Abstract
Expression of the Drosophila segment polarity genes is initiated by a pre-pattern of pair-rule gene products and maintained by a network of regulatory interactions throughout several stages of embryonic development. Analysis of a model of gene interactions based on differential equations showed that wild-type expression patterns of these genes can be obtained for a wide range of kinetic parameters, which suggests that the steady states are determined by the topology of the network and the type of regulatory interactions between components, not the detailed form of the rate laws. To investigate this, we propose and analyse a Boolean model of this network which is based on a binary ON/OFF representation of mRNA and protein levels, and in which the interactions are formulated as logical functions. In this model the spatial and temporal patterns of gene expression are determined by the topology of the network and whether components are present or absent, rather than the absolute levels of the mRNAs and proteins and the functional details of their interactions. The model is able to reproduce the wild-type gene expression patterns, as well as the ectopic expression patterns observed in overexpression experiments and various mutants. Furthermore, we compute explicitly all steady states of the network and identify the basin of attraction of each steady state. The model gives important insights into the functioning of the segment polarity gene network, such as the crucial role of the wingless and sloppy paired genes, and the network’s ability to correct errors in the pre-pattern.
Keywords: Drosophila segmentation, Segment polarity genes, Gene regulatory network, Boolean model
1. Introduction
Understanding how genetic information is translated into proteins in the correct temporal sequence and spatial location to produce the various cell types in an adult organism remains a major challenge in developmental biology. The sequencing of the genome of various organisms provides the first step toward this understanding, but it is equally important to understand the patterns of control involved in gene regulation. Gene products often regulate their own production or that of other proteins at any of a number of steps, and frequently the result is a complex network of regulatory interactions. Recent experimental progress in dissecting the qualitative structure of many signal transduction and gene control networks (see, for example, Davidson et al., 2002) has produced a surge of interest in the quantitative description of gene regulation. Broadly speaking, the modeling approaches can be divided into two main groups. In the “discrete-state” approach each network node (mRNA or protein) is assumed to have a small number of discrete states and the regulatory interactions between nodes are described by logical functions similar to those used in programming. Typically time is also quantized, and the network model that describes how gene products interact to determine the state at the next time gives rise to a discrete dynamical system (Bodnar, 1997; Mendoza et al., 1999; Bodnar and Bradley, 2001; Sánchez and Thieffry, 2001; Yuh et al., 2001). A more detailed level of description is used in the “continuous-state” approach, in which the level of mRNAs, proteins, and other components are assumed to be continuous functions of time, and the evolution of these components within a cell is modeled by differential equations with mass-action kinetics or other rate laws for the production and decay of all components (Reinitz and Sharp, 1995; von Dassow et al., 2000; Gursky et al., 2001). While it is widely thought that merely specifying the topology of a control network (i.e. the connections between nodes) places few constraints on the dynamics of the network, our purpose here is to demonstrate that in one well-characterized system, knowledge of the interactions together with their signatures, by which we mean whether an interaction is activating or inhibiting, is enough to reproduce the main characteristics of the network dynamics.
The genes involved in embryonic pattern formation in the fruit fly Drosophila melanogaster, as well as the majority of the interactions between them, are known (for recent reviews see Ingham and McMahon, 2001; Sanson, 2001; Hatini and DiNardo, 2001). As in other arthropods, the body of the fruit fly is composed of segments, and determination of the adult cell types in these segments is controlled by about 40 genes organized in a hierarchical cascade comprising the gap genes, the pair-rule genes, and the segment polarity genes (Hooper and Scott, 1992). These genes are expressed in consecutive stages of embryonic development in a spatial pattern that is successively more precisely defined, the genes at one-step initiating or modulating the expression of those involved in the next step of the cascade. While most of these genes act only transiently, the segment polarity genes are expressed throughout the life of the fly. The segment polarity genes refine and maintain their expression through the network of intra- and intercellular regulatory interactions shown in Fig. 1. The stable expression pattern of these genes (specifically the expression of wingless and engrailed) defines and maintains the borders between different parasegments (the embryonic counterparts of the segments) and contributes to subsequent developmental processes, including the formation of denticle patterns and of appendage primordia (Hooper and Scott, 1992; Wolpert et al., 1998). Homologs of the segment polarity genes have been identified in vertebrates, including humans, which suggests strong evolutionary conservation of these genes.
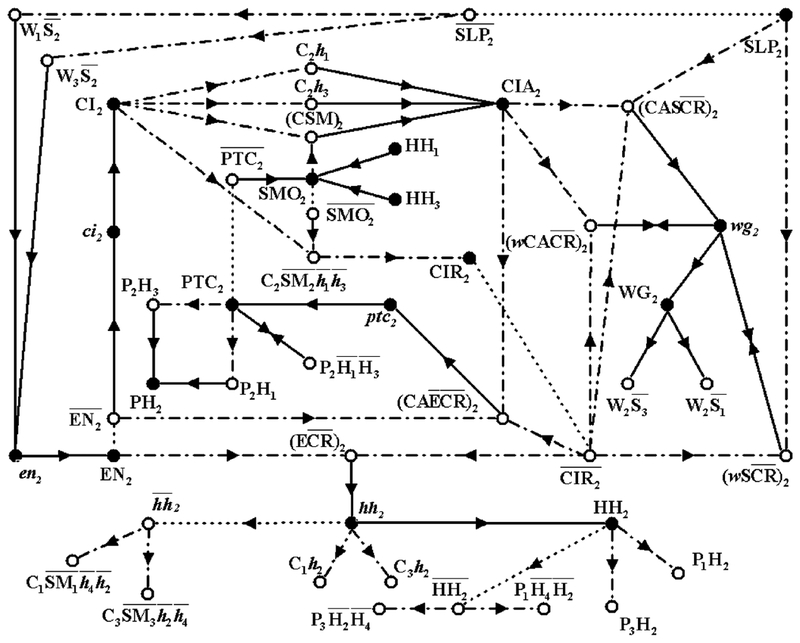
Fig. 1.
The network of interactions between the segment polarity genes. The shape of the nodes indicates whether the corresponding substances are mRNAs (ellipses), proteins (rectangles) or protein complexes (octagons). The edges of the network signify either biochemical reactions (e.g. translation) or regulatory interactions (e.g. transcriptional activation). The edges are distinguished by their signatures, i.e. whether they are activating or inhibiting. Terminating arrows (→) indicate translation, post-translational modifications (in the case of CI), transcriptional activation or the promotion of a post-translational modification reaction (e.g. SMO determining the activation of CI). Terminating segments (⊣) indicate transcriptional inhibition or in the case of SMO, the inhibition of the post-translational modification reaction CI→CIR.
The segment polarity genes encode for the transcription factor engrailed (EN), the cytosolic protein cubitus interrupters (CI), the secreted proteins wingless (WG) and hedgehog (HH), and the transmembrane receptor proteins patched (PTC) and smoothened (SMO) involved in transduction of the HH signal.
The pair-rule gene sloppy paired (slp) is activated before the segment polarity genes and expressed constitutively thereafter (Grossniklaus et al., 1992; Cadigan et al., 1994). slp encodes two forkhead domain transcription factors with similar functions that activate wg transcription and repress en transcription, and since they are co-expressed we designate them both SLP. The wg gene encodes a glycoprotein that is secreted from the cells that synthesize it (Hooper and Scott, 1992; Pfeiffer and Vincent, 1999), and can bind to the Frizzled (FZ) receptor on neighboring cells. Binding of WG to the FZ receptors on adjacent cells initiates a signaling cascade leading to the transcription of engrailed (en) (Cadigan and Nusse, 1997). EN, the homeodomain-containing product of the en gene, promotes the transcription of the hedgehog gene (hh) (Tabata et al., 1992). In addition to the homeodomain, en contains a separate repression domain (Han and Manley, 1993) that affects the transcription of ci (Eaton and Kornberg, 1990) and possibly ptc (Hidalgo and Ingham, 1990; Taylor et al., 1993). The hedgehog protein (HH) is tethered to the cell membrane by a cholesterol linkage that is severed by the dispatched protein (Ingham, 2000), freeing it to bind to the HH receptor PTC on a neighboring cell (Ingham and McMahon, 2001). The intracellular domain of PTC forms a complex with smoothened (SMO) (van den Heuvel and Ingham, 1996) in which SMO is inactivated by a post-translational conformational change (Ingham, 1998). Binding of HH to PTC removes the inhibition of SMO, and activates a pathway that results in the modification of CI (Ingham, 1998). CI contains at least three distinct domains: an NH2 terminal region characteristic of transcriptional repressors, a zinc finger domain, and a COOH region typical of activation domains in transcription factors (Alexandre et al., 1996). The CI protein can be converted into one of two transcription factors, depending on the activity of SMO. Several proteins have been implicated in this conversion, including Fused, Suppressor of Fused, Costal-2, Protein kinase A, Slimb and the CREB-binding protein (Aza-Blanc and Kornberg, 1999; Lefers et al., 2001). When SMO is inactive, CI is cleaved to form CIR, a transcriptional repressor that represses wg, ptc (Aza-Blanc and Kornberg, 1999) and hh transcription (Ohlmeyer and Kalderon, 1998; Méthot and Basler, 1999). When SMO is active, CI is converted to a transcriptional activator CIA that promotes the transcription of wg and ptc (Alexandre et al., 1996; von Ohlen and Hooper, 1997; Méthot and Basler, 1999; Aza-Blanc and Kornberg, 1999).
