Abstract
As the world population ages, primary prevention of age-related cognitive decline and disability will become increasingly important. Prevention strategies are often developed from an understanding of disease pathobiology, but models of biological success may provide additional useful insights. Here, we studied 224 older adults, some with superior memory performance (n=41), some with normal memory performance (n=109), and some with mild cognitive impairment (MCI) or Alzheimer’s disease (AD) (n=74) to understand metabolomic differences which might inform future interventions to promote cognitive health. Plasma metabolomics revealed significant differential abundance of 12 metabolites in those with superior memory relative to controls (ROC AUC = 0.89) and the inverse abundance pattern in the MCI, AD (AUC = 1.0) and even preclinical AD groups relative to controls (AUC = 0.97). The 12 metabolites are components of key metabolic pathways regulating oxidative stress, inflammation, and nitric oxide bioavailability. These findings from opposite ends of the cognitive continuum highlight the role of these pathways in superior memory abilities and whose failure may contribute to age-related memory impairment. These pathways may be targeted to promote successful cognitive aging.
Keywords: Memory, Metabolomics, Alzheimer’s Disease, Oxidative Stress, Nitric Oxide, Arginine Metabolism
1. Introduction
Aging is characterized by the accumulation of life experiences that present opportunities and challenges for continued growth and development. Aging research has historically focused on what happens when we fail to negotiate these challenges and relatively less attention has been paid to understanding characteristics of successful aging. While there is no generally accepted definition of successful aging, most include some concept of freedom from physical and cognitive disability (Depp and Jeste, 2006) which optimize functional capacity and quality of life. Maintaining cognitive abilities in the face of age-related physiological changes represents a significant challenge especially given the diversity of individual life experience and complexity of brain organization which interact to produce individual cognitive trajectories (Albert, 1997). From the fourth decade of life onward, the most common cognitive trajectory is characterized by subtle decline in many abilities, most frequently those requiring rapid transfer of information across widespread brain networks (Salthouse, 2009), but this is not invariant and relative stability and improvement in complex cognitive abilities such as memory is occasionally encountered into old age (Rowe and Kahn, 1987, Gefen et al., 2014). These alternate paths may be supported by resistance to age-related accumulation of pathologies (Balasubramanian et al., 2012) or cognitive reserve (Stern, 2012) or enhanced neuroplasticity (Gutchess, 2014).
The search for lower cost, minimally invasive, high-throughput biosignatures of cognitive dysfunction has driven technological advances in metabolomic platforms (Li et al., 2010, Quinones and Kaddurah-Daouk, 2009). For example, peripheral blood metabolomic analyses allow qualitative and quantitative assessment of circulating small molecules representing central metabolic pathways (Voyle et al., 2016). Together with genomics, transcriptomics, and proteomics, metabolomics is helping expand our detailed appreciation of systems biology. The biofluid matrix (e.g., plasma or serum) being interrogated via metabolomics, along with the molecular separation methods used (e.g., gas versus liquid chromatography) coupled with mass spectrometry for molecular identification, determines the specific yield of molecular species that can be used as a phenotypic readout. These data provide direct and/or indirect evidence for altered biochemical pathways linked to pathobiology (Pan et al., 2016) and brain structural (Ciavardelli et al., 2016) and functional (Cansev, 2016) integrity. The scope and depth of such molecular perturbations defined through metabolomics may ultimately empower individualized molecular phenotyping and our understanding of disease-specific mechanisms. Herein, we report an analysis of the plasma metabolome of older adults with superior memory. Through this investigation we sought new information about the biochemical processes that support successful cognitive aging trajectories and may provide insights into age-related cognitive disorders, such as Alzheimer’s disease (AD), where memory impairment is the cardinal feature.
2. Methods
2.1 Participants
All participants in this study were recruited from the communities of Rochester, NY or Irvine, CA as part of the Rochester/Orange County Aging Study (R/OCAS). Inclusion criteria included age over 70 years, good overall physical health, visual acuity and hearing sufficient for cognitive testing, and proficiency with the English language. Exclusion criteria included major neurological or psychiatric illness including a known diagnosis of any phenotype of Mild Cognitive Impairment (MCI) or AD, current or recent (<1 month) use of anticonvulsants, neuroleptics, highly active anti-retroviral therapy (HAART), antiemetics, and antipsychotics for any reason, and serious blood disorder including chronic abnormalities in complete blood count and anemia requiring therapy and/or transfusion. All R/OCAS participants gave written informed consent and all procedures in this study were approved by Research Studies Review Boards at the University of Rochester, University of California Irvine, and Georgetown University.
2.2 Cognitive assessment and classification
As part of the R/OCAS, all study participants underwent yearly cognitive testing and provided a yearly blood sample. Cognitive testing was performed at each yearly visit following the blood draw and breakfast. The cognitive battery consisted of commonly used measures administered in the standardized manner (Mapstone et al., 2014) (Supplemental Table 1). The verbal episodic memory measure was the Rey Auditory Verbal Learning Test (RAVLT) (Rey, 1964). We classified the subjects in this study using composite Z-scores based on the group characteristics adjusted for age, education, sex, and visit. Adjustment for visit allowed us to account for putative practice effects over the multi-year study. The five composite cognitive domain Z-scores included: attention (Zatt); executive (Zexe); language (Zlan); memory (Zmem); and visuospatial (Zvis)(Mapstone et al., 2014) (Supplemental Table 2). To reduce the effect of cognitively impaired participants on the mean and SD, age-, education-, sex-, and visit-adjusted residuals from each domain Z-score model were robustly standardized to have median 0 and robust SD=1, where the robust SD=IQR/1.35, as 1.35 is the IQR (Inter-Quartile Range) of a standard normal distribution.
A total of 525 participants were enrolled in the R/OCAS and 497 participants had complete blood and cognitive data. From this group of 497, we identified 41 participants (8% of the total sample) showing superior memory abilities using the above criteria. Superior memory for the supernormal (SN) group was defined as Zmem > 1.35 SD and corresponds to the 90th%ile. To further isolate successful cognitive aging in the SN group, all other domain composite Z-scores were required to be > −1.35 SD or greater than the 10th%ile. After defining the SN participants we used frequency matching to select in a pseudo random manner an age-, education-, and sex-matched normal control group of 41 participants (NCs) for the SN group. In order to enhance the specificity of our analyses, all normal control participants in this study were conservatively defined with Zmem ±1 SD (15th %ile – 85th%ile) of the cohort median rather than simply non-impaired or ≥ −1.35, and all other Z-scores ≥ −1.35 SD (Supplemental Figure 1).
