Abstract
The Prader-Willi syndrome (PWS) usually results from a paternal deletion of 15q11-q13 or maternal disomy for chromosome 15. Reduced pigmentation of skin, hair, and eyes is common in PWS and was suggested previously to be associated with the 15q11-q13 deletion. The P gene, located in this same region, is associated with OCA2, an autosomal recessive disorder that is the most frequent form of tyrosinase-positive oculocutaneous albinism. We studied 28 individuals with PWS and found that hemizygosity for the P gene was significantly correlated with the occurrence of hypopigmentation among PWS patients. However, we found little or no relationship between the occurrence of hypopigmentation and the polymorphism haplotype of the intact P allele. Thus, our results indicate that hypopigmentation is likely the result of deletion of the P gene in the context of PWS but do not support the linked hypothesis that hypopigmentation results from hemizygosity for variant P alleles with reduced function.
Keywords: Prader-Willi syndrome, Angelman syndrome, P gene, hypopigmentation, OCA2, oculocutaneous albinism
INTRODUCTION
The Prader-Willi syndrome (PWS) most frequently results from paternal deletions of 15q11-q13 or maternal uniparental disomy (UPD) of chromosome 15 [reviewed in Nicholls, 1993]. Individuals with PWS typically exhibit infantile hypotonia, early childhood obesity, short stature, small hands and feet, hypogonadism, mental deficiency, and characteristic facial abnormalities [Butler, 1990], In addition, about half of PWS patients exhibit mild to moderate hypopigmentation in comparison to unaffected relatives [Butler et al., 1986; Wiesner et al., 1987; Butler, 1989]. Hypopigmentation is also common in Angelman syndrome (AS) [Fryburg et al., 1991; King et al., 1993; Nicholls, 1993], which results from maternal deletions of 15q11-q13 or paternal UPD of chromosome 15 and has a clinical phenotype quite different from PWS [reviewed in Nicholls, 1993]. Some patients with PWS and AS have manifested oculocutaneous albinism [Wallis and Beighton, 1989; Fryburg et al., 1991]. These observations led to the suggestion that a gene involved in melanin pigmentation is located in proximal 15q [Butler et al., 1986]. Ramsay et al. [1992] subsequently mapped a gene for tyrosinase-positive oculocutaneous albinism (OCA2) to chromosome segment 15q11.2-ql2, and we showed that OCA2 results from either deletion or point mutation of the P gene located in this region [Rinchik et al., 1993; Lee et al., 1994a,b].
Hypopigmentation in PWS and AS is most frequent in deletion cases [Butler, 1989], and we have suggested that hypopigmentation in these patients might result from hemizygosity for the P gene [Rinchik et al., 1993]. We have identified pathologic mutations of the P gene in patients with a wide range of clinical OCA phenotypes, from severe OCA2 to so-called “autosomal recessive ocular albinism” [Rinchik et al., 1993; Lee et al., 1994a,b]. In surveys of large numbers of normal individuals we identified 28 different non-pathologic polymorphisms of the P gene, of which 5 result in amino acid substitutions: R305W, R419Q, L440F, H615R, and I722T [Lee et al., 1995a], and we speculated that functional differences among variant P alleles might account for part of the normal variation of human pigmentation. Pigmentary differences resulting from functional differences among the corresponding variant P polypeptides might be most evident in persons who are hemizygous for P, perhaps accounting for hypopigmentation in PWS. To test this hypothesis, we characterized the P genotypes of 28 PWS patients, and we attempted to correlate these genotypes with their pigmentation phenotypes. We found that the occurrence of hypopigmentation in PWS is highly correlated with hemizygosity for P. However, we found little or no apparent relationship between the occurrence of hypopigmentation and the polymorphism haplotype of the intact P allele.