The gene networks governing embryonic segmentation in Drosophila have been modeled using either the discrete-state approach (Bodnar, 1997; Bodnar and Bradley, 2001; Sánchez and Thieffry, 2001), or the continuous-state approach (Reinitz and Sharp, 1995; Gursky et al., 2001), but the first modeling work focusing on the segment polarity gene network was done by von Dassow and collaborators (von Dassow et al., 2000; von Dassow and Odell, 2002). von Dassow et al., developed a continuous-state model of the core network of five genes (en, wg, ptc, ci and hh) and their proteins. The initial choice of network topology failed to reproduce the observed expression patterns for these genes, but the introduction of two additional interactions led to surprisingly robust patterning with respect to variations in the kinetic constants in the rate laws. The robustness of this model with respect to the changes in the reaction parameters suggests that the essential features involved are the topology of the segment polarity network and the signatures of the interactions in the network (i.e. whether they are activating or inhibiting), and our first objective is to test this prediction.
The Boolean model we develop here is not based on continuous concentrations of mRNAs and proteins; only two states are admitted for each component corresponding to whether or not they are present. This choice is motivated by the ON/OFF character of the experimentally observed gene expression patterns.1 We show that this model reproduces the wild-type gene expression pattern, as well as the ectopic patterns corresponding to various mutants. We determine all the steady states of the model analytically, and we find that there is a surprisingly small number (6) of distinct ones, half of which are observed experimentally. We find the domains of attraction of these steady states by a systematic search in the space of possible initial conditions. Furthermore, we determine the minimal pre-patterning necessary to produce the wild-type spatial expression pattern, and thereby show that the majority of non-initiation errors in the pre-pattern can be corrected during the temporal evolution from such states. The model demonstrates the remarkable robustness of the segment polarity network, and shows that the robustness resides in the topology of the network and the signature of the interactions.
2. Description of the Boolean model
In the model, each mRNA or protein is represented by a node of a network, and the interactions between them are encoded as directed edges (see Fig. 1).2 The state of each node is 1 or 0, according as the corresponding substance is present or not. The states of the nodes can change in time, and the next state of node i is determined by a Boolean (logical) function of its state and the states of those nodes that have edges incident on it. In general, a Boolean or logical function is written as a statement acting on the inputs using the logical operators “and”, “or” and “not” and its output is 1(0) if the statement is true (false). The rules governing the transcription of a gene, for example, are determined by a Boolean function of the state of its transcriptional activators and inhibitors. Transcription will only commence if the activators are expressed and the inhibitors are not, thus the Boolean function will have the form “ACT and not INH”, where ACT represents the activators and INH the inhibitors. Moreover, if this mRNA is translated into a protein, its state enters the Boolean function of the protein. This type of network modeling is rooted in the pioneering work of Kauffman (1969, 1993) on random Boolean networks and of Thomas (Thomas, 1973; Thomas and D’Ari, 1990) on Boolean models describing generic gene networks. The novelty of our work lies in the fact that we base the Boolean model on the known topology of the Drosophila segment polarity network.
Expression of the segment polarity gene occurs in stripes that encircle the embryo, and therefore we can treat the two-dimensional pattern as one-dimensional. We consider a line of 12 cells corresponding to three parasegment primordia (i.e. the spatial regions that will become the parasegments), and impose periodic boundary conditions on the ends. We use four cells per parasegment primordium because when expression of the segment polarity genes begins, a given gene is expressed in every fourth cell. The use of 12 cells rather than 4 allows better illustration of the patterns, but throughout the analysis we assume that the initial pattern of the segment polarity genes is repeated every four cells. We concentrate on the network shown in Fig. 1, but do not include FZ and smo in our base network, because these nodes are not regulated by other nodes in the network. Consequently, the total number of nodes in the model system is n N = 15 × 12 or 180.
The states of the nodes evolve in discrete time steps under the following algorithm. We choose a time interval that is larger or equal to the duration of all transcription and translation processes, and we use this interval as the length of a unit time step. We then prescribe an initial state for each node, and the state at the next time step is determined by the Boolean function for that node. If is the state of the i-th node in the j-th cell at time t, and x = (x11, … xnN), then the next state of the network is , which defines a discrete dynamical system whose iteration determines the evolution of the state of all nodes. A fixed point of is a time-invariant state of the system, whereas a fixed point of for a minimal p > 1 represents a state that repeats periodically with least period p. Since the objective is to obtain the experimentally-observed stable expression pattern of the segment polarity genes starting from wild-type initial conditions, one test of the model is whether the state evolves to a fixed point that corresponds to this pattern.
The Boolean interaction functions are constructed from the interactions between nodes displayed in Fig. 1. In order to focus on the effects of the topology of the segment polarity network and the signature of the interactions on the predicted steady states, the model is based on the simplest assumptions concerning the interactions between nodes. These assumptions can be summarized as follows:
the effect of transcriptional activators and inhibitors is never additive, but rather, inhibitors are dominant;
transcription and translation are ON/OFF functions of the state;
if transcription/translation is ON, mRNAs/proteins are synthesized in one time step;
mRNAs decay in one time step if not transcribed;
transcription factors and proteins undergoing post-translational modification decay in one time step if their mRNA is not present.
For example, en is translated from en, and therefore if Since en is a transcription factor, it is assumed that its expression will decay sufficiently rapidly that if , then . These two assumptions mean that does not depend on , only on the expression of en, and therefore
| (1) |
Table 1 gives an overview of the Boolean functions for each node, which hereafter are labeled by their biochemical symbol. In each case, subscripts signify spatial position (i.e. cell number) and superscripts signify time. A detailed rationale for our choice of Boolean rules used for updating the state of each node is presented in the appendix.
Table 1.
The Boolean functions used in the model
| Node | Boolean updating function |
|---|---|
| SLPi | |
| wgi | |
| WGi | |
| eni | |
| ENi | |
| hhi | |
| HHi | |
| ptci | |
| PTCi | |
| PHi | |
| SMOi | |
| cii | |
| CIi | |
| CIAi | |
| CIRi |
The functions are based on the known interactions between mRNAs and proteins shown in Fig. 1, and on the temporal assumptions listed above. In general, the updating rule gives the expression of a node at time t + 1 as a function of the expression of its effector nodes at time t. However, there are three exceptions: we assume that the expression of SLP does not change, and that the activation of SMO and the binding of PTC to HH are instantaneous.
3. Functional topology of the segment polarity network
The rules for advancing the current state of the network given in Table 1 can be used to construct an expanded graph that reflects the function of the network. Consider the transcription of the hh gene. Fig. 1 shows that hh has two incoming edges, one from en and one from CIR, and Table 1 shows that transcription of the hh gene requires both the presence of the en protein and the absence of the CIR protein. To represent this conjunction in a graphical form, one can say that hht+1 depends on the current state of a “pseudo-node” that we denote by (see Fig. 2). The state of this new node is one whenever ENt = 1 and CIRt = 0, and zero otherwise. We can also represent the dependence of the pseudo-node on EN and CIR in a graphical form by introducing a “complementary” pseudo-node, , that is expressed whenever CIR is not. In order to take into account the inter-dependence of and CIR we connect them with a symmetrical (non-directed) edge. Finally, we draw two directed edges starting from EN and and ending in , to represent the dependence of on the expression of EN and (see Fig. 2).
Fig. 2.
Illustration of the network expansion process used to construct the functional topology. To express the logical rule governing the transcription of hh graphically, we introduce the complementary node and the composite node . The expanded network contains real nodes (filled circles) and pseudo-nodes (open circles), an interdependence relation between CIR and (dotted line), edges corresponding to the activation of (dash-dotted lines) and a single edge expressing the activation of hh transcription.