The same cognitive assessment and Z-score methods were used to define the 74 amnestic MCI (aMCI), AD and preclinical AD (ConverterpreAD), and their 73 matched control participants (NCo) detailed in our previous work(Mapstone et al., 2014). We chose to include the amnestic phenotype of MCI rather than include other behavioral phenotypes in order to conservatively restrict our analysis to a group of MCI with the highest likelihood of common underlying pathobiology. Thus, our combined aMCI/AD group ostensibly represents a relatively homogenous group of individuals with nascent AD pathobiology. We combined the aMCI and AD subjects into a single group for all analyses. In order to preserve non-overlapping normal control samples for the SN and aMCI/AD groups, five of the 73 NCo participants reported in the previous study(Mapstone et al., 2014) were included as NCs for the SN group. Thus 68 of the original 73 remained as NCo for the aMCI/AD group (Supplemental Figure 1). As defined, the participant groups were not significantly different from each other based on age, sex, and education (Table 1). There was a significant main effect of sex on education level when comparing the SN and aMCI/AD groups (MANOVA F= 4.85, p = 0.003) such that the SN males were more highly educated than the aMCI/AD females. As defined, the groups did differ on the cognitive Z-scores (Supplemental Figure 2).
Table 1.
Demographic Details
| Group | n | Mean Age years (SD) | Number Male (%) | % ApoE4 | Mean BMI (SD) | Mean Education years (SD) | Mean MMSE (SD) |
|---|---|---|---|---|---|---|---|
| SN | 41 | 83.22 (3.37) | 20 (48.8) | 5/41 (12%) | 26.55 (4.94) | 16.41 (2.68) | 29.05 (1.07) |
| Discovery | 26 | 82.69 (3.50) | 13 (50.0) | 0/26 (0%) | 26.10 (4.61) | 15.92 (2.37) | 29.15 (1.08) |
| Validation | 15 | 84.13 (3.04) | 7 (46.7) | 5/15 (33%) | 27.34 (5.54) | 17.27 (3.06) | 28.87 (1.06) |
| NCS | 41 | 83.29 (3.82) | 20 (48.8) | 4/41 (9%) | 25.53 (3.21) | 16.24 (2.47) | 28.66 (1.37) |
| Discovery | 26 | 82.88 (3.34) | 18 (69.2) | 4/26 (15%) | 25.08 (2.91) | 16.50 (2.63) | 28.68 (1.29) |
| Validation | 15 | 84.50 (4.42) | 2 (14.3) | 0/15 (0%) | 26.29 (3.63) | 15.86 (2.25) | 28.62 (1.27) |
| aMCI/AD | 74 | 81.93 (4.37) | 20 (27.0) | 24/74 (32%) | 25.93 (3.97) | 15.36 (2.45) | 26.33 (2.77) |
| ConverterpreAD# | 28 | 80.21 (4.02) | 12 (42.9) | 5/28 (17%) | 26.51 (4.54) | 15.04 (2.74) | 28.61 (2.49) |
| NCO | 68 | 81.69 (3.40) | 26 (38.2) | 16/68 (23%) | 27.03 (4.77) | 15.43 (2.40) | 28.19 (3.75) |
Note: SN = Supernormal; NCs = Normal control for supernormal sample; aMCI/AD = amnestic mild cognitive impairment and Alzheimer’s disease group; ConverterpreAD = Preclinical AD; NCo = Normal control for ConverterpreAD and aMCI/AD sample. The aMCI/AD, ConverterpreAD, and NCo participants were included in our previous study(Mapstone et al., 2014). MMSE = Mini Mental State Examination. BMI = Body Mass Index.
Baseline data for subjects who converted to aMCI/AD within 2.1 years.
2.3 Blood samples
All study participants provided a blood sample on the same day as the cognitive testing. Because certain chronobiological factors including circadian(Panda et al., 2002, Storch et al., 2002, Reddy et al., 2006), seasonal(Reinberg et al., 1988, Walker et al., 1997) and diurnal (Bollard et al., 2005, Walsh et al., 2006) rhythms are known to affect metabolism and presumably ephemeral metabolites such as lipids, our group has implemented strict standardization of blood collection and handling methods (Mapstone et al., 2014, Fiandaca et al., 2015). In this study, the blood draw was performed as close as possible to the same time of day and day of the year to control for circadian, seasonal, and other chronobiological effects on the blood metabolomics. All study participants underwent phlebotomy between 8am and 10am, while fasting and withholding their morning medications. Blood specimens were collected in EDTA vacutainers and, after thorough mixing, placed on wet ice immediately after collection and remained on ice until the blood components were separated within 24 hours, in order to retard degradation of metabolites (Hammad et al., 2010). Each sample yielded multiple 100 uL plasma aliquots that were frozen immediately thereafter at −80°C until undergoing metabolomic analyses. The smaller plasma aliquots allowed specimen use following a single freeze-thaw cycle prior to metabolomic processing for all specimens.
Metabolomic analyses of the aMC/AD, ConverterpreAD, and NCO plasma samples were completed in September 2013 and on the SN and NCS samples in November 2014. One-way ANOVA on plasma sample storage length with subject group as the independent variable showed that the mean storage length was significantly different across groups (F=22.31, p<0.001). Post-hoc analysis showed that the plasma storage time of the two main groups under study here, the SN and NCS groups were not significantly different (SN mean storage = 49.7 months, NCS mean storage = 49.3 months), nor were the aMCI/AD and NCO groups different from each other (aMCI/AD mean storage = 38.2 months, NCO mean storage = 37.1 months). However, the ConverterpreAD samples had been stored for significantly longer than all other groups (ConverterpreAD mean storage = 59.8 months) (Supplemental Table 3).
2.4 Reagents
LC/MS-grade acetonitrile (ACN), Isopropanol (IPA), water and methanol were purchased from Fisher Scientific (New Jersey, USA). High purity formic acid (99%) was purchased from Thermo-Scientific (Rockford, IL). Debrisoquine, 4-Nitrobenzoic acid (4-NBA), Pro-Asn, Glycoursodeoxycholic acid, Malic acid were purchased from Sigma (St. Louis, MO, USA). All lipid standards including 14:0 LPA (lysophosphatidic acid), 17:0 Ceramide, 12:0 LPC, 18:0 Lyso phosphatidylinositol (PI), and 22:6 phosphatidylcholine (PC) were procured from Avanti Polar Lipids Inc. (USA).
