary expression pattern in the medullary stroma more distant from the collecting ducts (Fig. 5 F–H). These results further confirm the identities of the cells in the distinct clusters and provide novel compartment specific marker genes. Tables of genes with compartment enriched expression are provided in supplementary data (Table S4).
Figure 5. Cell type specific gene expression validations.
A: Rprm, UT: ureteric tip B: Pcp4, Pod: Podocytes, C: Spock2, CM: cap mesenchyme, D: Gpc3, NZS: Nephrogenic zone stroma, E: CRABP1, NZS, F: Penk, CS: Cortical Stroma, G: Alx1, CS, H: Col6a1, MS: Medullary stroma. A, B, D, F, G, and H are from the Allen Brain Atlas, and C from the GUDMAP database. E was validated using immunofluorescence.
Global analysis of receptor/ligand interactions during kidney development
The global analysis of gene expression patterns in the different developing kidney cell types allows the definition of potential compartment cross talk. To this end we filtered for the top scoring population-specific receptor and ligand genes and examined their potential interactions (Fig. 6). There are several limitations to this analysis, as spatial contiguity is not factored in and only population specific markers are considered, and hence some predicted interactions are unlikely and some known interactions, such as WNT9b from the UB tips driving CM differentiation, are missed. However, this analysis reveals a number of interesting possible cell-cell signaling predictions. The endothelial cells, for example, show potential for cross talk with a large number of flanking compartments. In addition, the Cap Mesenchyme nephron progenitors show possible cross talk with the comma and S-shaped bodies through Rspondin1,3/Lgr4,5 interactions. Notably, while many of these cell-cell interactions are novel, multiple of these predicted interactions have been previously demonstrated. For example the interaction between SLIT2 in the collecting ducts and ROBO2 in the CM has been shown to be necessary for proper kidney development (Grieshammer et al., 2004). In Slit2 null mice multiple ureteric budding events occur as opposed to a normal single budding event. It has also been shown that Robo2 is expressed within the CM and Slit2 is expressed within the collecting ducts (Grieshammer et al., 2004).
Figure 6. Possible cell-cell interactions identified through receptor-ligand interaction analysis.
A circos plot displaying prior annotated receptor ligand interactions shared between all possible cell-type combinations. Cell clusters are labeled on the outer most edge of the circle. This analysis was restricted to MarkerFinder cell-state specific genes (Pearson correlation >0.3). Receptors are labeled in red and ligands are labeled in blue. Interconnecting lines are color coded to match the heat map cell clusters. MCD: medullary collecting duct, CD: collecting duct, UT: ureteric tip, LOH: loop of Henle, DCSB, distal comma shaped body, Pod: podocyte, MSSB: mid S-shaped body, PT: proximal tubule, CM: cap mesenchyme, Endo: Endothelium, NZS: nephrogenic zone stroma, CS: cortical stroma, MS: medullary stroma.
Stromal cells express Gdnf
Surprisingly, from the cell-interaction analyses, Gdnf is predicted to have the most consistent and specific expression in nephrogenic zone stromal cells. The receptor-ligand interaction analysis further predicts possible GDNF interaction with the RET/GRFA1 receptors of the ureteric tips, which could theoretically help drive branching morphogenesis. This was surprising because the Cap Mesenchyme has historically been considered the sole source of GDNF (Dressler, 2009; McMahon, 2016). We confirmed this unexpected finding in several ways. First, the three scRNA-Seq technologies, Drop-Seq, Fluidigm and Chromium, all showed expression of Gdnf in stromal cells, as well as cap mesenchyme, providing cross platform validation (Fig. 4C). An expanded view heatmap focusing on selected stromal and cap mesenchyme genes further illustrates the robust Gdnf expression in some stromal cells, often exceeding levels seen in cap mesenchyme cells (Fig. 7).
Figure 7. Gdnf expression in the nephrogenic zone stroma.
Heatmaps using data from all three scRNA-seq platforms show Gdnf expression by stromal cells. Six2, Cited1 and Crym are markers of cap mesenchyme (CM), while Meis1, Foxd1, Crabp1 and Aldh1a2 are expressed in stroma. As expected, cells that clustered with cap mesenchyme as determined by these markers as well as complete gene expression signatures often showed expression of Gdnf. Surprisingly, many cells that strongly clustered with the stromal cell compartment also showed robust Gdnf expression.