In general, we define “complementary” pseudo-nodes for any nodes whose negated expression enters Table 1, and “composite” pseudo-nodes for any combinations of node expressions that are connected by the “and” operator in Table 2. In effect, we expand the initial network into a kind of bipartite network composed of real and pseudo-nodes; the real nodes are the original nodes while the pseudo-nodes correspond to the regulation rules between the nodes.3 The total number of nodes in the network for 12 cells is 37 × 12 = 444. Of the 37 nodes per cell, 28 are cell-autonomous, and nine composite nodes represent signaling between neighboring cells. Fig. 3 shows the nodes and edges corresponding to the mRNAs and proteins in the second cell of the parasegment (15 real nodes, drawn as filled circles) together with the pseudo-nodes these mRNAs and proteins influence, both cell-autonomously (seven complementary and six composite pseudo-nodes, drawn as open circles), and in the neighboring cells (12 composite pseudo-nodes, drawn as open circles). To illustrate the symmetrical nature of the intercellular signaling, we also include on the figure the pseudo-nodes in neighboring cells that affect the expression of mRNAs and proteins in the second cell (six composite pseudo-nodes, drawn as open circles).
Table 2.
Definition of the symbols for pseudo-nodes used in Fig. 3
| Symbol of pseudo-node | Relation to parent node(s) |
|---|---|
| Complementary nodes | |
| not EN | |
| not hh | |
| not HH | |
| not PTC | |
| not SLP | |
| not SMO | |
| Composite nodes corresponding to a single cell | |
| CIA2 and and | |
| CIA2 and SLP2 and | |
| (CSM)2 | CI2 and SMO2 |
| EN2 and | |
| wg2 and CIA2 and | |
| wg2 and SLP2 and | |
| Composite nodes corresponding to intercellular interactions | |
| Cihj | CIi and hhj |
| CIi and and and | |
| PTCi and and | |
| PiHj | PTCi and HHj |
| WiSj | WGi and SLPj |
The state of each composite node is determined from the logical function giving its relation to the state of its “parent” nodes.
Fig. 3.
Functional topology of the network affecting the second cell of the parasegment. Filled circles represent real nodes, open circles represent pseudo-nodes, denoted according to Table 2. Pseudo-nodes with multiple indexes correspond to intercellular interactions and either receive some of their inputs from the neighboring cells, or contribute to the expression of the nodes in the neighboring cells (not shown). Symmetrical edges between nodes and their complementaries are drawn with dotted lines; edges determining the expression of composite nodes are drawn with dash-dotted lines; edges determining the expression of real nodes in the next time step are continuous. Double arrows denote a pair of oppositely directed edges.
Although the introduction of the pseudo-nodes increases the number of nodes in the network, it eliminates the distinction between edges based on their signatures; all directed edges in Fig. 3 now signify activation. However, there are differences in the way multiple activating edges are taken into account: multiple edges ending in composite pseudo-nodes (dash-dotted lines in Fig. 3) are added by the operator “and”, while multiple edges starting at pseudo-nodes and ending in real nodes are cumulated by the operator “or”. The presence of inhibitory control in Fig. 1 is reflected in activation by complementary nodes in Fig. 3. Although all directed edges signify activation, there are temporal differences in the underlying processes. The expression of the pseudo-nodes at time t is set by the expression of their parent nodes at time t, while the expression of most real nodes at time t is set by the expression of pseudo- and/or real nodes at time t – 1.
Fig. 3 illustrates the heterogeneous functional topology of the segment polarity network. The majority of nodes have few edges, but there are key nodes with a large number of incoming or outgoing edges. For example, has five outgoing edges, while HH has four. The important role of HH in the network is reflected in the fact that it affects the future expression of four other proteins (CIA, CIR, PTC and PH) in the neighboring cells, i.e. eight nodes. Other nodes such as CIA have several incoming edges, indicating that they can be activated in many ways. A single node, SLP, has only outgoing edges because it is constitutively present; all others have both incoming and outgoing edges (the apparent exceptions interact with nodes in the neighboring cells).
The functional network of Fig. 3 gives insight into the time evolution of the expression of the segment polarity genes. Given an initial state for all 15 real nodes in the network at a time t, the state of the complementary nodes is the negation of the state of the corresponding real node. The state of a composite pseudo-node is 1 if all of its parent nodes have state 1, otherwise it is 0. The state of real and pseudo-nodes at time t determines the state of real nodes at time t + 1. If a real node has multiple incoming edges, its state is 1 whenever at least one node at the origin of these edges is expressed. This way the general dynamical system can be decomposed into
| (2) |
where the operators Tor and Tand correspond to the updating processes described above. It is tempting to imagine these operators as matrices, but unfortunately the “and” and “or” operators cannot be linearized (i.e. “A and B” cannot be written as a linear combination of A and B).
While it is not possible to follow the evolution of the network’s expression analytically, we can gain insight into it by studying the cluster (group) of nodes that can be activated by the expression of a given node. We can estimate the size of this cluster by successively following the directed edges starting from the node, then those starting from the endpoints of these edges and continuing until the edges leave Fig. 3. This method gives us an upper bound for the number of activated nodes, since in most cases additional conditions must be satisfied in order for a node to be activated. Note that in this method we cannot follow the symmetrical edges, since their endpoints have opposite expressions. The absence of EN (or conversely the presence of ) gives the largest activated cluster, containing ci, CI, CIA, ptc, PTC, PH, wg, and WG. A separate activated cluster starts with the presence of en, and contains EN, hh and HH. These activating clusters indicate that the stripe of en and hh expression never overlaps with those of wg, ptc and ci. This separation into anterior and posterior compartments expressing different genes is well-known, in fact, it is the basis for calling these genes “segment polarity genes” (Wolpert et al., 1998).
While the majority of the activating effects propagate outside the cell, there are three cases in which an activation can return to its source. In other words, three short cycles exist in the network of Fig. 3. The first two cycles connect wg2 with or and the third connects PTC2 with . These cycles ensure the maintenance of wg and PTC if all the conditions for the expression of the pseudo-node in the cycle are met (i.e. wg is maintained if SLP or CIA is present and CIR is absent, PTC is maintained if HH is not expressed in the neighboring cells). The successful activation of the wg cycle can induce the stable expression of en and hh in those neighboring cells where neither SLP nor CIR is expressed, and stable expression of PTC leads to stable CIR expression two cells removed from en expression. The results presented in the following sections confirm the special role of these cycles in the dynamics of the segment polarity gene expression pattern.
4. Comparison between numerical and experimental results
The segment polarity genes are activated by the pair-rule genes in the cellular blastoderm phase (stage 5 according to the classification of Campos-Ortega and Hartenstein, 1985) of Drosophila embryogenesis, and maintain the parasegment borders and later the polarity of the segments from the end of gastrulation through germ-band elongation (stages 8–11, see Wolpert et al., 1998). The parasegment borders form between the wg and en/hh expressing cells (Hooper and Scott, 1992; Wolpert et al., 1998). Since our model is intended to describe the effect of the segment polarity genes in maintaining the parasegment border, the patterns of segment polarity genes formed before stage 8 can be considered as given initial states, and the final stable state should coincide with the wild-type patterns maintained during stages 9–11.
The initial state of each parasegment primordium, based on the experimental observations of stage 8 embryos, includes a two-cell-wide SLP stripe in the posterior half (Cadigan et al., 1994), a single-cell-wide wg stripe in the most posterior part (Hooper and Scott, 1992), single-cell-wide en and hh stripes in the most anterior part (Tabata et al., 1992; Hooper and Scott, 1992), and ci and ptc expressed in the posterior three-fourths (Hidalgo and Ingham, 1990; Hooper and Scott, 1992; Taylor et al., 1993). Since the proteins are translated after the mRNAs are transcribed, we assume that the proteins are not expressed in the initial state. The one-dimensional representation of the mRNA and protein patterns is shown in Fig. 4(a).
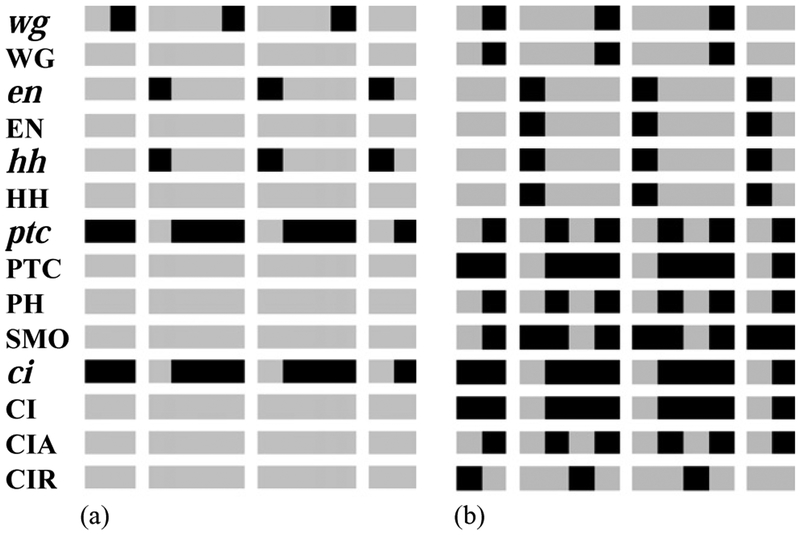
Fig. 4.