2.5 Targeted metabolomics using stable isotope dilution – multiple reaction monitoring- mass spectrometry (SID-MRM-MS)
In this study, targeted metabolomic analysis of plasma samples was performed using the Biocrates Absolute-IDQ P180 (BIOCRATES, Life Science AG, Innsbruck, Austria). This validated targeted assay allows for simultaneous detection and quantification of metabolites in plasma samples (10uL) in a high throughput manner. The plasma samples were processed as per the instructions by the manufacturer and analyzed on a triple quadrupole mass spectrometer (Xevo TQ-S, Waters Corporation, USA) operating in the MRM mode. The measurements were made in a 96 well format for a total of 82 samples, seven calibration standards and three quality control samples were integrated in the kit. Briefly, the flow injection analysis (FIA) tandem mass spectrometry (MS/MS) method was used to quantify a panel of 144 lipids simultaneously by MRM. The other metabolites are resolved on the UPLC and quantified using scheduled MRMs. The kit facilitates absolute quantitation of 21 amino acids, hexose, carnitine, 39 acylcarnitines, 14 sphingomyelins, 87 phosphatidylcholines and 21 biogenic amines. The abundance is calculated from an area under the curve (AUC) by normalizing to the respective isotope labeled internal standard and differential abundance between different participant groups was computed based on relative ratios of normalized response. The concentration is expressed as nmol/L. Human EDTA plasma samples spiked with metabolite standards were used as quality control samples to assess reproducibility of the assay. The mean coefficient of variation (CV) for the 180 metabolites was 0.08 and 95% of the metabolites had a CV of <0.15 and all had CVs < 0.2. The data were pre-processed using the MetIDQ software (BIOCRATES, Life Science AG) prior to statistical consideration. Raw abundance of each metabolite for each group is reported in the Supplemental Materials (Supplemental Table 6). Summary statistics for metabolites were completed using MetaboAnalyst 3.0 (Xia and Wishart, 2011, Xia et al., 2015)
2.6 Statistical Analysis
The primary analysis focused on creating a logistic regression model from the targeted metabolomic data elements to classify the SN from the NCs. In addition, we wished to test suitability of this model derived from participants with superior memory in the aMCI/AD participants; a group characterized by impaired memory. We also wished to apply the 10-lipid panel developed in our previous study (Mapstone et al., 2014) to the SN participants; a group without clinical evidence for neurodegenerative disease. Finally, we wished to create a comprehensive model based on the metabolomic features of 1) memory function from the SN model and 2) neurodegenerative disease from our previously published model.
The procedure for metabolite selection was similar to our previous report(Mapstone et al., 2014). In the metabolomic discovery phase, we performed targeted analysis using the Biocrates Absolute IDQ p180 kit on plasma from 2/3 of the SN and NCs participants (n=26 in each group) while the remaining 1/3 of the samples from each group were reserved for an internal validation phase. The abundance measurements for the metabolites were initially transformed using natural log transformation and normalized via quantile normalization. We developed group classification models using the least absolute shrinkage selection operator (LASSO)(Tibshirani, 1996) and emphasizing selection of annotated metabolites which classified the two groups (SN vs NCs) with the greatest accuracy. To evaluate the predictive power of the metabolite panel, we fit deLong’s test of the receiver operating characteristic (ROC) regularized logistic regression model based on the LASSO penalty for the discovery cohort (26 SN vs. 26 NCs). We first obtained the regularization path over a grid of values for the optimizing parameter λ through N fold cross-validation to generate stable estimates. The optimal value of the tuning parameter λ was then used to estimate the penalty regression coefficients in the model. Models were fit using the “glmnet” package in R, which uses cyclical coordinate descent in a path-wise fashion. All of the individual metabolites with nonzero coefficients were retained for subsequent analysis. Logistic regression was used to create a classifier model and the classification performance of the model was assessed using deLong’s test of area under the ROC curve (AUC), measuring the predictive accuracy separately for the discovery and validation stages. In order to validate the model from the discovery stage, we performed ROC analyses with the validation set of SN (n = 15) and their matched NCs (n=15) as an internal validation. Positive predictive value (PPV) and negative predictive value (NPV) for the optimal sensitivities and specificities were calculated using an estimated prevalence of 5%. We conservatively estimated this figure from our statistical definition of supernormal which requires memory performance above one robust standard deviation factoring in normal performance in other cognitive domains.
3. Results
3.1 A panel of 12 metabolites distinguishes cognitively superior from control participants
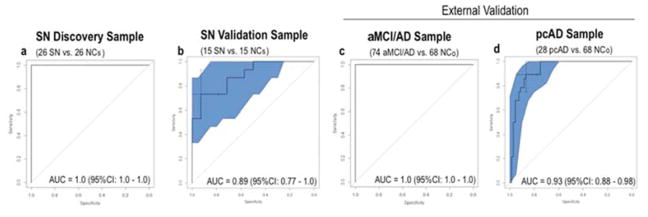
The LASSO procedure selected twelve metabolites (Aspartate, Hydroxyhexadecadienylcarnitine (C16:2-OH), 3-Hydroxypalmitoleylcarnitine (C16:1-OH), Lyso PC a C28:1, Arginine, Valerylcarnitine (C5), Lyso PC a C17:0, Asparagine, Citrulline, Nitrotyrosine, PC aa C38:5, and Histamine) which met the specific criteria for the classification model (Table 2). One of the 12 metabolites, 16:1-OH was featured in our previously reported panel of ten plasma lipids(Mapstone et al., 2014). The logistic regression classifier model constructed with this set of metabolites produced a ROC AUC of 1.0 [95% CI: 1.0 – 1.0] (Figure. 1A) indicating error-free classification of the SN and NCs groups. At the optimal threshold, sensitivity was 1.0, specificity was 1.0, positive predictive value (PPV) was 1.0, and negative predictive value (NPV) was 1.0 (Supplemental Table 4).
Table 2.