MATERIALS AND METHODS
Subjects
We studied 28 Caucasian patients with PWS [Holm et al., 1993], 27 of whom have been described previously [Butler, 1989; Wagner-Mascari, 1991; Mascari et al., 1992; Butler et al., 1996]. Patients were clinically diagnosed with PWS on the basis of hypotonia, hypogenitalism, early childhood obesity, small hands and feet, short stature, a characteristic facial appearance, and delayed psychomotor development and/or mental retardation. The diagnosis of PWS was confirmed by cytogenetic demonstration of a deletion and/or molecular analysis in all cases. During the physical examination detailed attention was given to pigmentation status (skin, hair, and eye color) and eye examination of each PWS patient and his or her first-degree relatives, as described previously [Butler, 1989], and when possible was documented by photographs.
Molecular Cytogenetic Analyses
All patients were studied by high-resolution chromosome analysis as described [Butler et al., 1986]. Patients were genotyped by use of PCR at several microsatellites in the 15q11-q13 region [Malcolm and Donlon, 1994], including D15S541, D15S542, D15S543, D15S219, and D15S165 (Fig. 1). Quantitative Southern blot hybridization was carried out using 5 μg of patient genomic DNA digested by HindIII, BamHI, TaqI, or EcoRI as described [Mbikay et al., 1988; Tantravahi et al., 1989; Butler et al., 1993]. A 0.8 kb BamHI-HindIII fragment of IR10-1 [Nicholls et al. 1989; Tantravahi et al., 1989], corresponding to exon 19 of the human P gene [Lee et al., 1995a], or a 680 bp TaqI-EcoRI fragment of DN10 [Rinchik et al., 1993], corresponding to exon 25 of the human P gene, were used as probes, and filters were co-hybridized with H2-26 or H2-42, derived from chromosome 13, as internal controls. Genomic DNA was digested with ScaI for use with IR10-1, which detects a 3-allele RFLP [Nicholls et al., 1989]. Autoradiographic bands were scanned using an LKB Ultrascan soft laser densitometer. Relative band signal intensities of at least two lanes were computed and normalized as described [Tantravahi et al., 1989].
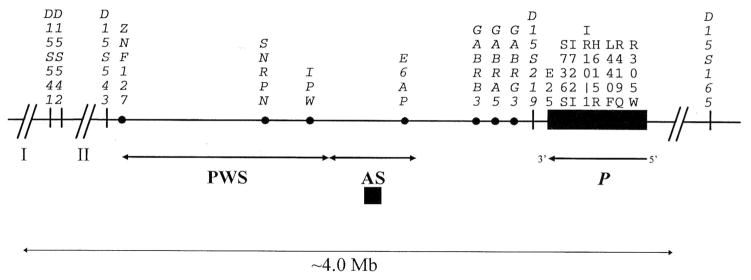
Fig. 1.
Map of markers used in this study. Marker order and approximate distances are as described [Malcolm and Donlon, 1994; Christian et al., 1995; Butler et al., 1996]. Additionally, markers D15S165 and D15S219 are not contained within human P phage clones or YACs 93C9 and 149A9 [Lee et al., 1995a] (R.A. Spritz, unpublished data), supporting the locations of these markers distal and proximal to P, respectively. The approximate PWS and AS critical intervals and the P gene are indicated.
Analysis of P and TYR Gene Polymorphisms
P exon segments containing the R305W, R419Q, L440F, H615R, and I722T polymorphisms were amplified by the PCR as described [Lee et al., 1995a] and analyzed by two methods. Genotypes were assigned on the basis of PCR-based simultaneous single-strand polymorphism (SSCP)/heteroduplex analysis [Lee et al., 1995b] of characteristic bandshifts associated with these base substitutions [Lee et al., 1995a]. However, in some cases SSCP/HDX patterns were complicated by the occurrence of additional polymorphisms in the same exon [Lee et al., 1995a]. Therefore, the genotypes for all five polymorphisms were also determined by allele-specific oligonucleotide hybridization analysis, essentially as described [Kogan and Gitschier, 1990] by using 32[P] 5′ end-radiolabeled 19-mer oligonucleotides corresponding to codons 305R (5′-ATCAGCATCCGGGCCTCCC-3′), 305W (5′-ATCAGCATCTGGGCCTCCC-3′), 419R (5′-GGCTCTCCCGGGGACGGGT-3′), 419Q (5′-GGCTCTCCCAGGGACGGGT-3′), 440L (5′-TGCCTTCTTGGACAACGTC-3′), 440F (5′-TGCCTTCTTCGACAACGTC-3′), 615H (5′-TCTTACAGCATAGGATATC-3′), 615R (5′-TCTTACAGCGTAGGATATC-3′), 722I (5′-CAGCGCCTCATAGCCGCCATT-3′), and 722T (5′-CAGCGCCTC ACAGCCGCCATT-3′).