To further confirm the expression of Gdnf in stromal cells we next examined public gene expression resources. Using the Allen Brain Atlas public resource (http://www.brain-map.org) in situ hybridization dataset we observed, as expected, Six2 expression primarily in the cap mesenchyme cell population located overlying and immediately flanking the branching ureteric tips at E13.5 (Fig. 8). Gdnf showed an overlapping gene expression pattern, but with weaker expression in the cells overlying the ureteric bud and stronger expression in the cells flanking the ureteric bud. Further, the expression domain of Gdnf appeared to extend deeper than that of Six2, in some cases likely including stromal cells, based on position (Fig. 8).
Figure 8. Gdnf and Six2 in situ hybridization.
A,B: Gdnf and C,D: Six2 in situ hybridization images of E13.5 embryonic kidneys from the Allen Brain atlas. Arrows point to regions of Gdnf expression that are deeper in the kidney than normally seen for Six2, suggesting that they are not cap mesenchyme cells. Arrowheads point to regions overlying ureteric tips, showing low levels of Gdnf expression.
This result was further confirmed using the GUDMAP gene expression resource (GUDMAP.org)(Brunskill et al., 2008). Of interest, the GUDMAP dataset showed the strongest Gdnf expression in the Meis1-GFP FACS purified stromal cell population, with cap mesenchyme cells showing the second highest expression. GUDMAP therefore further verifies that Gdnf is expressed in both cap mesenchyme and stromal cells.
To still further validate the expression of Gdnf by stromal cells we carried out double Gdnf in situ hybridization coupled with SIX2 or MEIS1 IHC. Consistent with the Drop-Seq, Fluidigm and Chromium scRNA-Seq data, the Allen Brain Atlas in situ hybridization data, and the GUDMAP FACS/RNA-Seq data, we observed stromal cell expression of Gdnf in cells that were not SIX2 positive, and were therefore likely stromal cells (Fig. 9D,E,E′). We further confirmed this by Gdnf in situ hybridization coupled with IHC for MEIS1, a specific marker of the stromal lineage. We observed that some of the MEIS1 positive cells indeed showed robust expression of Gdnf (Fig. 9F,G,G′). As predicted by the Allen Brain Atlas, these Gdnf expressing stromal cells were often located near the cap mesenchyme, but extending deeper into the kidney. The stromal cells flanking and immediately beneath the branching UB tips showed the strongest Gdnf expression.
Figure 9. Gdnf expression in stromal cells.
A: Gdnf in situ hybridization (ISH) showing Gdnf expression in the nephrogenic zone. B: SIX2 immunohistochemistry (IHC) showing expression in the cap mesenchyme and its early derivatives. C: MEIS1 IHC showing expression in the stroma. D: Gdnf ISH alone, higher magnification (20X). E: Same Gdnf ISH region as in panel D, but with added SIX2 IHC, highlighting the presence of Gdnf positive, SIX2 negative cells, indicating expression of Gdnf by cells that are not cap mesenchyme. E′: 40X view of Gdnf positive, SIX2 negative cells. F: Gdnf ISH alone (20X). G: Gdnf ISH and MEIS1 (IHC) double staining, showing the presence of Gdnf, MEIS1 double positive stroma cells. G′: 40x view of Gdnf positive, MEIS1 positive cells. Arrowheads point to cells positive for Gdnf and negative for SIX2. Dashed lines outline the SIX2 positive CM. Black double arrowheads point to Gdnf negative, MEIS1 positive cells below the nephrogenic zone. Arrows point to Gdnf positive, MEIS1 double positive cells within the nephrogenic zone.
These combined results suggest that stromal cells might contribute to GDNF signaling. GDNF interacts with the Ret/Gfra1 receptors on the ureteric tips to help drive its branching morphogenesis. Germline mutation of Gdnf, Ret, or Gfra1 results in renal agenesis, with failure of ureteric bud (Olsson et al.) formation (Costantini and Shakya, 2006; Skinner et al., 2008). It is interesting to note that mutation of the transcription factor Foxd1, expressed in the stromal lineage, results in severely reduced ureteric bud branching (Hatini et al., 1996). It was hypothesized that the cause could be an “as yet unidentified ligand for the Ret receptor…” produced by the stroma. Our results, however, suggest that the Foxd1 mutation could result in reduced GDNF expression by the stroma. It would be interesting, in the future, to determine the effects of stromal lineage specific deletion of Gdnf expression.