Natural Log Transformed Data of the Discovery and Internal Validation Samples for the 12-Metabolite Panel.
| Discovery sample | Validation sample | Total sample | ||||||
|---|---|---|---|---|---|---|---|---|
| Metabolite | Group | Mean | SD | t test* (p) | Mean | SD | t test* (p) | Beta, SE (p) # |
| Arginine | SN | 4.16 | 0.37 | −0.84 (.41) | 4.34 | 0.51 | −1.22 (.23) | −0.17, 0.96 (.10) |
| NCs | 4.25 | 0.40 | 4.53 | 0.32 | ||||
| Hydroxyhedadecadienylcarnitine (C16:2-OH) | SN | −4.68 | 0.24 | −1.02 (.32) | −4.68 | 0.23 | −1.74 (.10) | −0.25, 2.49 (.01) |
| NCs | −4.61 | 0.24 | −4.56 | 0.15 | ||||
| 3-Hydroxypalmitoleylcarnitine (C16:1-OH) | SN | −4.59 | 0.25 | −0.38 (.70) | −4.62 | 0.25 | −2.17 (.04) | −0.05, 2.08 (.90) |
| NCs | −4.57 | 0.18 | −4.43 | 0.20 | ||||
| Lyso PC a C17:0 | SN | 0.84 | 0.27 | −1.02 (.31) | 0.78 | 0.27 | −1.31 (.20) | −0.34, 1.70 (.004) |
| NCs | 0.91 | 0.27 | 0.93 | 0.37 | ||||
| Asparagine | SN | 4.19 | 0.22 | −1.03 (.31) | 4.20 | 0.18 | −2.19 (.01) | −0.67, 0.88 (.001) |
| NCs | 4.26 | 0.22 | 4.36 | 0.21 | ||||
| Lyso PC a C28:1 | SN | −0.81 | 0.29 | 1.77 (.08) | −0.75 | 0.33 | −0.90 (.38) | −0.01, 1.42 (.99) |
| NCs | −0.93 | 0.20 | −0.64 | 0.28 | ||||
| Nitrotyrosine | SN | −0.30 | 0.18 | 0.14 (.89) | −0.18 | 0.13 | 2.86 (.007) | 0.94, 1.42 (.009) |
| NCs | −0.31 | 0.26 | −0.52 | 0.42 | ||||
| Valerylcarnitine (C5) | SN | −1.99 | 0.49 | 0.90 (.37) | −2.15 | 0.34 | −0.32 (.75) | 0.10, 0.78 (.08) |
| NCs | −2.12 | 0.50 | −2.11 | 0.39 | ||||
| Histamine | SN | −0.39 | 2.30 | −0.29 (.78) | −0.59 | 2.17 | 0.17 (.87) | 0.03, 0.13 (.81) |
| NCs | −0.19 | 2.65 | −0.76 | 3.41 | ||||
| PC aa C38:5 | SN | 4.16 | 0.30 | 2.00 (.05) | 4.14 | 0.28 | 0.23 (.82) | 0.41, 1.87 (.001) |
| NCs | 3.98 | 0.32 | 4.12 | 0.28 | ||||
| Aspartate | SN | 3.04 | 0.48 | 2.34 (.02) | 3.02 | 0.42 | −1.42 (.18) | 0.35, 0.88 (.15) |
| NCs | 2.73 | 0.48 | 3.19 | 0.23 | ||||
| Citrulline | SN | 4.15 | 0.32 | 1.28 (.21) | 4.10 | 0.26 | −1.72 (.10) | 0.80, 1.91 (.009) |
| NCs | 4.04 | 0.27 | 4.28 | 0.28 | ||||
Note: SN = Supernormal; NCs = Normal control for supernormal sample;
Significant difference between the groups for individual metabolites is not required for inclusion in the logistic regression classifier model.
Beta weights are from the overall classifier model with SN vs. NCs as the outcome and 12 metabolites collectively as the predictors for the Discovery and Validation samples. Negative Beta weights indicate lower abundance of the metabolite in the SN group compared to the NCs group, while positive Beta weights reflect greater abundance in the SN group.
Figure 1. Results of ROC analysis using the 12-metabolite panel.
This figure shows plots of SN vs NCs ROC analysis using the 12-metabolite panel in targeted discovery (a) and validation (b) phases and application of the 12-metabolite panel to the external aMCI/AD (c) and ConverterpreAD (d) samples. 95% confidence intervals shaded in blue. Crosshair on ROC plot represents optimal ROC threshold. SN = Supernormal, NCs = Normal control for supernormal sample; aMCI/AD = amnestic mild cognitive impairment and Alzheimer’s disease; ConverterpreAD = Preclinical AD; NCo = Normal control for ConverterpreAD and aMCI/AD samples.
Because this procedure results in overfitting by design, we applied the model to the reserved validation group samples whose group membership was blinded to the statistical team. Here, the classifier model produced a ROC AUC of 0.89 [95% CI: 0.77 – 1.0] indicating very good separation of the SN and NCs (Figure. 1B). We further confirmed model fit using the Hosmer-Lemeshow test run at 10 folds in the discovery and validation groups separately, which showed good calibration (p values > 0.05). In the validation phase, sensitivity was 0.93, specificity was 0.73, PPV was 0.92 and NPV was 0.76.
Five of the selected metabolites were not concordantly expressed in the discovery and validation cohorts. Given the small sample size and lack of statistical significance, we did not exclude the five non-concordant metabolites from our final model. In the validation dataset, post-hoc analyses using only the seven concordant metabolites resulted in a non-significant decline in AUC compared to the 12-metabolite panel (deLong’s test: |Z| = 1.28, p=0.20) lending support to the inclusion of the non-concordant metabolites (Supplemental Figure 3).
3.2 Twelve metabolite panel distinguishes cognitively impaired from control participants
We then sought to determine whether the 12-metabolite panel, reflecting superior memory function, could also discriminate individuals with impaired cognition. We reversed the signs on the coefficients in our 12-metabolite classifier model and applied this to the aMCI/AD and NCo groups. The reversed 12 metabolite classifier model produced a ROC AUC of 1.0 [95% CI: 1.0 – 1.0] (Fig, 1C) indicating error-free classification of the memory impaired aMCI/AD group from their cognitively normal controls.
3.3 Twelve metabolite panel distinguishes preclinical AD from control participants
We then sought to examine the utility of the reversed 12-metabolite classifier model in preclinical AD by applying it to 28 ConverterpreAD participants, who phenoconverted from normal cognition at entry in the study to aMCI or AD on average 2.1 years later, and their cognitively normal controls. The reversed 12-metabolite classifier model produced a ROC AUC of 0.97 [95% CI: 0.92 – 1.0] for the 28 ConverterpreAD participants compared to their controls (Fig. 1D). This is particularly interesting as the ConverterpreAD participants did not, by definition, demonstrate memory impairment, but did so within the next several years, suggesting the 12 metabolites may reflect early memory-related biochemical alterations that precede threshold for clinical detection.