Genotyping of patient DNAs for the R402Q polymorphism in exon 4 of the TYR gene [Tripathi et al., 1991] was carried out as described [Fukai et al., 1995].
RESULTS
Correlation of Hypopigmentation With P Gene Deletion
A total of 28 patients with PWS was studied (Table I). Sixteen (57%) had karyotypic evidence of 15q11-q13 deletion and one had a Robertsonian translocation involving chromosomes 13 and 15. Of the 11 patients with no apparent cytogenetic abnormalities, PCR analyses of 15q11-q13 microsatellite markers and dosage analysis using P gene probes showed that three (patients 13, 14, and 17) had deletions of the paternal 15q11-q13 not apparent in 550 band level karyotypes [Wagner-Mascari, 1991; Mascari et al., 1992], one (patient 20) had a microdeletion of the imprinting center [Butler et al., 1996] in central 15q11-q13 and not including any part of the P gene, and seven (patients 21–26, and 28) had maternal UPD of chromosome 15. The patient (27) with the t(13;15) was also found to have maternal UPD for markers in proximal 15q. In two patients (18 and 19) with 15q11-q13 deletions, molecular analyses showed that at least the 3′ half of the P gene was intact.
TABLE I.
PWS Patient, Cytogenetic, and Marker Data*
| Patient | Phenotype | Pigment | Chromosome | D15S541 | D15S542 | D15S543 | D15S219 | PE25 | PS736S | PI722T | IR10-1 | PH615R | PL440F | PR419Q | PR305W | D15S165 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | PWS | ↓ | del(15q) | h | h | h | h | ND | h | h | 1 | h | h | h | h | h |
| 2 | PWS | ↓ | del(15q) | h | h | d | d | 1 | h | h | 1 | h | h | h | h | H |
| 3 | PWS | ↓ | del(15q) | h | h | h | d | 1 | h | h | 1 | h | h | h | h | H |
| 4 | PWS | ↓ | del(15q) | h | h | h | d | 1 | h | h | 1 | h | h | h | h | H |
| 5 | PWS | ↓ | del(15q) | h | h | h | h | ND | h | h | 1 | h | h | h | h | H |
| 6 | PWS | ↓ | del(15q) | H | h | h | d | 1 | h | h | 1 | h | h | h | h | H |
| 7 | PWS | ↓ | del(15q) | H | h | h | d | 1 | h | h | 1 | h | h | h | h | H |
| 8 | PWS | ↓ | del(15q) | H | H | h | d | 1 | h | h | 1 | h | h | h | h | H |
| 9 | PWS | ↓ | del(15q) | H | H | h | d | 1 | h | h | 1 | h | h | h | h | H |
| 10 | PWS | ↓ | del(15q) | H | ND | d | d | 1 | h | h | 1 | h | h | h | h | H |
| 11 | PWS | ↓ | del(15q) | H | H | h | d | ND | h | h | 1 | h | h | h | h | H |
| 12 | PWS | ↓ | del(15q) | H | H | h | d | ND | h | h | 1 | h | h | h | h | H |
| 13 | PWS | ↓ | del(15q) | h | H | h | h | ND | h | h | 1 | h | h | h | h | H |
| 14 | PWS | ↓ | del(15q) | h | H | d | h | ND | h | h | 1 | h | h | h | h | h |
| 15 | PWS | Nl | del(15q) | d | d | h | h | ND | h | h | 1 | h | h | h | h | h |
| 16 | PWS | Nl | del(15q) | H | h | h | d | ND | h | h | 1 | h | h | h | h | H |
| 17 | Atyp | Nl | del(15q) | H | h | h | h | ND | h | h | 1 | h | h | h | h | h |
| 18 | PWS | ↓ | del(15q)a | H | h | h | h | 2 | H | h | 2 | h | h | h | h | H |