Wild-type expression patterns of the segment polarity genes. Here and hereafter left corresponds to anterior and right to posterior in each parasegment. Horizontal rows correspond to the pattern of individual nodes—specified at the left side of the row—over two full and two partial parasegments. Each parasegment is assumed to be four cells wide. A black (gray) box denotes a node that is ON (OFF). (a) The experimentally observed initial state before stage 8. en, wg and hh are expressed in one-cell-wide stripes, while the broad ptc and ci stripes are complementary to en. (b) The steady state of the model when initialized with the pattern in (a). This pattern is in agreement with the observed gene expression patterns during stages 9–11 (see text).
We iterate the dynamical system defined by the rules in Table 1 starting from the initial state described above. We find that after only six time steps, the expression pattern stabilizes in a time-invariant spatial pattern (see Fig. 4(b)) that coincides with the experimentally observed wild-type expression of the segment polarity genes during stages 9–11. Indeed, wg and WG are expressed in the most posterior cell of each parasegment (Ingham et al., 1991), while en, EN, hh and HH are expressed in the most anterior cell of each parasegment, as is observed experimentally (Ingham et al., 1991; Tabata et al., 1992), ptc is expressed in two stripes of cells, one stripe on each side of the en-expressing cells, the anterior one coinciding with the wg stripe (Hidalgo and Ingham, 1990; Hooper and Scott, 1992). SMO is present in broad stripes whose anterior border coincides with the anterior border of the wg stripe and whose posterior border extends about one cell further from the en stripe (Alcedo et al., 2000). ci is expressed almost ubiquitously, with the exception of the cells expressing en (Eaton and Kornberg, 1990; Alexandre et al., 1996). CIA is expressed in the neighbors of the HH-expressing cells, while CIR is expressed far from the HH-expressing cells (Aza-Blanc and Kornberg, 1999). Note that while the majority of the proteins are expressed in the same cells as their mRNAs, this is not the case for PTC. Indeed, while the ptc stripe separates into two by stage 11 (Taylor et al., 1993), the PTC stripe remains broad. There are indications that the level of PTC decreases in the middle of the stripe (Taylor et al., 1993), but the existence of PTC in those cells is very important because their signaling maintains the production of CIR (see Fig. 4(b)).
We have also done a systematic analysis of the patterns obtained when the initial expression of individual genes or groups of genes differs from the wild-type initial condition. In principle, the attractor for some initial conditions could be periodic in time, but we have found that the only stable attractors are steady states. Since the purpose of the segment polarity network is to stabilize and maintain the parasegment borders, this result is biologically realistic.
We first concentrate on the overexpressed initial patterns, as these provide direct comparison with heat-shock experiments. In these experiments the initial expression of a selected gene is ubiquitously induced following a heat shock. According to these experiments, the wg and ptc stripes expand anteriorly when hh is ubiquitously induced (Gallet et al., 2000). When the same induction is done on en, broadened en stripes result (Heemskerk et al., 1991), and narrower ci stripes emerge after a transient decay of ci (Schwartz et al., 1995). Our model indicates that these two cases lead to the same steady-state expression pattern that incorporates all experimental observations: broad en, wg, ptc and hh stripes, and narrower ci stripes (see Fig. 5(a)). We can understand the process leading to this state from the functional topology of the network (Fig. 3): ubiquitous hh means that is not expressed in any of the cells, nor is any , thereby removing the only path to the production of CIR. This means that CIA is produced in each ci -expressing cell, and it activates the transcription of wg and ptc in a wider domain.
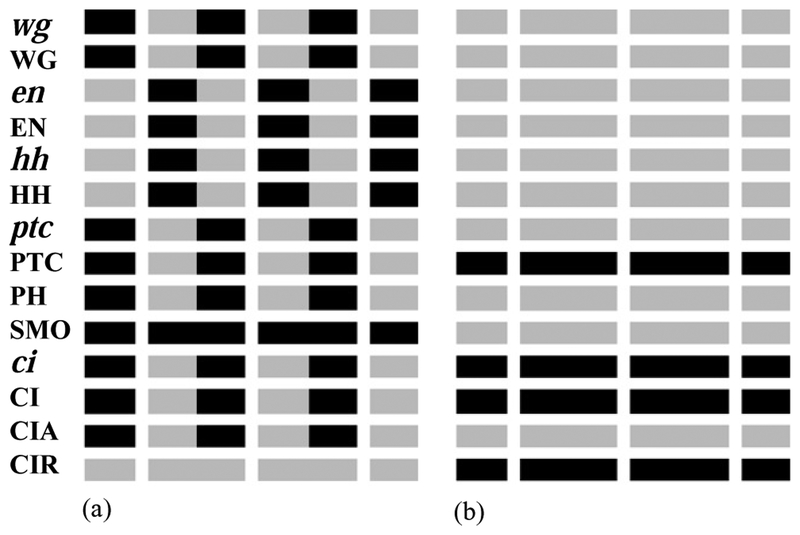
Fig. 5.
Ectopic expression patterns of the segment polarity genes obtained from the model by varying the initial conditions or inactivating certain nodes. (a) Broad-type expression pattern. The stripes of en, wg, ptc and hh are broader than normal, while the ci stripe narrows and CIR is not expressed. The anterior broadening of the en stripe together with the posterior broadening of the wg stripe induces an ectopic “border” in the middle of the parasegment. This state arises if wg, en or hh is initiated in broader stripes than wild type. This pattern is in perfect agreement with the experimentally observed gene expression after heat shock experiments on en and hh (see text). A similar pattern, only without ptc, PTC and PH expression, is obtained from the model when the expression of ptc is kept OFF, in agreement with observations on ptc mutants (see text). (b) Stable pattern with no stripes for wg, en, hh and ptc. This pattern arises if any of wg, en or hh is kept OFF in the model, when wg initiation is substantially delayed, or when intercellular interactions are disrupted.
Another method for inferring gene interactions experimentally is to silence selected genes by mutations. These inactive genes can be simulated by setting the expression of the transcript to zero and not updating it during the evolution of the system. Our results indicate that if any of en, wg or hh are blocked, while the other genes are initiated in the wild-type pattern (see Fig. 4(a)), the steady state is a pattern with no en, wg, ptc or hh, as in Fig. 5(b). We can see from Fig. 3 that each of these mutations disrupts intercellular signaling, causing ubiquitous expression of CIR, which in turn leads to ubiquitous repression of transcription. In agreement with this result, it has been observed experimentally that the hh expression in en null embryos starts normally, but disappears before stage 10 (Tabata et al., 1992). In wg null embryos, en is initiated normally but fades away by stage 9, as observed by DiNardo et al. (1988), while ci is ubiquitously expressed (Schwartz et al., 1995). In hh mutant embryos the wg expression disappears by stage 10 (Hidalgo and Ingham, 1990), as does the expression of ptc, and there is no segmentation (Gallet et al., 2000). All these experimental results are in excellent agreement with the numerically obtained pattern shown in Fig. 5(b).
If the ptc gene is blocked, we obtain a pattern with broad wg, en and hh stripes, which differs from the pattern of Fig. 5(a) only in the fact that there is no ptc, PTC and PH expression. Indeed, the network in Fig. 3 shows that the role of ptc is to restrict ectopic expressions of en, wg and hh, if ptc is deactivated, will be ubiquitous, causing all CI to be transformed into CIA, which leads to broadening of the wg and en/hh stripes. This pattern agrees with the experimental results on ptc mutants. These results indicate broad en, wg and hh stripes (Tabata et al., 1992; Gallet et al., 2000); Martinez-Arias et al. (1988) and Gallet et al. (2000) find that a new ectopic groove forms at the second en–wg interface at the middle of the parasegment. Also, ci is not expressed at this ectopic groove (Schwartz et al., 1995). Moreover, our results are in agreement with all experimental observations of double mutants as well (DiNardo et al., 1988; Ingham et al., 1991; Tabata et al., 1992; Bejsovec and Wieschaus, 1993).