Multilineage priming
We previously carried out scRNA-Seq on early stage developing kidneys, at E11.5 and E12.5, as well as FACS isolated renal vesicle cells from P4 kidneys (Brunskill et al., 2014). This study used Fluidigm C1 96 cell microfluidics/robotics technology, examining a relatively small number of cells. Of interest, we observed that the early cap mesenchyme cells already showed stochastic expression of genes associated with differentiated cell types. For example a small percent of cap mesenchyme cells expressed Mafb, a marker of differentiated podocytes. These cells did not, however, appear committed to the podocyte lineage. Many of the cap mesenchyme cells expressed a small number of podocyte associated genes, but none showed a strong podocyte gene expression signature. In addition, cells expressing Mafb also showed apparently stochastic expression of markers of other lineages. There was an interesting progression at the later renal vesicle (RV) stage of nephrogenesis. In the RV some cells showed stronger podocyte gene expression signatures, with robust expression of multiple podocyte lineage genes, while other RV cells expressed very few, if any podocyte related genes. The RV cells appeared more lineage committed. Nevertheless, many of the cells showing a podocyte signature simultaneously expressed multiple genes associated with proximal tubules. It appeared, therefore, that while potential lineage directions had narrowed, there were still multiple options available.
In the current study, we used three high throughput scRNA-Seq technologies to examine the total E14.5 kidney, with the results confirming and extending our previous multilineage priming observations. The cap mesenchyme progenitors show stochastic expression of genes associated with podocyte, loop of Henle and proximal tubule lineages (Fig. 10). This expression is robust and not near noise levels. It is impossible to carry out a time course gene expression study for an individual cell using scRNA-Seq, as the cell is destroyed in the process. Nevertheless, the data is consistent with the expression of these different lineage associated genes being sporadic and bursting in nature. As differentiation proceeds it appears there is a progressive repression of genes associated with rejected lineages and simultaneous expression of additional genes related to remaining potential developmental directions. For example in the mid S-shaped body, with cells now largely committed to form proximal tubules, there is more abundant expression of proximal tubule genes and reduced expression of podocyte lineage genes (Fig. 10). In conclusion, as we previously observed, only now looking at far more cells and more developmental stages, nephron progenitors show multi-lineage priming.
Figure 10. Multi-lineage priming.
Heatmap with representative cells from the CM: cap mesenchyme, PA: pretubular aggregate, DCSB: distal comma shaped body, MSSB: mid S-shaped body, PT: proximal tubule, LOH: loop of Henle, and Pod: Podocyte clusters from Drop-seq, Chromium 10X Genomics, and Fluidigm 800-cell. The early progenitor CM cells show stochastic expression of markers of multiple lineages. The MSSB cells are more committed and show elevated expression of proximal tubule associated genes and reduced expression of podocyte marker genes.
Blurred lineage boundaries
According to the classic Waddington landscape model a cell makes a series of binary differentiation decisions during development, excluding one lineage direction while choosing another. The current dogma is that differentiating kidney cells make a similar binary decision early in development, choosing either the stromal or nephrogenic lineage. This dogma is based largely on lineage tracing studies, which show that Six2-Cre labels the nephron lineage (Kobayashi et al., 2008) while Foxd1-Cre labels the stromal lineage (Kobayashi et al., 2014). Of interest it was recently shown that mutation of the Pax2 gene in cap mesenchyme breaks this lineage boundary, causing the nephron progenitor cells to convert to the stromal lineage (Naiman et al., 2017). In this report we show that the nephrogenic zone stroma, as well as cap mesenchyme, both express Gdnf, blurring the functional boundary between these two compartments.
The scRNA-Seq data for E14.5 wild type developing kidneys consistently shows some clouding of the nephron/stromal lineage boundary. For example, a fraction of cells that otherwise show very strong stromal type gene expression patterns nevertheless also show expression of Six2, a nephron progenitor marker, and similarly a significant fraction of cells that cluster well with nephron progenitors nevertheless express Foxd1, a classic marker of the stromal lineage (Fig. 7). This cross-compartment expression extends to other stroma/cap mesenchyme marker genes including Cited1 and Crym (cap mesenchyme/nephron), as well as Meis1 and Crabp1 (stroma) (Fig. 7). This is not the simple result of artifact drops or chambers with two cells, as these single barcodes do not exhibit a full nephron/stroma dual gene expression profile, which would be generated by a stromal cell and a nephron progenitor cell both occupying a single drop.
This observed cross compartment expression can be viewed as an extension of multilineage priming. Progenitors show stochastic expression of genes associated with their multiple potential future developmental directions. The differentiation process then requires activation of additional genes associated with the chosen developmental direction, as well as further repression of genes associated with the rejected developmental directions. Residual cross compartment expression may be the result of remaining incomplete repression of inappropriate genes.