| 19 | PWS | Nl | del(15q)a | H | H | h | H | 2 | ND | h | 2 | h | h | h | h | H |
| 20 | Atyp | Nl | microdel(15q)a,b | h | H | h | h | ND | ND | h | 2 | h | h | h | h | H |
| 21 | PWS | Nl | UPD | h | h | h | h | 2 | ND | h | 2 | h | h | h | h | H |
| 22 | PWS | Nl | UPD | h | h | h | H | 2 | ND | h | 2 | h | h | H | h | h |
| 23 | PWS | Nl | UPD | H | h | H | h | 2 | ND | h | 2 | h | h | h | h | h |
| 24 | PWS | Nl | UPD | H | H | H | h | 2 | ND | h | 2 | h | h | h | H | H |
| 25 | PWS | Nl | UPD | h | h | h | h | ND | ND | h | 2 | h | h | h | h | h |
| 26 | PWS | Nl | UPD | H | h | H | h | ND | H | h | 2 | h | h | h | h | H |
| 27 | PWS | Nl | t(13;15),UPD | H | H | H | h | ND | ND | h | 2 | h | h | h | h | h |
| 28 | Atyp | Nl | UPD | H | H | h | H | ND | H | h | 2 | h | h | h | h | H |
Specific allele data are obtainable from the authors. PWS, Prader-Willi syndrome; Atyp, atypical PWS phenotype; Nl, normal; H, heterozygous; d, deleted based on DNA studies of the family; h, single-allele pattern (either homozygous or deleted); 1, DNA studies indicate one copy; 2, DNA studies indicate two copies; ND, not determined.
DNA studies indicate that at least the 3′ part of the P gene is intact.
DNA studies show a ~200 kb deletion around SNRPN [Butler et al, 1996].
Of the 28 patients, 15 (54%) were judged to be hypopigmented, most of whom had deletions that included the P gene. Of the 17 PWS patients with a deletion of 15q11-q13 that included P, 14 (82%) were judged hypopigmented. In contrast, of the three patients in whom the deletion did not include P (including the imprinting center microdeletion), only one was hypopigmented, and of the eight PWS patients with no apparent deletion of 15q, none were hypopigmented. These data demonstrate a very high correlation between hemizygosity for the P gene and the occurrence of hypopigmentation in the context of PWS (P < 0.00027 by Fisher’s exact test; two-tailed). The overall prevalence of hypopigmentation among PWS patients hemizygous for the P gene is thus about 82%.
Hypopigmentation Is Not Related to the Haplotype of the Intact P Allele
The R305W, R419Q, L440F, H615R, and I722T polymorphisms of P are all relatively uncommon, with respective allele frequencies among normal Caucasians of approximately 0.17, 0.09, 0.10, 0.0, and 0.26; moreover, the L440F polymorphism has only been observed among Ashkenazim [Lee et al., 1995a], Therefore, the predominant haplotype for these five polymorphisms is 305R/419R/440L/615H/722I (abbreviated RRLHI). Of the 28 PWS patients studied here almost all were homozygous or hemizygous for the RRLHI haplotype. The only exceptions were two patients who were heterozygous and one hemizygous for the RQLHI haplotype, and one patient who was heterozygous for the WRLHI haplotype. Of these, only the patient who was hemizygous for RQLHI was hypopigmented.