3.4 Combined 10-lipid and twelve metabolite panels accurately classifies all groups
We then explored the utility of our previously reported 10 lipid panel(Mapstone et al., 2014), which shares a single common metabolite (C16:1-OH) with the 12 metabolite panel, to distinguish the SN from the NCs group. We found only moderate evidence that the former, a proposed marker of early neurodegeneration, is associated with the physiology of superior memory ability (ROC AUC = 0.71, 95% CI: 0.59 – 0.82) (Supplemental Table 4). The combination of the 10 lipid panel, putatively representing early neurodegeneration, with the 12 metabolite panel, putatively representing memory function, into a 21 metabolite panel (with C16:1-OH overlapped), however, accurately classified the SN and the NCs groups (ROC AUC = 1.0, 95%CI: 1.0 – 1.0), the aMCI/AD and NCo groups (ROC AUC = 1.0 95% CI = 1.0 – 1.0), and the ConverterpreAD and NCo groups (ROC AUC = 0.99, 95% CI: 0.97 – 1.0) (Supplemental Table 4).
3.5 Twelve metabolite panel specific to memory ability
Finally, we developed a plasma 12-metabolite index, using the standardized coefficient (Beta) of each metabolite in the SN (n=41) vs NCs (n=41) logistic regression classifier model (Table 2) to weigh the natural log transformed metabolite abundance and create a single 12 metabolite-index for all participants in the study (SN n= 41, aMC/AD n=74, and combined NC n=109) (Figure. 2A). Linear regression models of the 12-metabolite index and the five cognitive domains (Zatt, Zexe, Zlan, Zmem, Zvis) controlling for group (SN, aMCI/AD, NC) showed a significant relationship between the 12-metabolite index and memory composite Z-scores in the aMCI/AD, NC (combined NCs and NCo), and SN groups (Beta = 0.09, t = 2.30, p = 0.022) when adjusting for group (Figure. 2B). Importantly, the 12-metabolite index was not associated with other cognitive domains supporting its specificity to memory processes (Supplemental Table 5).
Figure 2. 12-metabolite index and relationship with memory performance.
This figure shows the derived 12-metabolite index for the three groups (SN, NC, and aMCI/AD) (a). Box-plot for index values between groups. Using multinomial logistic regression, the 12-metabolite index significantly differentiated the SN, NC, and aMCI/AD groups (all p < .013). The relationship between the 12-metabolite index and memory composite z-score is shown for each group of participants in the study (b). Zmem = Memory composite score; SN = Supernormal; NC = Normal control (combined NCs and NCo); aMCI/AD = amnestic mild cognitive impairment and Alzheimer’s disease. Note. ** p < .01; *** p < .001.
4. Discussion
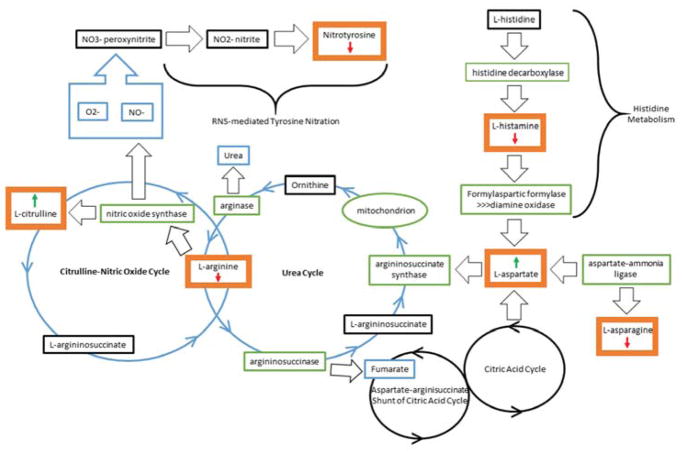
Here we report a set of plasma-derived metabolites that characterize a state of successful cognitive aging in a limited clinical cohort. The strong association of these metabolites (phospholipids, acylcarnitines, amino acids, and biogenic amines) with the composite memory score, and the specificity for memory, support the use of this molecular-phenotyping approach in the discovery of biologically relevant pathways associated with successful cognitive aging. While the precise molecular network of interaction for these 12 metabolites remains to be elucidated, their apparent connection to superior memory performance is provocative (Figure 3). We found lower levels of L-arginine in our participants with superior memory. A recent plasma metabolomic study showed elevated L-arginine levels in stable MCI subjects, as well as in MCI subjects who converted to AD when compared to controls (Graham et al., 2015). In addition, other groups have shown evidence of altered transcript and protein levels of arginase, together with reduced ornithine decarboxylase and polyamine levels in AD brain tissue, suggesting a link between arginine metabolism and AD (Morrison et al., 1995, Morrison et al., 1998, Colton et al., 2006, Hansmannel et al., 2010). In contrast, reduction of L-arginine in our SN group, while yet to be fully dissected, might reflect rapid turnover of the substrate to form L-citrulline through either the urea cycle or the nitric oxide (NO) cycle(Liu et al., 2014). Up-regulation of the urea cycle decreases nitrosative stress, which is consistent with the reduced levels of nitrotyrosine noted in our SN group. Further, reduction of nitrotyrosine and histamine in our SN participants may reflect a state of lower overall oxidative stress and systemic inflammation (Tohgi et al., 1999, Alvarez, 2009) in this model of successful cognitive aging.
Figure 3.
Biological pathways implicated in superior memory performance. This diagram shows the pathways implicated by the metabolomic results of the SN vs NCs comparisons. The bold orange boxes show the metabolites which were significantly altered in the analysis and the arrows associated with the metabolite indicate the level of the metabolite in the SN group relative to the NCS group. For the aMCI/AD vs NCO comparison, the arrows would be reversed indicating the opposite relationship between the levels of each metabolite relative to the NCO.
These metabolomic results reveal a unique set of potential physiological markers for a diverse range of memory abilities (aMCI/AD < preclinical AD < normal < SN) and implicate several memory-related physiological processes. The upregulation of aspartate, a potent N-methyl-D-aspartate receptor agonist may support synaptic plasticity and superior memory (Shimizu et al., 2000) characterizing SN participants. In addition, putatively increased bioavailability of NO in SN participants may mechanistically enhance long term potentiation(Schuman and Madison, 1991) and promote synaptic plasticity(Nikonenko et al., 2013, Chakroborty et al., 2015) and cognitive reserve(Lores-Arnaiz et al., 2006). In the aMCI and AD participants however, dysregulation of these memory-relevant processes may contribute to the characteristic memory loss of these conditions. We also found evidence of these metabolic disruptions in the preclinical state of AD where, by definition, memory ability is not impaired, but the antecedent pathobiology of future memory loss may be present. This observation in particular suggests metabolic disruption occurs and can be detected early in the disease process and may be related to the emergence of tau pathology and neurodegeneration characterizing stage 2 preclinical AD(Sperling et al., 2011). We find support for this notion in the strong classification performance of the combined memory and neurodegeneration 21-metabolite model for both aMCI/AD and preclinical AD; a model which may more comprehensively reflect AD pathobiology. In the cognitively successful SN brain, efficient information transfer in memory and executive brain networks(LePort et al., 2012) may reflect successful adjustments to age-related neurophysiological decline(Grady, 2008). In contrast, the memory impairment associated with the AD brain may reflect inadequate or failed compensation to cumulative pathobiologic events(Mesulam, 2000).