Single-cell RNA-Seq only provides data on RNA levels, and not protein. One can then ask if the observed expression of RNAs that are compartment inappropriate extends to the protein level. In this regard, cells that dual express both SIX2 and FOXD1 protein appear to be very rare, at least ten fold less frequent than cells with both Foxd1 and Six2 RNA (Brunskill et al., 2014; Naiman et al., 2017). Perhaps the cross-compartment transcription is pulsatile in nature, with the resulting transient RNA giving only low level and hence difficult to detect amounts of protein. It is also possible that there are post-transcriptional regulatory mechanisms that limit the quantity of protein produced from these cell type inappropriate RNAs (Li et al., 2017; Liu et al., 2016).
Conclusions
In this report we carry out scRNA-Seq analysis of the E14.5 mouse kidney using three independent technologies. Importantly, this is to our knowledge the first documented combined use of three independent single-cell profiling technologies along with associated methods for their harmonization and side-by-side comparison. As such, we believe the presented bioinformatics workflow has the strong potential to be applied in numerous ongoing large-scale efforts to map and compare sample heterogeneity across diverse samples, laboratories and analytical technologies. The results provide a global definition of the gene expression patterns of the progenitor and multiple differentiating cell types present. Surprisingly, we find that stromal cells, as well as cap mesenchyme nephron progenitors, express Gdnf, a key driver of branching morphogenesis of the collecting duct system. We also confirm and extend earlier studies of multilineage priming during kidney development, examining more cells and additional stages of cell type differentiation. As a resource for the developmental biology research community, we have provided the comparative data from these complementary datasets via a new online interactive web-portal along with the underlying processed and raw gene expression data. In the future, we aim to improve upon these bioinformatics methods, computationally remove doublet cell expression profiles and enhance the prediction of novel rare kidney cell states.
Supplementary Material
Highlights.
Single cell RNA-seq of over 8,000 E14.5 developing kidney cells
Stromal cells, as well as cap mesenchyme, express GDNF
Comparison of 10X Genomics, Drop-Seq and Fluidigm 800 cell IFC single cell RNA-Seq
Multilineage priming at many stages of nephrogenesis
Acknowledgments
We thank Bruce J. Aronow for help with initial stages of data analysis. This work was supported by NIH RO1 DK099995 (Potter) and the NHLBI Progenitor Cell Biology Consortium, Administrative Coordinating Center (U01HL099997) (Salomonis).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Brunskill EW, Aronow BJ, Georgas K, Rumballe B, Valerius MT, Aronow J, Kaimal V, Jegga AG, Yu J, Grimmond S, McMahon AP, Patterson LT, Little MH, Potter SS. Atlas of gene expression in the developing kidney at microanatomic resolution. Dev Cell. 2008;15:781–791. doi: 10.1016/j.devcel.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunskill EW, Georgas K, Rumballe B, Little MH, Potter SS. Defining the molecular character of the developing and adult kidney podocyte. PLoS One. 2011;6:e24640. doi: 10.1371/journal.pone.0024640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunskill EW, Park JS, Chung E, Chen F, Magella B, Potter SS. Single cell dissection of early kidney development: multilineage priming. Development. 2014;141:3093–3101. doi: 10.1242/dev.110601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunskill EW, Potter SS. Gene expression programs of mouse endothelial cells in kidney development and disease. PLoS One. 2010;5:e12034. doi: 10.1371/journal.pone.0012034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunskill EW, Potter SS. Changes in the gene expression programs of renal mesangial cells during diabetic nephropathy. BMC Nephrol. 2012a;13:70. doi: 10.1186/1471-2369-13-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunskill EW, Potter SS. RNA-Seq defines novel genes, RNA processing patterns and enhancer maps for the early stages of nephrogenesis: Hox supergenes. Dev Biol. 2012b;368:4–17. doi: 10.1016/j.ydbio.2012.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini F, Shakya R. GDNF/Ret signaling and the development of the kidney. Bioessays. 2006;28:117–127. doi: 10.1002/bies.20357. [DOI] [PubMed] [Google Scholar]
- Dave V, Wert SE, Tanner T, Thitoff AR, Loudy DE, Whitsett JA. Conditional deletion of Pten causes bronchiolar hyperplasia. Am J Respir Cell Mol Biol. 2008;38:337–345. doi: 10.1165/rcmb.2007-0182OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressler GR. Advances in early kidney specification, development and patterning. Development. 2009;136:3863–3874. doi: 10.1242/dev.034876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieshammer U, Le M, Plump AS, Wang F, Tessier-Lavigne M, Martin GR. SLIT2-mediated ROBO2 signaling restricts kidney induction to a single site. Dev Cell. 2004;6:709–717. doi: 10.1016/s1534-5807(04)00108-x. [DOI] [PubMed] [Google Scholar]