Lack of Effect of a Functionally Significant Polymorphism of Tyrosinase (R402Q)
We also considered that hypopigmentation in PWS might relate to the co-occurrence of hemizygosity for the P gene and a polymorphism of the tyrosinase gene, R402Q, which we have previously shown results in a thermolabile enzyme with reduced activity at 37°C [Tripathi et al., 1991]. To test this hypothesis we genotyped all 28 PWS patients for the TYR R402Q polymorphism. We observed no apparent correlation between the pigmentation phenotype and the TYR R402Q genotype (data not shown).
DISCUSSION
We have investigated the relationship between the moderate hypopigmentation frequently observed in patients with PWS and the structure of the P gene in these patients. We find that hypopigmentation in PWS is very highly correlated with deletion of chromosome 15q11-q13, and with hemizygosity for the P gene in particular. In contrast, most patients in whom P is intact appear normally pigmented, consistent with the report of two other normally-pigmented PWS patients whose 15q11-q13 deletions did not include IR10-1 [Tantravahi et al., 1989; Wagner-Mascari, 1991; Rinchik et al., 1993]. However, deletion of the P gene is neither necessary nor sufficient for hypopigmentation in PWS; three patients in whom the P gene was deleted were judged normally pigmented and one in whom P was intact was judged hypopigmented. The occurrence of such occasional discrepancies is not surprising given the subjective nature of clinical assessment of pigmentary status and the likelihood of epistatic modifiers that affect pigmentation.
We initially hypothesized that hemizygosity for the P gene in PWS might unmask functionally significant polypeptide variation in the intact P allele, resulting in moderate hypopigmentation in some individuals. Clearly, our data disprove this hypothesis. Among those PWS patients with P gene deletions, we found no correlation between the occurrence or apparent degree of hypopigmentation and the polymorphism haplotype of the intact P allele. Even the three patients with P gene deletion who were judged normally pigmented (patients 15–17) all shared the common RRLHI haplotype on the hemizygous P allele, as did most of the patients who were judged to be hypopigmented. We also found no apparent relationship to the R402Q polymorphism of tyrosinase, which is known to moderately reduce catalytic activity of this key enzyme of melanin biosynthesis [Tripathi et al., 1991].
Though our results demonstrate a high correlation between hypopigmentation in PWS and deletion of P, they do not explain why hemizygosity for the P gene in the context of PWS (and presumably also AS) is associated with hypopigmentation, whereas simple heterozygosity for OCA2 mutant alleles of the P gene is not. OCA2 behaves as a pure autosomal recessive in humans, as does pink-eyed dilution (p) in mice [Lyon and Searle, 1989; Rinchik et al., 1993]; OCA2 carriers are not hypopigmented. Although hypopigmentation might be subtle in Caucasian OCA2 carriers (but no more so than in patients with PWS), hypopigmentation should be readily apparent among African and African-American OCA2 carriers, among whom null alleles of P are prevalent [Durham-Pierre et al., 1994; Lee et al., 1994a, Spritz et al., 1995; Stevens et al., 1995]. Even partial deletions of the P gene that are associated with OCA2 but not PWS do not result in hypopigmentation in heterozygotes [Rinchik et al., 1993; Durham-Pierre et al., 1994; Lee et al., 1994a].