These results may have important implications for promoting successful cognitive aging, which can lead to improved functional independence and quality of life for millions of older adults, however, there are several limitations to the inferences we can draw from this study. First, like other peripheral blood biomarker studies of brain-related conditions, the link between central and peripheral metabolomic reflections is likely to be indirect. This is less of a problem when investigating animal models, where central and peripheral biochemistry can be directly measured, or when using matrices such as human cerebrospinal fluid, which is in direct contact with the brain parenchyma. Although certain explanations (Fiandaca et al., 2015) provide plausible support for direct peripheral metabolomic manifestations resulting from an active central brain process, the direct proof for such postulates is currently lacking.
Second, as with many other investigations utilizing peripheral biosignatures of brain-related conditions, replication/validation of our results is required. To do so, investigators must not only utilize similar study designs and analytic methods, but also investigate similar subject groups (demographics) and matrices (plasma, and not serum), as in our study. In addition, our small cohort of analyzed subjects makes generalization of our results quite challenging. Future analyses of larger demographically different subject cohorts are necessary to investigate the applicability of our results beyond our limited group. Expanded studies, using similar metabolomic methods and comparably defined subjects, provide opportunities for novel metabolomic discovery that may support, critique, or go beyond our current observations.
Third, we do not know the impact of plasma sample storage duration on measurement of these metabolites. The ConverterpreAD samples had been stored for significantly longer duration (approximately 23 months longer) than the other groups and we do not know for sure if there was any effect on sample quality and ultimately relative metabolite abundances due to this. Plasma sample quality very likely deteriorates over extended storage (>10 years) even at −80C, but recent work suggests that short storage periods of up to 2.5 years has negligible effects on metabolite measurements (Pinto et al., 2014). Furthermore, we believe that differences in sample storage duration did not significantly affect the metabolomic results here because the same control group (NCO) was used as comparison for both the aMCI/AD and ConverterpreAD groups. The NCO and aMCI/AD samples had very similar mean storage durations. If sample storage duration significantly influences metabolite abundance we would expect to have seen differences in these comparisons, but the ROC AUC results for both groups were nearly identical for the 12-metabolite (aMCI/AD vs NCO = 1.0, ConverterpreAD vs NCO = 0.97) and the 21-metabolite panels (aMCI/AD vs NCO = 1.0, ConverterpreAD vs NCO = 0.99) suggesting that storage duration, at least the 23 month difference in our study, had minimal effect. Certainly, greater storage duration (e.g., 10 or more years) may have a much greater impact on sample quality and metabolite measurements.
Finally, there is a lack of consensus on how to define the construct of successful cognitive aging and we recognize that our operational definition focusing on the singular feature of memory ability may be limited. We chose superior memory as an exemplar of successful cognitive aging in order to 1) discretely operationalize the construct, 2) recognize the relative complexity of memory above other cognitive abilities (e.g., attention), and 3) to provide a common dimension for comparison with a more commonly used model of unsuccessful cognitive aging, AD where memory impairment is the cardinal feature. We do not believe that superior memory alone defines the entirety of successful aging and recognize that other attempts at operationalizing are equally valid and may provide additional useful insights into aging success. We also recognize that interventions designed to directly improve memory or mitigate memory decline may not lead to beneficial effects on functional capacity or quality of life in older adults. However, we anticipate that an improved understanding of both health- and disease-related metabolomic characteristics may ultimately lead to preventative strategies that not only maximize general health and longevity, but also reduce likelihood of age-related cognitive decline.
Supplementary Material
Highlights.
We examined plasma metabolomics of older adults with superior memory
12 metabolites were differentially abundant in these subjects compared to controls
These 12 metabolites were abundant in the opposite direction in Alzheimer’s disease
The metabolites are involved in inflammation, oxidative stress, and NO availability
Modulation of these pathways may promote successful cognitive aging trajectories
Acknowledgments
The authors thank Eileen Johnson, RN for collecting and Robert Padilla, MS for processing the blood samples.
Funding: This work was funded by NIH R01AG030753 and DOD W81XWH-09-1-0107 to H.J.F. Mike Nalls and Andrew Singleton’s participation was supported in part by the Intramural Research Program of the National Institute on Aging, National Institutes of Health, Department of Health and Human Services, project Z01 AG000950-06.
Abbreviations
- ROC
Receiver Operating Characteristic
- AUC
Area under the curve
- EDTA
ethylene diamine tetraacetic acid
- LC/MS
liquid chromatography/mass spectrometry
- UPLC
Ultra Performance Liquid Chromatography
- MRM
Multiple reaction monitoring
Footnotes
Data and materials availability: All metabolomic data from this study are freely available in the European Bioinformatics Institute MetaboLights database with accession code: MTBLS72
- Disclosures: Drs. Mark Mapstone, Feng Lin, Amrita K. Cheema, Massimo S. Fiandaca, and Howard J. Federoff have patents filed on their behalf through Georgetown University. These authors are named as co-inventors on a provisional patent application filed by Georgetown University and the University of Rochester related to the specific biomarker technology described in this manuscript. Dr. Mike A. Nalls has no disclosures. Dr. Andrew B. Singleton has no disclosures.
- Financial Support: This work was funded by NIH R01AG030753 and DOD W81XWH-09-1-0107 to Howard J. Federoff. Mike Nalls and Andrew Singleton’s participation was supported in part by the Intramural Research Program of the National Institute on Aging, National Institutes of Health, Department of Health and Human Services, project Z01 AG000950-06.
- The authors verify that the data presented in this manuscript have not been previously published nor are they being considered for publication at another journal at this time.
- All subjects gave informed consent prior to their participation in this study and all protocols reported here were approved by the institutional review boards at the University of Rochester, Georgetown University and the University of California, Irvine.