If all the other genes are initiated normally, we find that a ci deletion does not affect the en, wg and hh patterns. However, ptc expression disappears, while PTC is expressed in a single cell wide stripe. Indeed, Fig. 3 shows that the deactivation of ci leads to the disappearance of CIA and CIR, but wild-type wg can still be maintained by SLP, and the interactions between wg, en and hh can maintain the en/hh stripe as well. In this case experiments indicate that the segmental grooves are present and wg is expressed until stage 11, but ptc expression decays (Gallet et al., 2000). Our results confirm the experimental result that the deletion of ci is able to counter the broadening effect of heat-shock-induced ubiquitous hh expression (von Ohlen and Hooper, 1997). Correct segmentation is maintained even for the other extreme, in ci–hh double mutants, in agreement with the results of Gallet et al. (2000). These findings indicate that the role of ci is to provide additional control over a functional sub-network formed by en, wg, hh and SLP. This sub-network can maintain an initial wild-type pattern, but loses expression in the case of initiation errors. The existence of the dual transcription factors CIA and CIR suggests a mechanism to correct both under- and overexpression errors in the initiation of wg and en.
5. Determination of the steady states and their domains of attraction
The fact that the Boolean model reproduces the results of numerous experiments remarkably well suggests that the structure of the model is essentially correct, and warrants exploration of problems that have not been studied experimentally. For example, we can determine the complete set of stable steady-state patterns of segment polarity gene expression, and estimate the domain of attraction of these states. The former can be done analytically by noting that these are fixed points of the discrete dynamical system, and so . Thus a steady state is the solution of the system of equations obtained from Table 1 by simply removing the time indices. This leads to the following equations.
| (3) |
Since the parasegments are assumed to be identical, we can restrict attention to one parasegment, and assume that i can run only from 1 to 4, i.e. the width of one parasegment. The majority of the variables in Eq. (3) can be eliminated, with the exception of wgi and PTCi. The expression of these nodes appears on both sides of the equations, reflecting the cycles of wg and PTC in Fig. 3. The final set of equations to be solved to find the steady states is
| (4) |
The asymmetry of these equations with respect to the anterior and posterior parts of the parasegments is due to the effect of SLP. Note that this set of equations shows that specifying compatible expressions for wg and PTC is sufficient for the complete description of a stable pattern. We solve this system of equations by examining all 26 = 64 possible states to determine whether they remain unchanged under the transformations given in Table 1. We obtain 10 solutions, pictured in Fig. 6. The complete patterns can be obtained from the solutions in Fig. 6 by using Eq. (3) to determine the expression of the other nodes in the network.
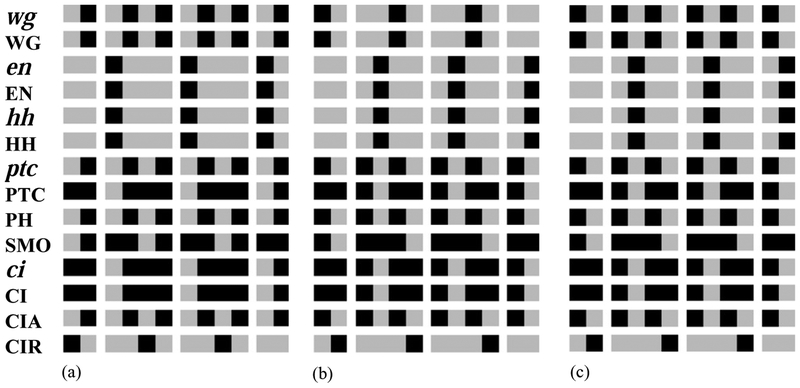
Fig. 6.
Patterns of wg and PTC obtained as solutions of Eq. (4). (a) Solution leading to the non-segmented pattern of Fig. 5(b). (b) Solution corresponding to the wild-type pattern of Fig. 4(b). (c) Solution leading to a variant of the wild-type pattern of Fig. 4(b) differing from it only in the expression of PTC that in this case becomes ubiquitous. (d), (e) Ectopic solutions that lead to patterns with no parasegment borders. The only difference between the two patterns is in the width of the PTC stripe (three- and four-cells-wide, respectively). (f) Solution leading to the broad type pattern of Fig. 5(a). (g), (h) Almost wild-type solutions with two wg stripes. The two patterns differ only in the width of the PTC stripe. (i), (j) Ectopic solutions similar to (d) and (e), but with two wg stripes.
The first steady state, resulting from solution 6(a), is the pattern with no segmentation first presented in Fig. 5(b). The solution 6(b) corresponds to the wild-type pattern shown in Fig. 4(b). There is also a variant of this solution, shown on Fig. 6(c), that has ubiquitous PTC expression rather than the posterior three quarters of the parasegment. This solution leads to a pattern that differs from the wild-type pattern only in the expression of PTC, thus we do not classify it as a distinct steady state.
In the third distinct steady state, corresponding to the solution 6(d), the wg stripe is displaced anteriorly from its wild-type position, while the en/hh stripe is displaced posteriorly (see Fig. 7(b)). This expression pattern corresponds to an ectoderm with no parasegmental grooves, since the end of the wg stripe does not meet the beginning of the en stripe. The solution 6(e) corresponds to the ubiquitous PTC variant of this pattern. The solution with broadened wg and PTC stripes shown in Fig. 6(f) leads to the broad type pattern first presented on Fig. 5(a).
Fig. 7.
Various stable patterns of the segment polarity genes obtained from Eq. (3). (a) Almost wild-type expression pattern with two wg stripes. This state corresponds to the solution of Eq. (3) presented in Fig. 6(g). (b) Ectopic pattern with displaced wg, en and hh stripes. This state arises from the solution on Fig. 6(d). (c) Variant of the ectopic pattern shown in (b) with two wg stripes. This state is determined from the solution 6(h).
The next solution, 6(g), leads to a steady state that differs from the wild type only in the fact that it contains two wg stripes, one in its normal position and one in a symmetrical position in the anterior part of the parasegment. The solution 6(h) leads to a variant with ubiquitous PTC. The last distinct steady state, corresponding to the solution 6(i), is similar to the ectopic pattern of Fig. 7(b), but it has two wg stripes, and an ectopic parasegment border between the third and fourth cell of the parasegment (see Fig. 7(c)). The solution 6(j) leads to a ubiquitous PTC variant of this state.
In summary, the ten solutions of Eq. (4) lead to six distinct steady states. Three of these steady states are well-known experimentally, corresponding to the wild-type pattern and two mutant patterns with either no stripes or broadened stripes. The existence of three additional states not observed experimentally suggests that the network can also produce patterns beyond those required in Drosophila embryogenesis. One of these states, corresponding to solution 6(g), coincides with the wild-type steady state in the expression of all nodes except for wg, but this divergence from wild type implies the emergence of a second, ectopic parasegment border. The second ectopic steady state (Fig. 7(b)) has displaced stripes for all nodes, and no parasegment borders, while the last ectopic state is a two-wg variant of this state with ectopic parasegment borders. None of these patterns have been observed experimentally, probably because they require ectopic initial conditions (see below). Additionally, if we relax the assumption of identical parasegments, the steady-state patterns corresponding to individual parasegments can be combined to form diverse patternings of the whole ectoderm. For example, the wild-type steady state and its double wg variant can be seamlessly integrated.
While each of the steady states can be obtained starting from suitable nearby states, the number of initial conditions leading to a chosen stable pattern, i.e. its domain of attraction, can be very different.4 Consider first the number of initial states that lead to the wild-type steady state. If we fix all nodes but one in their wild-type pattern, there are 24 = 16 distinct initial patterns corresponding to the four cells of the parasegment. We do this for each of the 14 variable nodes in turn (we do not change the expression of SLP) and find that the number of initial patterns leading to the wild-type steady state is three for wg or WG variation, four for en, EN, hh or HH variation, eight for ptc, PTC, CI or CIA variation, and 16 for PH, SMO, ci or CIR variation. We then use initial states in which the expression of pairs and triplets of nodes is different from the wild-type pattern. We find that the wild-type steady state is reachable from all initial conditions for which each variable node has one of the patterns that were previously found to lead to wild type when only the expression of that node was varied. If we now extend this result and assume that the attractor is the wild-type pattern if the pre-pattern for each node taken separately is one that led to wild-type expression when only the pattern of that node was varied, the number of network-wide wild-type inducing pre-patterns can be calculated by multiplying the numbers of individual wild-type inducing pre-patterns i.e. nwg × nWG × ⋯ × nCIR, where nx is the number of wild-type inducing pre-patterns of node x. There are 32 44 84 164 ~6 × 1011 such pre-patterns for each parasegment, which is a fraction of 8 × 106 of the total number of initial states Nst = 1614.
This fraction seems small, but we should not forget that not all initial conditions are biologically realistic. Indeed, it is almost impossible to initiate wg, en or hh in a broad stripe as their initiation depends on complex interactions between different pair-rule gene products (see Wolpert et al., 1998). It is much more realistic to focus on underexpression errors corresponding, for example, to delays in initiation. We find that the network is very robust with respect to missing initial expression of nodes. We have determined that the minimal prepatterning that leads to wild-type stable expression is as follows.
wg is wild type,
en and hh are not expressed,
ptc is expressed in the third cell of the parasegment primordium,
ci and the proteins are not expressed.