- Hatini V, Huh SO, Herzlinger D, Soares VC, Lai E. Essential role of stromal mesenchyme in kidney morphogenesis revealed by targeted disruption of Winged Helix transcription factor BF-2. Genes Dev. 1996;10:1467–1478. doi: 10.1101/gad.10.12.1467. [DOI] [PubMed] [Google Scholar]
- James RG, Kamei CN, Wang Q, Jiang R, Schultheiss TM. Odd-skipped related 1 is required for development of the metanephric kidney and regulates formation and differentiation of kidney precursor cells. Development. 2006;133:2995–3004. doi: 10.1242/dev.02442. [DOI] [PubMed] [Google Scholar]
- Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi A, Mugford JW, Krautzberger AM, Naiman N, Liao J, McMahon AP. Identification of a multipotent self-renewing stromal progenitor population during mammalian kidney organogenesis. Stem Cell Reports. 2014;3:650–662. doi: 10.1016/j.stemcr.2014.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi A, Valerius MT, Mugford JW, Carroll TJ, Self M, Oliver G, McMahon AP. Six2 defines and regulates a multipotent self-renewing nephron progenitor population throughout mammalian kidney development. Cell Stem Cell. 2008;3:169–181. doi: 10.1016/j.stem.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CJ, Hong T, Tung YT, Yen YP, Hsu HC, Lu YL, Chang M, Nie Q, Chen JA. MicroRNA filters Hox temporal transcription noise to confer boundary formation in the spinal cord. Nat Commun. 2017;8:14685. doi: 10.1038/ncomms14685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Beyer A, Aebersold R. On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell. 2016;165:535–550. doi: 10.1016/j.cell.2016.03.014. [DOI] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A, McCarroll SA. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon AP. Development of the Mammalian Kidney. Curr Top Dev Biol. 2016;117:31–64. doi: 10.1016/bs.ctdb.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugford JW, Yu J, Kobayashi A, McMahon AP. High-resolution gene expression analysis of the developing mouse kidney defines novel cellular compartments within the nephron progenitor population. Dev Biol. 2009;333:312–323. doi: 10.1016/j.ydbio.2009.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naiman N, Fujioka K, Fujino M, Valerius MT, Potter SS, McMahon AP, Kobayashi A. Repression of Interstitial Identity in Nephron Progenitor Cells by Pax2 Establishes the Nephron-Interstitium Boundary during Kidney Development. Dev Cell. 2017;41:349–365. e343. doi: 10.1016/j.devcel.2017.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson A, Venkatasubramanian M, Chaudhri VK, Aronow BJ, Salomonis N, Singh H, Grimes HL. Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature. 2016;537:698–702. doi: 10.1038/nature19348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raines AM, Magella B, Adam M, Potter SS. Key pathways regulated by HoxA9,10,11/HoxD9,10,11 during limb development. BMC Dev Biol. 2015;15:28. doi: 10.1186/s12861-015-0078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramilowski JA, Goldberg T, Harshbarger J, Kloppmann E, Lizio M, Satagopam VP, Itoh M, Kawaji H, Carninci P, Rost B, Forrest AR. A draft network of ligand-receptor-mediated multicellular signalling in human. Nat Commun. 2015;6:7866. doi: 10.1038/ncomms8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roedder S, Sigdel T, Salomonis N, Hsieh S, Dai H, Bestard O, Metes D, Zeevi A, Gritsch A, Cheeseman J, Macedo C, Peddy R, Medeiros M, Vincenti F, Asher N, Salvatierra O, Shapiro R, Kirk A, Reed EF, Sarwal MM. The kSORT assay to detect renal transplant patients at high risk for acute rejection: results of the multicenter AART study. PLoS Med. 2014;11:e1001759. doi: 10.1371/journal.pmed.1001759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigdel TK, Salomonis N, Nicora CD, Ryu S, He J, Dinh V, Orton DJ, Moore RJ, Hsieh SC, Dai H, Thien-Vu M, Xiao W, Smith RD, Qian WJ, Camp DG, 2nd, Sarwal MM. The identification of novel potential injury mechanisms and candidate biomarkers in renal allograft rejection by quantitative proteomics. Mol Cell Proteomics. 2014;13:621–631. doi: 10.1074/mcp.M113.030577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MA, Safford SD, Reeves JG, Jackson ME, Freemerman AJ. Renal aplasia in humans is associated with RET mutations. Am J Hum Genet. 2008;82:344–351. doi: 10.1016/j.ajhg.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambon AC, Gaj S, Ho I, Hanspers K, Vranizan K, Evelo CT, Conklin BR, Pico AR, Salomonis N. GO-Elite: a flexible solution for pathway and ontology over-representation. Bioinformatics. 2012;28:2209–2210. doi: 10.1093/bioinformatics/bts366. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.