- All authors have reviewed the content of this manuscript and approve of its contents and validate the accuracy of the data.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albert MS. The ageing brain: normal and abnormal memory. Philos Trans R Soc Lond B Biol Sci. 1997;352(1362):1703–9. doi: 10.1098/rstb.1997.0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez EO. The role of histamine on cognition. Behav Brain Res. 2009;199(2):183–9. doi: 10.1016/j.bbr.2008.12.010. [DOI] [PubMed] [Google Scholar]
- Balasubramanian AB, Kawas CH, Peltz CB, Brookmeyer R, Corrada MM. Alzheimer disease pathology and longitudinal cognitive performance in the oldest-old with no dementia. Neurology. 2012;79(9):915–21. doi: 10.1212/WNL.0b013e318266fc77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollard ME, Stanley EG, Lindon JC, Nicholson JK, Holmes E. NMR-based metabonomic approaches for evaluating physiological influences on biofluid composition. NMR Biomed. 2005;18(3):143–62. doi: 10.1002/nbm.935. [DOI] [PubMed] [Google Scholar]
- Cansev M. Synaptogenesis: Modulation by Availability of Membrane Phospholipid Precursors. Neuromolecular Med. 2016;18(3):426–40. doi: 10.1007/s12017-016-8414-x. [DOI] [PubMed] [Google Scholar]
- Chakroborty S, Kim J, Schneider C, West AR, Stutzmann GE. Nitric oxide signaling is recruited as a compensatory mechanism for sustaining synaptic plasticity in Alzheimer’s disease mice. J Neurosci. 2015;35(17):6893–902. doi: 10.1523/JNEUROSCI.4002-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciavardelli D, Piras F, Consalvo A, Rossi C, Zucchelli M, Di Ilio C, Frazzini V, Caltagirone C, Spalletta G, Sensi SL. Medium-chain plasma acylcarnitines, ketone levels, cognition, and gray matter volumes in healthy elderly, mildly cognitively impaired, or Alzheimer’s disease subjects. Neurobiol Aging. 2016;43:1–12. doi: 10.1016/j.neurobiolaging.2016.03.005. [DOI] [PubMed] [Google Scholar]
- Colton CA, Mott RT, Sharpe H, Xu Q, Van Nostrand WE, Vitek MP. Expression profiles for macrophage alternative activation genes in AD and in mouse models of AD. J Neuroinflammation. 2006;3:27. doi: 10.1186/1742-2094-3-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depp CA, Jeste DV. Definitions and predictors of successful aging: a comprehensive review of larger quantitative studies. Am J Geriatr Psychiatry. 2006;14(1):6–20. doi: 10.1097/01.JGP.0000192501.03069.bc. [DOI] [PubMed] [Google Scholar]
- Fiandaca MS, Zhong X, Cheema AK, Orquiza MH, Chidambaram S, Tan MT, Gresenz CR, FitzGerald KT, Nalls MA, Singleton AB, Mapstone M, Federoff HJ. Plasma 24-metabolite Panel Predicts Preclinical Transition to Clinical Stages of Alzheimer’s Disease. Front Neurol. 2015;6:237. doi: 10.3389/fneur.2015.00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gefen T, Shaw E, Whitney K, Martersteck A, Stratton J, Rademaker A, Weintraub S, Mesulam MM, Rogalski E. Longitudinal neuropsychological performance of cognitive SuperAgers. J Am Geriatr Soc. 2014;62(8):1598–600. doi: 10.1111/jgs.12967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grady CL. Cognitive neuroscience of aging. Ann N Y Acad Sci. 2008;1124:127–44. doi: 10.1196/annals.1440.009. [DOI] [PubMed] [Google Scholar]
- Graham SF, Chevallier OP, Elliott CT, Holscher C, Johnston J, McGuinness B, Kehoe PG, Passmore AP, Green BD. Untargeted metabolomic analysis of human plasma indicates differentially affected polyamine and L-arginine metabolism in mild cognitive impairment subjects converting to Alzheimer’s disease. PLoS One. 2015;10(3):e0119452. doi: 10.1371/journal.pone.0119452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutchess A. Plasticity of the aging brain: new directions in cognitive neuroscience. Science. 2014;346(6209):579–82. doi: 10.1126/science.1254604. [DOI] [PubMed] [Google Scholar]
- Hammad SM, Pierce JS, Soodavar F, Smith KJ, Al Gadban MM, Rembiesa B, Klein RL, Hannun YA, Bielawski J, Bielawska A. Blood sphingolipidomics in healthy humans: impact of sample collection methodology. J Lipid Res. 2010;51(10):3074–87. doi: 10.1194/jlr.D008532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansmannel F, Sillaire A, Kamboh MI, Lendon C, Pasquier F, Hannequin D, Laumet G, Mounier A, Ayral AM, DeKosky ST, Hauw JJ, Berr C, Mann D, Amouyel P, Campion D, Lambert JC. Is the urea cycle involved in Alzheimer’s disease? J Alzheimers Dis. 2010;21(3):1013–21. doi: 10.3233/JAD-2010-100630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LePort AK, Mattfeld AT, Dickinson-Anson H, Fallon JH, Stark CE, Kruggel F, Cahill L, McGaugh JL. Behavioral and neuroanatomical investigation of Highly Superior Autobiographical Memory (HSAM) Neurobiol Learn Mem. 2012;98(1):78–92. doi: 10.1016/j.nlm.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li NJ, Liu WT, Li W, Li SQ, Chen XH, Bi KS, He P. Plasma metabolic profiling of Alzheimer’s disease by liquid chromatography/mass spectrometry. Clin Biochem. 2010;43(12):992–7. doi: 10.1016/j.clinbiochem.2010.04.072. [DOI] [PubMed] [Google Scholar]
- Liu P, Fleete MS, Jing Y, Collie ND, Curtis MA, Waldvogel HJ, Faull RL, Abraham WC, Zhang H. Altered arginine metabolism in Alzheimer’s disease brains. Neurobiol Aging. 2014;35(9):1992–2003. doi: 10.1016/j.neurobiolaging.2014.03.013. [DOI] [PubMed] [Google Scholar]
- Lores-Arnaiz S, Bustamante J, Arismendi M, Vilas S, Paglia N, Basso N, Capani F, Coirini H, Costa JJ, Arnaiz MR. Extensive enriched environments protect old rats from the aging dependent impairment of spatial cognition, synaptic plasticity and nitric oxide production. Behav Brain Res. 2006;169(2):294–302. doi: 10.1016/j.bbr.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Mapstone M, Cheema AK, Fiandaca MS, Zhong X, Mhyre TR, MacArthur LH, Hall WJ, Fisher SG, Peterson DR, Haley JM, Nazar MD, Rich SA, Berlau DJ, Peltz CB, Tan MT, Kawas CH, Federoff HJ. Plasma phospholipids identify antecedent memory impairment in older adults. Nat Med. 2014;20(4):415–8. doi: 10.1038/nm.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesulam MM. A plasticity-based theory of the pathogenesis of Alzheimer’s disease. Ann N Y Acad Sci. 2000;924:42–52. doi: 10.1111/j.1749-6632.2000.tb05559.x. [DOI] [PubMed] [Google Scholar]