In summary, it is enough to initiate the expression of two genes in two cells per parasegment primordium, and the interactions between the segment polarity genes will initiate the others. While this result might not be completely realistic due to the assumptions of our model, it suggests a remarkable error-correcting ability for the segment polarity gene network.
Note that the minimal pre-pattern contains the wild-type stripe of wg. If wg is not expressed initially we find that the final pattern has no stripes, as shown in Fig. 5(b), regardless of the initial pattern of the other nodes. Consequently, a fraction of at least 1/16-th of the initial states leads to the pattern of Fig. 5(b). This finding suggests that wg has a special role in the functioning of the segment polarity network, and has to be activated at a specific time and specific cells in order to obtain wild-type gene expression.
In the other limit, broader than wild-type initial expression of any node except PH, SMO, ci and CIR leads to the pattern with broad stripes as in Fig. 5(a). This pattern is obtained in the vast majority of pre-patterns, comprising about 90% of the total number of initial states.
The almost wild-type steady-state pattern with two wg stripes can only appear if CIA is initialized at the same time as wg, and wg is pre-patterned with this ectopic pattern. Since the activation of CI to CIA requires HH signaling, and the HH stripes were found to appear later than the hh stripes (Taylor et al., 1993), the probability of simultaneous wg and CIA pre-expression is small.
The minimal pre-pattern needed for the ectopic pattern with displaced wg and en stripes (Fig. 5(c)) is wg expression in the third cell of the parasegment primordium (the same as its steady pattern), and ptc expression in the last cell of the parasegment primordium, where the wild-type stripe of wg would normally be. Note that this minimal initial condition is simply a shifted version of the minimal condition for the wild-type steady state. Indeed, the number of initial patterns leading to this steady state is the same as the number of wild-type inducing initial states, i.e. a fraction of 8 × 106 of the total number of initial states. In practice the simultaneous ectopic initiation of several nodes is very improbable, and indeed, this steady state has never been observed. The variant with two wg stripes is even more improbable, requiring simultaneous ectopic pre-patterning for wg, ptc and CIA. Finally, all ubiquitous-PTC variants of distinct steady states (corresponding to the solutions 6(c), 6(e), 6(h) and 6(j)) require a ubiquitous PTC pre-pattern in addition to the pre-pattern required for their striped-PTC counterparts.
6. A two-step model
While the steady states of the model reproduce experimentally-observed expression patterns, the temporal evolution may not reflect the in vivo evolution of expression patterning. We assume that the expression of mRNAs/proteins decays in one time step if their transcriptional activators/mRNAs are switched off, and this assumption can induce transient on-off flickering in the expression pattern. Such flickering can be eliminated by relaxing the assumptions of immediate switch-off, and assume more realistic, longer decay times.
As an illustration we consider a variant of our model incorporating slower decay of proteins. In this model we assume that protein expression in the (t + 1)st step depends on the expression of their mRNAs in the previous two steps, t and t – 1. According to this assumption, if the transcript is not present at time t, but it was expressed at time t – 1, we assume that the protein’s expression (initiated at time t) is maintained at time t + 1. In other words, the expression of a protein is maintained for at least two steps following the appearance of the transcript. The minimum value of two steps is obtained when the transcript appears and then disappears. According to the two-step assumption, the logical rules describing the expression of EN, WG, CI and HH become
The logical rule governing the expression of PTC (Eq. (A.9)) has an additional term enabling the maintenance of PTC if no HH is expressed in the neighboring cells. This term will not be modified by the two-step assumption, only the dependence on the transcript, such that the new rule for PTC is
| (5) |
The modification of CI into CIA and CIR is not directly affected by the two-step assumption, since it does not involve the ci transcript. Nevertheless, it is no longer necessary to assume that the presence of the hh transcript affects the outcome of the modification, since the two-step maintenance of HH prevents the propagation of transient hh decays. Thus the rules of creation of CIA and CIR simplify to
Finally, the rule for the production of the PH complex is unmodified by the two-step assumption, as are the rules governing the transcription of wg, en, hh, ptc and ci.
Since we do not introduce new steps, but only delay certain steps in the two-step model compared with the one-step model, the former leads to exactly the same steady states as the latter. One way to explain this result is that the system of equations to be solved in order to find a fixed point of the Boolean rules is identical to Eq. (3), since in the fixed point the expression of any node. Another way of illustrating the identity of the steady states in the two models is by noting that the functional topology of the network changes very little by the two-step assumption, and the wg and PTC cycles are unmodified.
In addition, we find that the two-step model reaches the same wild-type steady state as that shown in Fig. 4(b) if it is started from the initial pattern of Fig. 4(a). The number of intermediate states is slightly lower than in the one-step case, five compared to six. These states arise mostly because the lack of proteins in the initial state leads to the decay of the transcription of several genes, and are not affected by the increased stability of the proteins. When the initial expression of a single gene is varied while the others are wild type, the two-step model yields the same steady states as the one-step model, with one notable exception: In the case when wg is not expressed initially, the one-step model leads to the state with no segmentation shown on Fig. 5(b). However, the two-step model settles into a periodic temporal repetition of 11 states, only five of them being devoid of an engrailed/hedgehog–wingless boundary. This is the only case in which the final expression pattern of the segment polarity genes is not time invariant, and while this difference between the end states is intriguing, it does not change the conclusion that wg has to be pre-patterned in order to reach the wild-type steady state. Accordingly, the minimal pre-patterning leading to the wild-type final state remains the same as for the one-step model.
Our analysis shows that all the results presented in Section 4 are preserved if we use the two-step assumption. Gene mutations and ectopic pre-patternings lead to the same steady states as in the original model. The only change is in the intermediate states visited en route to the final state: both the wild-type and the broad type pattern stabilizes on average 30% faster using the two-step assumption. On the other hand, the pattern with no segmentation is reached at a slightly lower rate than in the original model. In conclusion, the two-step assumption provides a more realistic modeling of the decay of the proteins without changing the conclusions of the model.
7. Expression of the segment polarity genes after a round of cell division
When defining the Boolean functions characterizing the interactions between nodes, we have assumed that the transport of WG and HH can be disregarded. One of the reasons for doing so is the fact that we assume that the parasegments are four cells wide, as they are at the beginning of germ band elongation. Our results indicate that if we allow the spreading of WG and HH further than the membrane of the nearest neighbors of the cells producing them, ectopically broad wg, en, and hh stripes result. This suggests that during this stage the signaling of these secreted proteins is limited to the interaction with the neighboring cells. This conclusion is in agreement with the preference of the von Dassow model for low WG transport rates (von Dassow et al., 2000; von Dassow and Odell, 2002). However, in later stages the parasegment is enlarged due to two rounds of cell division (Wolpert et al., 1998). While the wg stripe remains a single cell wide, the en stripe widens to three cells. The maintaining of this en requires WG transport, and, indeed, wingless protein is seen to diffuse over a distance of 2–3 cell diameters in stage 10 embryos (Bejsovec and Martinez-Arias, 1991). It is possible that HH transport is also stronger at this stage, although it is still limited by PTC.
In order to determine if our model is able to describe the segment polarity gene patterns in later stages, we applied it to the transition between a four- and eight-cell-wide parasegment. We started with the wild-type pattern of Fig. 4(a) and assumed that each cell divides into two identical cells, with the same genes expressed in each of the two. We also assumed that WG and HH can be transported through the nearest neighbors of the cells expressing their mRNAs, and can interact with their receptors in the membrane of the second neighbor cells. The model leads to the steady state represented in Fig. 8, with a single cell wide wg stripe, three cell wide en and hh stripes, and two ptc stripes flanking the en domain. This steady state agrees perfectly with the wild-type pattern observed in stage 11 embryos (Hidalgo and Ingham, 1990; Eaton and Kornberg, 1990; Hooper and Scott, 1992; Alexandre et al., 1996; Alcedo et al., 2000). Note that in this state the WG stripe is broader than the wg stripe (as observed experimentally by Bejsovec and Wieschaus, 1993), but HH is expressed in the same cells as hh, its transport being restricted by the broad PTC domain.
Fig. 8.
Stable expression pattern of the segment polarity genes after a round of cell division, as obtained from our model. We assume that at this stage WG and HH can be transported through the neighboring cells. This pattern is in good agreement with experimental observations of stage 11 embryos.