We can suggest several possible explanations for hypopigmentation in deletional PWS. First, it may be that functionally significant variation in the P promoter region, not looked for in this study, accounts for reduced transcription of the intact allele. This seems unlikely, as hypopigmentation in PWS is almost completely correlated with deletion, not with haplotype, and in any case this explanation would predict that OCA2 heterozygotes would also appear hypopigmented. Second, it might be that an as-yet unknown, non-imprinted pigmentation gene might exist in this region of proximal 15q and thus is deleted along with P in PWS. The human P gene is quite large, spanning 250–600 kb [Lee et al., 1995a] (Fig. 1), large enough to include several undiscovered genes. Arguing against this explanation, however, several large deletions that include the p gene do not result in hypopigmentation in heterozygous mice [Johnson et al., 1995]; thus, a putative unknown pigmentation gene would have to have been introduced into this region in the human lineage subsequent to the mammalian radiation. Third, and we believe most likely, hypopigmentation in PWS and AS may result from reduced expression of the intact P allele in the specific context of large proximal 15q deletions. LaSalle and Lalande [1996] recently observed preferential association of the maternal and paternal chromosome 15 homologues in the imprinted q11–q13 regions during late S-phase, which they suggested may mediate allele-specific transcription of genes in this region. This association was disrupted in PWS and AS patients, particularly those with 15q deletions [LaSalle and Lalande, 1996]. Perhaps this results in disruption of the complex pattern of imprinted and non-imprinted gene regulation throughout this chromosomal region, with consequent reduced expression of the intact P allele and thus in hypopigmentation in hemizygous individuals. If disrupted gene regulation in an imprinted chromosomal domain can influence regulation of imprinted and non-imprinted genes on the other homologue, this could have major implications for our understanding of gene interaction in other human genetic diseases.
Accordingly, we suggest that hypopigmentation in PWS and AS patients results either from reduced expression of the intact P allele in the specific context of large 15q deletions as the result of complex gene regulation throughout this chromosomal region or, alternatively, that hypopigmentation in PWS and AS may result from deletion of an unknown gene or genes located between P and the PWS/AS imprinted domain.
Acknowledgments
Contract grant sponsor: The March of Dimes Birth Defects Foundation; Contract grant number: 6-408; Contract grant sponsor: National Institutes of Health; Contract grant numbers: AR39892, HD30329, AR39750, SDRC-P8F57; Contract grant sponsor: Non-Directed Research Fund of the Korea Research Foundation; Contract grant sponsor: Pew Scholar Program in the Biomedical Sciences.
We thank C. Denniston for statistical assistance. This work was supported by Clinical Research Grant 6-408 from the March of Dimes Birth Defects Foundation (R.A.S.), National Institutes of Health Grants AR39892 (RAS), HD30329 (MGB), AR39750, and SDRC-P8F57 (RDN), and the Non-Directed Research Fund of the Korea Research Foundation (S.-T.L.). R.D.N. was supported by the Pew Scholar Program in the Biomedical Sciences. This is paper 3444 from the Laboratory of Genetics, University of Wisconsin.
References
- Butler MG. Hypopigmentation: A common feature of Prader-Labhart-Willi syndrome. Am J Hum Genet. 1989;45:140–146. [PMC free article] [PubMed] [Google Scholar]