- Morrison LD, Becker L, Ang LC, Kish SJ. Polyamines in human brain: regional distribution and influence of aging. J Neurochem. 1995;65(2):636–42. doi: 10.1046/j.1471-4159.1995.65020636.x. [DOI] [PubMed] [Google Scholar]
- Morrison LD, Cao XC, Kish SJ. Ornithine decarboxylase in human brain: influence of aging, regional distribution, and Alzheimer’s disease. J Neurochem. 1998;71(1):288–94. doi: 10.1046/j.1471-4159.1998.71010288.x. [DOI] [PubMed] [Google Scholar]
- Nikonenko I, Nikonenko A, Mendez P, Michurina TV, Enikolopov G, Muller D. Nitric oxide mediates local activity-dependent excitatory synapse development. Proc Natl Acad Sci U S A. 2013;110(44):E4142–51. doi: 10.1073/pnas.1311927110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Nasaruddin MB, Elliott CT, McGuinness B, Passmore AP, Kehoe PG, Holscher C, McClean PL, Graham SF, Green BD. Alzheimer’s disease-like pathology has transient effects on the brain and blood metabolome. Neurobiol Aging. 2016;38:151–63. doi: 10.1016/j.neurobiolaging.2015.11.014. [DOI] [PubMed] [Google Scholar]
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109(3):307–20. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- Pinto J, Domingues MR, Galhano E, Pita C, do Almeida MC, Carreira IM, Gil AM. Human plasma stability during handling and storage: impact on NMR metabolomics. Analyst. 2014;139(5):1168–77. doi: 10.1039/c3an02188b. [DOI] [PubMed] [Google Scholar]
- Quinones MP, Kaddurah-Daouk R. Metabolomics tools for identifying biomarkers for neuropsychiatric diseases. Neurobiol Dis. 2009;35(2):165–76. doi: 10.1016/j.nbd.2009.02.019. [DOI] [PubMed] [Google Scholar]
- Reddy AB, Karp NA, Maywood ES, Sage EA, Deery M, O’Neill JS, Wong GK, Chesham J, Odell M, Lilley KS, Kyriacou CP, Hastings MH. Circadian orchestration of the hepatic proteome. Curr Biol. 2006;16(11):1107–15. doi: 10.1016/j.cub.2006.04.026. [DOI] [PubMed] [Google Scholar]
- Reinberg A, Gervais P, Levi F, Smolensky M, Del Cerro L, Ugolini C. Circadian and circannual rhythms of allergic rhinitis: an epidemiologic study involving chronobiologic methods. J Allergy Clin Immunol. 1988;81(1):51–62. doi: 10.1016/0091-6749(88)90220-5. [DOI] [PubMed] [Google Scholar]
- Rey A. L’examen clinique en psychologie. Paris: Presses Universitaires de France; 1964. [Google Scholar]
- Rowe JW, Kahn RL. Human aging: usual and successful. Science. 1987;237(4811):143–9. doi: 10.1126/science.3299702. [DOI] [PubMed] [Google Scholar]
- Salthouse TA. When does age-related cognitive decline begin? Neurobiol Aging. 2009;30(4):507–14. doi: 10.1016/j.neurobiolaging.2008.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuman EM, Madison DV. A requirement for the intercellular messenger nitric oxide in long-term potentiation. Science. 1991;254(5037):1503–6. doi: 10.1126/science.1720572. [DOI] [PubMed] [Google Scholar]
- Shimizu E, Tang YP, Rampon C, Tsien JZ. NMDA receptor-dependent synaptic reinforcement as a crucial process for memory consolidation. Science. 2000;290(5494):1170–4. doi: 10.1126/science.290.5494.1170. [DOI] [PubMed] [Google Scholar]
- Sperling RA, Aisen PS, Beckett LA, Bennett DA, Craft S, Fagan AM, Iwatsubo T, Jack CR, Jr, Kaye J, Montine TJ, Park DC, Reiman EM, Rowe CC, Siemers E, Stern Y, Yaffe K, Carrillo MC, Thies B, Morrison-Bogorad M, Wagster MV, Phelps CH. Toward defining the preclinical stages of Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7(3):280–92. doi: 10.1016/j.jalz.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern Y. Cognitive reserve in ageing and Alzheimer’s disease. The Lancet Neurology. 2012;11(11):1006–1012. doi: 10.1016/S1474-4422(12)70191-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417(6884):78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- Tibshirani R. Regression Shrinkage and Selection via the Lasso. J R Statist Soc B. 1996;58(1):267–288. [Google Scholar]
- Tohgi H, Abe T, Yamazaki K, Murata T, Ishizaki E, Isobe C. Alterations of 3-nitrotyrosine concentration in the cerebrospinal fluid during aging and in patients with Alzheimer’s disease. Neurosci Lett. 1999;269(1):52–4. doi: 10.1016/s0304-3940(99)00406-1. [DOI] [PubMed] [Google Scholar]
- Voyle N, Kim M, Proitsi P, Ashton NJ, Baird AL, Bazenet C, Hye A, Westwood S, Chung R, Ward M, Rabinovici GD, Lovestone S, Breen G, Legido-Quigley C, Dobson RJ, Kiddle SJ Alzheimer’s Disease Neuroimaging I. Blood metabolite markers of neocortical amyloid-beta burden: discovery and enrichment using candidate proteins. Transl Psychiatry. 2016;6:e719. doi: 10.1038/tp.2015.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BR, Best R, Noon JP, Watt GC, Webb DJ. Seasonal variation in glucocorticoid activity in healthy men. J Clin Endocrinol Metab. 1997;82(12):4015–9. doi: 10.1210/jcem.82.12.4430. [DOI] [PubMed] [Google Scholar]
- Walsh MC, Brennan L, Malthouse JP, Roche HM, Gibney MJ. Effect of acute dietary standardization on the urinary, plasma, and salivary metabolomic profiles of healthy humans. Am J Clin Nutr. 2006;84(3):531–9. doi: 10.1093/ajcn/84.3.531. [DOI] [PubMed] [Google Scholar]
- Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0--making metabolomics more meaningful. Nucleic Acids Res. 2015;43(W1):W251–7. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Wishart DS. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat Protoc. 2011;6(6):743–60. doi: 10.1038/nprot.2011.319. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.