8. Discussion and outlook
Since this work focuses on the regulatory network of the same genes as in von Dassow et al. (2000), it is worthwhile to discuss the common points and differences between our results. Our model, based on the topology and signature of the interactions between the segment polarity genes, confirms their suggestion that the topology of the network has a dominant role in its function. But it should be noted that the topology used in the von Dassow et al. (2000) model and the present work is different. Since the first reconstruction of von Dassow et al. did not capture the asymmetrical activation of en by WG, they introduced two additional interactions corresponding to WG autoactivation and the inhibition of en by CIR. The inhibition of en was not observed experimentally, instead, the asymmetrical en activation is thought to be caused by the transcription factors encoded by the sloppy paired gene (Cadigan et al., 1994), as taken into account in our model. To test whether the activity of the SLP proteins is necessary, we have studied the effects of inactivated and overexpressed SLP. We obtain seven possible steady states for inactivated SLP, but none of them corresponds to the wild-type pattern. The closest state, obtained when we start from wild-type initial conditions, has a wild-type wg and ptc pattern, but en and hh have an ectopic stripe anterior to the wg stripe, ci separates into two thin stripes, and CIR is not expressed (Fig. 9). At this point this state is a theoretical prediction that can be verified by conditional SLP mutants (i.e. mutants that have normal pair-rule activity, but no segment polarity activity). We also find that ubiquitously expressed SLP leads to the state with no segmentation presented on Fig. 5(b). This finding is in agreement with the experimental results of Cadigan et al. (1994). Based on these results we conclude that the SLP proteins play a vital role in this network.
Fig. 9.
The pattern obtained from our model when we start from wild-type initial conditions, but SLP is not functional. Note that en is expressed in two stripes, on both sides of the wg stripe.
Another difference between the von Dassow model and our model is the relative importance of inhibitory and activating regulation. von Dassow et al. consider inhibition as a modulation of the strength of activation (see the equations in the Supplement to von Dassow et al., 2000), while we assume that inhibition is dominant. The relatively weak role of inhibition in the former probably contributes to the large number of patterns with very broad en or wg stripes found even for wild-type pre-patterns, some of these patterns appearing much more frequently (i.e. for a larger number of random parameter combinations) than the wild-type pattern.
Our model is in very good agreement with known experimental results on the wild-type and mutant expression of the segment polarity genes. This gives numerous insights into the internal robustness of the regulatory network, insights that are important both from an experimental and theoretical perspective. First, we conclude that the wingless gene plays a key role in the system, and it is imperative that it be initiated at the right time in the right pattern. However, non-initiation of engrailed and hedgehog can be rescued by the activity of the network. Experiments with conditional mutants defective in initiation could verify these predictions. Second, we find that the segment polarity genes can evolve into an ectopic pattern with displaced stripes if initiated in a certain way. While this ectopic initiation is difficult, it should be possible. Finally, the isolation of the segment polarity- and pair-rule roles of the SLP proteins would permit further insights into the functioning of the segment polarity gene network.
Our goal was to remain as close to the experimentally known facts as possible, and to keep the number of additional assumptions and unknown parameters at a minimum. In this spirit, we have assumed that the majority of the interactions follow a single time-scale. Two exceptions to this rule are the assumptions of the persistence of existent wg and PTC expression in Eqs. (A.2) and (A.9). The stability of these nodes has a major role in stabilizing the expression of the segment polarity genes. Indeed, these assumptions are reflected in the existence of the cycles in the functional topology of the network (Fig. 3) and in the fact that the steady states are completely determined by the pattern of wg and PTC. It is therefore important to check what happens if these assumptions are not used. If, instead of Eq. (A.2) we require that both CIA and SLP be expressed in order that wg be transcribed, i.e. we assume that
| (6) |
it is still possible to arrive at the wild-type steady state, but only for much more restricted initial states. Furthermore, the resilience of the network to mutations in ci is destroyed, as all initial states lead to the steady state with no segmentation. Since it is observed experimentally that ci null mutants still display almost normal segmentation (Gallet et al., 2000), we can conclude that the stability of wg is required for the functioning of the segment polarity genes. If we do not assume the maintenance of initial PTC expression, the pattern of PTC will follow that of its transcript and split into two stripes. This will cause the complete disappearance of CIR, normally expressed in the cells in the middle of the PTC stripe, and the activation of all CI into CIA. As a consequence, the wild-type steady state will be impossible to reach, instead, the only steady state will be the pattern with broad stripes as in Fig. 5(a). Thus both persistence assumptions are necessary, meaning that the expression of wg and PTC does not decay during the interval of functioning of the segment polarity network.
The two-step model represents a step forward towards reproducing the transition from the initial state to the final steady state of the segment polarity network. A more realistic model would assume different time intervals (expressed in number of steps) for the decay for mRNAs and proteins. While this extension would involve unknown parameters, the condition of reaching the same steady states as the original model would provide constraints on the variability of the decay rates. Another direction where our model could be extended is to consider a two-dimensional array of cells. It is known experimentally that the stripes of segment polarity genes are not initiated as straight lines, but have jagged borders (Wolpert et al., 1998). During the functioning of the segment polarity network these stripes straighten, and the parasegment borders become sharp. A two-dimensional simulation of our model could lead to important insights into this process.
While we have focused on the segment polarity genes here, the Boolean approach can readily be applied to any gene regulation network with relatively well-characterized interactions. The Boolean approach enables the integration of qualitative observations on gene interactions into a coherent picture, and provides an easy verification of the sufficiency of these interactions. The analysis of a Boolean model is more tractable than that of a model based on differential equations, which inevitably has numerous unknown parameters, and a Boolean model facilitates a more systematic study of the possible steady states and their attractors. We envision realistic topology-based Boolean modeling as an important first step in understanding the interplay between the topology and functioning of gene regulatory networks. While the segment polarity gene network was successfully modeled by a simple synchronous binary Boolean model, other networks might require more detailed models incorporating asynchronous updating and/or multi-level variables (especially relevant for systems incorporating long-range diffusion). Of course there are undoubtedly systems, such as metabolic networks, for which a Boolean approach might not be an appropriate first level of analysis.
Acknowledgements
This work was supported in part by NIH Grant #GM 29123.
Appendix. Detailed rationale for the choice of Boolean updating rules
The SLP proteins are translated from the sloppy paired (slp) gene that has both pair-rule and segment polarity roles (Grossniklaus et al., 1992; Cadigan et al., 1994). These authors show that slp is expressed in the posterior half of the parasegment primordium, and this expression does not change during the developmental stages corresponding to the functioning of the segment polarity network. Since none of the nodes of the segment polarity network influence the translation of the SLP proteins (see Fig. 1), we assume that their expression is maintained constant.
| (A.1) |
The transcription of wg is promoted by CIA (Aza-Blanc and Kornberg, 1999) and SLP (Cadigan et al., 1994), and repressed by CIR (Aza-Blanc and Kornberg, 1999) (see Fig. 1). According to the assumptions listed above, this means that in the cells where and . The transcription factors CIA and SLP act independently (Cadigan et al., 1994; von Ohlen and Hooper, 1997), thus we assume that the effect of one of them is enough to maintain existing wg expression (i.e. the transcriptional activation is stronger than the degradation of the mRNA). Therefore, if , it is enough if either CIAi or SLPi is present and CIRi is absent to obtain . Consequently, we have that
| (A.2) |
WG is translated from wg, thus if . WG is secreted from the wg-expressing cells, and it is transported to neighboring cells (Pfeiffer and Vincent, 1999; Hatini and DiNardo, 2001; von Dassow and Odell, 2002). The details of the transport mechanism are not clear, but its strength strongly depends on the developmental stage of the embryo. In general, WG protein spreads from the wg-expressing cells at a distance roughly equal to the width of the en stripe (Lecourtois et al., 2001). In the early stages of germ-band elongation WG is mainly associated with the cell membrane of the wg-expressing cells (von Dassow and Odell, 2002), and mosaic embryo experiments done by Vincent and Lawrence (1994) indicate that en expression is sustained only in the cells adjoining wg. In our model the presence of a substance implies the capability to influence others, and consequently, we incorporate the lack of long-range WG signaling in this stage by assuming that WG is not transported further than the wg-expressing cells. However, we take into account “degradation” (which may in part be due to spatial spreading) by assuming that WG expression is not maintained if wg expression ceases, i.e. if . Thus, the expression of WG depends only of the expression of wg, and we have that
| (A.3) |
The transcription of en is promoted by WG in neighboring cells (Cadigan and Nusse, 1997) and repressed by SLP (Cadigan et al., 1994). We assume that the presence of WG in either of the neighbor cells is enough to activate en transcription, thus
| (A.4) |
EN is translated from en, and therefore if . Since EN is a transcription factor, it is assumed that its expression will decay sufficiently rapidly that if , then , therefore
| (A.5) |
hh transcription is promoted by en (Tabata et al., 1992) and inhibited by CIR (Aza-Blanc and Kornberg, 1999), and therefore
| (A.6) |
HH is translated from hh, thus if . HH is secreted from the hh-expressing cells, and becomes bound to the cell membrane, where it can bind to PTC present in one of the neighboring cells, forming PH (Ingham and McMahon, 2001). It is also possible that HH is transported away from its source, as is the case in the wing imaginal disks (Chen and Struhl, 1998; The et al., 1999; Ingham, 2000). The proteoglycans involved in HH transport compete with PTC in binding to HH, and thus PTC expression restricts HH movement. Since the PTC protein is expressed everywhere except for the en/hh expressing cells in this stage of embryonic development, we do not take into account the signaling effects of HH transport beyond the membrane of neighboring cells in this model. In addition, we assume that the HH levels decay if hh is not expressed, due to binding with PTC, degradation of the protein, etc.:
| (A.7) |
ptc transcription is activated by CIA (Aza-Blanc and Kornberg, 1999) and repressed by CIR (Aza-Blanc and Kornberg, 1999). It is likely that en represses ptc transcription (Hidalgo and Ingham, 1990), and thus ptc is transcribed whenever CIA is present and CIR and en are absent:
| (A.8) |
Since PTC is translated from ptc, if . The HH protein from the neighboring cells binds to PTC, forming the PH complex (Ingham and McMahon, 2001). Since PTC is a transmembrane receptor protein, and the binding to HH is the only reaction PTC participates in, we assume existing PTC levels are maintained even in the absence of ptc if there is no HH in the neighboring cells:
| (A.9) |
The PH complex forms when HH from neighboring cells binds to PTC. This binding is much faster than a transcription or translation process, and we assume that it is instantaneous on the chosen time-scale. Since the PH complex may undergo endocytosis (Taylor et al., 1993), we assume that existing PH levels are not maintained.