- Butler MG. Prader-Willi syndrome: Current understanding of cause and diagnosis. Am J Med Genet. 1990;35:319–332. doi: 10.1002/ajmg.1320350306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler MG, Meaney FJ, Palmer CG. Clinical and cytogenetic survey of 39 individuals with Prader-Labhart-Willi syndrome. Am J Med Genet. 1986;23:793–809. doi: 10.1002/ajmg.1320230307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler MG, Dahir GA, Schwartz HS. Molecular analysis of transforming growth factor beta in giant cell tumor of bone. Cancer Genet Cytogenet. 1993;66:108–112. doi: 10.1016/0165-4608(93)90237-g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler MG, Christian SL, Kubota T, Ledbetter DH. A 5-year-old white girl with Prader-Willi syndrome and a submicroscopic deletion of chromosome 15q11q13. Am J Med Genet. 1996;65:137–141. doi: 10.1002/(SICI)1096-8628(19961016)65:2<137::AID-AJMG11>3.0.CO;2-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian SL, Robinson WP, Huang B, Mutirangura A, Line MR, Nakao M, Surti U, Chakravarti A, Ledbetter DH. Molecular characterization of two proximal deletion breakpoint regions in both Prader-Willi and Angelman syndrome patients. Am J Hum Genet. 1995;57:40–48. [PMC free article] [PubMed] [Google Scholar]
- Durham-Pierre D, Gardner JM, Nakatsu Y, King RA, Francke U, Ching A, Aquaron R, del Marmol V, Brilliant MH. African origin of an intragenic deletion of the human P gene in tyrosinase positive oculocutaneous albinism. Nat Genet. 1994;7:176–179. doi: 10.1038/ng0694-176. [DOI] [PubMed] [Google Scholar]
- Fryburg JS, Breg WR, Lindgren V. Diagnosis of Angelman syndrome in infants. Am J Med Genet. 1991;38:58–64. doi: 10.1002/ajmg.1320380114. [DOI] [PubMed] [Google Scholar]
- Fukai K, Holmes SA, Lucchese NJ, Siu VM, Weleber RG, Schnur RE, Spritz RA. Autosomal recessive ocular albinism associated with a functionally significant tyrosinase gene polymorphism. Nat Genet. 1995;9:92–95. doi: 10.1038/ng0195-92. [DOI] [PubMed] [Google Scholar]
- Holm VA, Cassidy SB, Butler MG, Hanchett JM, Greenswag LR, Whitman BY, Greenberg F. Prader-Willi syndrome: Consensus diagnostic criteria. Pediatrics. 1993;91:398–402. [PMC free article] [PubMed] [Google Scholar]
- Johnson DK, Stubbs LJ, Culiat CT, Montgomery CS, Russell LB, Rinchik EM. Molecular analysis of 36 mutations at the mouse pink-eyed dilution (p) locus. Genetics. 1995;141:1563–1571. doi: 10.1093/genetics/141.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RA, Wiesner GL, Townsend D, White JG. Hypopigmentation in Angelman syndrome. Am J Med Genet. 1993;46:40–44. doi: 10.1002/ajmg.1320460109. [DOI] [PubMed] [Google Scholar]
- Kogan SC, Gitschier J. Genetic prediction of hemophilia A. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR Protocols. San Diego: Academic Press; 1990. pp. 288–299. [Google Scholar]
- LaSalle JM, Laande M. Homologous association of oppositely imprinted chromosomal domains. Science. 1996;272:725–728. doi: 10.1126/science.272.5262.725. [DOI] [PubMed] [Google Scholar]
- Lee S-T, Nicholls RD, Schnur RE, Guida LC, Lu-Kuo J, Spinner NB, Zackai EH, Spritz RA. Diverse mutations of the P gene among African-Americans with type II (tyrosinase-positive) oculocutaneous albinism (OCA2) Hum Molec Genet. 1994a;3:2047–2051. [PubMed] [Google Scholar]
- Lee S-T, Nicholls RD, Bundey S, Laxova R, Musarella M, Spritz RA. Mutations of the P gene in oculocutaneous albinism, ocular albinism, and Prader-Willi syndrome plus albinism. New Engl J Med. 1994b;330:529–534. doi: 10.1056/NEJM199402243300803. [DOI] [PubMed] [Google Scholar]
- Lee S-T, Nicholls RD, Jong MTC, Fukai K, Spritz RA. Organization and sequence of the human P gene and identification of a new family of transport proteins. Genomics. 1995a;26:354–363. doi: 10.1016/0888-7543(95)80220-g. [DOI] [PubMed] [Google Scholar]
- Lee S-T, Park S-K, Lee K-H, Holmes SA, Spritz RA. A non-radioactive method for simultaneous detection of single-strand conformation polymorphisms (SSCPs) and heteroduplexes. Molec Cell. 1995b;5:668–672. [Google Scholar]
- Lyon MF, Searle AG. Genetic Variants and Strains of the Laboratory Mouse. Oxford: Oxford University Press; 1989. [Google Scholar]
- Malcolm S, Donlon TA. Report of the second international workshop on human chromosome 15 mapping. Cytogenet Cell Genet. 1994;67:2–14. doi: 10.1159/000133717. [DOI] [PubMed] [Google Scholar]
- Mascari MJ, Gottlieb W, Rogan P, Butler MG, Waller D’Armour J, Jeffreys A, Ladda R, Nicholls RD. The frequency of uniparental disomy in Prader-Willi syndrome: Implications for molecular diagnosis. N Engl J Med. 1992;326:1599–1607. doi: 10.1056/NEJM199206113262404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbikay M, Linard CG, Sirois F, Zazure C, Seidah NG, Chretien M. Tissue-specific expression of the prostatic secretory protein PSP94 in cyanomologous monkey (Macacca fascicularis) Cell Biol. 1988;34:387–398. [PubMed] [Google Scholar]
- Nicholls RD, Knoll JH, Glatt K, Hersh JH, Brewster TD, Graham JM, Jr, Wurster-Hill D, Wharton R, Latt SA. Restriction fragment length polymorphisms within proximal 15q and their use in molecular cytogenetics and the Prader-Willi syndrome. Am J Med Genet. 1989;33:66–77. doi: 10.1002/ajmg.1320330109. [DOI] [PubMed] [Google Scholar]
- Nicholls RD. Genomic imprinting and uniparental disomy in Angelman and Prader-Willi syndromes: A review. Am J Med Genet. 1993;46:16–25. doi: 10.1002/ajmg.1320460106. [DOI] [PubMed] [Google Scholar]
- Ramsay M, Colman M-A, Stevens G, Zwane E, Kromberg J, Farrall M, Jenkins T. The tyrosinase-positive oculocutnaeous albinism locus maps to chromosome 15q11.2-q12. Am J Hum Genet. 1992;51:879–884. [PMC free article] [PubMed] [Google Scholar]
- Rinchik EM, Bultman SJ, Horsthemke B, Lee S-T, Strunk KM, Spritz RA, Avidano KM, Jong MTC, Nicholls RD. A gene for the mouse pink-eyed dilution locus and for human type II oculocutaneous albinism. Nature. 1993;361:72–76. doi: 10.1038/361072a0. [DOI] [PubMed] [Google Scholar]
- Spritz RA, Fukai K, Holmes SA, Luande J. Frequent intragenic deletion of the P gene in Tanzanian patients with type II oculocutaneous albinism (OCA2) Am J Hum Genet. 1995;56:1320–1323. [PMC free article] [PubMed] [Google Scholar]
- Stevens G, van Beukering J, Jenkins T, Ramsay M. An intragenic deletion of the P gene is the common mutation causing tyrosinase-positive oculocutaneous albinism in southern African negroids. Am J Hum Genet. 1995;56:586–591. [PMC free article] [PubMed] [Google Scholar]
- Tantravahi U, Nicholls RD, Stroh H, Ringer S, Neve RL, Kaplan L, Wharton R, Wurster-Hill D, Graham JM, Jr, Cantu ES, Frias JL, Kousseff BG, Latt SA. Quantitative calibration and use of DNA probes for investigating chromosome abnormalities in the Prader-Willi syndrome. Am J Med Genet. 1989;33:78–87. doi: 10.1002/ajmg.1320330110. [DOI] [PubMed] [Google Scholar]
- Tripathi RK, Giebel LB, Strunk KM, Spritz RA. A polymorphism of the human tyrosinase gene is associated with temperature-sensitive enzymatic activity. Gene Expression. 1991;1:103–110. [PMC free article] [PubMed] [Google Scholar]
- Wagner-Mascari M. PhD thesis. Pennsylvania State University; 1991. Molecular Diagnosis of Prader-Willi Syndrome. [Google Scholar]
- Wallis CE, Beighton PH. Synchrony of oculocutaneous albinism, the Prader-Willi syndrome, and a normal karyotype. J Med Genet. 1989;26:337–339. doi: 10.1136/jmg.26.5.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesner GL, Bendel CM, Olds DP, White JG, Arthur DC, Ball DW, King RA. Hypopigmentation in the Prader-Willi syndrome. Am J Hum Genet. 1987;40:431–442. [PMC free article] [PubMed] [Google Scholar]