| (A.10) |
smo is transcribed ubiquitously throughout the segment, but its protein is deactivated post-translationally by PTC (Ingham, 1998). Since we only consider the active substances in our model, we assume that if . Binding of HH from neighboring cells to PTC relieves the inhibitory effect of PTC on SMO, leading to active SMO. We assume that the conformational changes involved in the deactivation and activation of SMO are instantaneous compared to the time needed for transcription or translation, and thus if . The complete condition for activation of SMO is therefore given as
| (A.11) |
We do not explicitly relate the activation of SMO to the existence of PH, as it is drawn in Fig. 1, because the formation of PH and the activation of SMO are the two outcomes of the same process involving PTCi, SMOi and HHi±1. While pictorially it is more suggestive to draw a single edge from PH to SMO, as in Fig. 1, the basic condition for the activation of SMO is the presence of HHi±1. Moreover, the PH-dependent logical function, or is mathematically equivalent to Eq. (A.11).
Since EN inhibits ci transcription, ci is expressed in the complement of cells expressing en (Eaton and Kornberg, 1990; Schwartz et al., 1995), and therefore if . Thus
| (A.12) |
CI is a cytoplasmic protein produced by ci and transformed into two nuclear proteins, CIA and CIR (Aza-Blanc and Kornberg, 1999). Thus its expression decays if ci is not expressed, i.e. if . These two assumptions imply that CI depends only on the expression of ci,
| (A.13) |
The CI protein is transformed to either CIA or CIR, depending on the state of SMO (Aza-Blanc and Kornberg, 1999, see also Fig. 1). If active SMO is present at time t, CI will be transformed into CIA. Assuming that the duration of this process is one time step, the condition for the existence of CIA at time t + 1 is that and . If all SMO is deactivated by PTC, CI will be cleaved to form CIR; assuming that this process takes one time step, if and . Our results indicate that these assumptions are not sufficient to reproduce the wild-type patterns of CIA and CIR; instead, ectopic expression of CIR appears in cells in which CIA should be expressed. The cause of this discrepancy might be that the time needed for the post-translational modification of CI is shorter than the duration of transcription and translation, and is in fact influenced by SMO activated later than at time t. One way to take this into account is to assume that the presence of hh at time t in the neighboring cells may activate SMOi and lead to the formation of CIAi at time t + 1. This is incorporated as follows:
| (A.14) |
CIR forms in the alternate pathway (see Fig. 1), and therefore relaxing the conditions for CIA formation will translate to a restriction of CIR, as reflected in the rule
| (A.15) |
Footnotes
Indeed, thresholds are required even in continuous-state models in order to decide whether a certain concentration corresponds to an ON or OFF state for comparison with the experimental patterns.
Note that it is necessary to designate different nodes for mRNAs and proteins corresponding to the same gene, because in several cases they are expressed in different cells.
In regular bipartite networks only edges connecting different kinds of nodes are allowed, whereas our network contains edges between nodes of the same kind.
A systematic method of computing state pre-images in random Boolean networks has been proposed by Wuensche (1997).
References
- Alcedo J, Zou Y, Noll M, 2000. Posttranscriptional regulation of smoothened is part of a self-correcting mechanism in the hedgehog signaling system. Mol. Cell 6, 457–465. [DOI] [PubMed] [Google Scholar]
- Alexandre C, Jacinto A, Ingham PW, 1996. Transcriptional activation of hedgehog target genes in Drosophila is mediated directly by the Cubitus interruptus protein, a member of the GLI family of zinc finger DNA-binding proteins. Genes Dev. 10, 2003–2013. [DOI] [PubMed] [Google Scholar]
- Aza-Blanc P, Kornberg TB, 1999. Ci a complex transducer of the Hedgehog signal. Trends Genet. 15, 458–462. [DOI] [PubMed] [Google Scholar]
- Bejsovec A, Martinez-Arias A, 1991. Roles of wingless in patterning the larval epidermis of Drosophila. Development 113, 471–485. [DOI] [PubMed] [Google Scholar]
- Bejsovec A, Wieschaus E, 1993. Segment polarity gene interactions modulate epidermal patterning in Drosophila embryos. Development 119, 501–517. [DOI] [PubMed] [Google Scholar]
- Bodnar JW, 1997. Programming the Drosophila Embryo. J. theor. Biol 188, 391–445. [DOI] [PubMed] [Google Scholar]
- Bodnar JW, Bradley MK, 2001. Programming the Drosophila Embryo 2. Cell Biochem. Biophys 34, 153–189. [DOI] [PubMed] [Google Scholar]
- Cadigan KM, Nusse R, 1997. Wnt signaling: a common theme in animal development. Genes Dev. 11, 3286–3305. [DOI] [PubMed] [Google Scholar]
- Cadigan KM, Grossniklaus U, Gehring WJ, 1994. Localized expression of sloppy paired protein maintains the polarity of Drosophila parasegments. Genes Dev. 8, 899–913. [DOI] [PubMed] [Google Scholar]
- Campos-Ortega JA, Hartenstein V, 1985. The Embryonic Development of Drosophila melanogaster. Springer, Berlin, Heidelberg, New York. [Google Scholar]
- Chen Y, Struhl G, 1998. In vivo evidence that Patched and Smoothened constitute distinct binding and transducing components of a Hedgehog receptor complex. Development 125, 4943–4948. [DOI] [PubMed] [Google Scholar]
- Davidson EH, et al. , 2002. A genomic regulatory network for development. Science 295, 1669–1678. [DOI] [PubMed] [Google Scholar]
- DiNardo S, Sher E, Heemskerk-Jongens J, Kassis JA, O’Farrell PH, 1988. Two-tiered regulation of spatially patterned engrailed gene expression during Drosophila embryogenesis. Nature 332, 45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton S, Kornberg TB, 1990. Repression of ci-D in posterior compartments of Drosophila by engrailed. Genes. Dev 4, 1068–1077. [DOI] [PubMed] [Google Scholar]
- Gallet A, Angelats C, Kerridge S, Thérond PP, 2000. Cubitus interruptus-independent transduction of the Hedgehog signal in Drosophila. Development 127, 5509–5522. [DOI] [PubMed] [Google Scholar]
- Grossniklaus U, Pearson RK, Gehring WJ, 1992. The Drosophila sloppy paired locus encodes two proteins involved in segmentation that show homology with mammalian transcription factors. Genes Dev. 6, 1030–1051. [DOI] [PubMed] [Google Scholar]
- Gursky VV, Reinitz J, Samsonov AM, 2001. How gap genes make their domains: an analytical study based on data driven approximations. Chaos 11, 132–141. [DOI] [PubMed] [Google Scholar]
- Han K, Manley JL, 1993. Functional domains of the Drosophila engrailed protein. EMBO J. 12, 2723–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatini V, DiNardo S, 2001. Divide and conquer: pattern formation in Drosophila embryonic epidermis. Trends Genet. 17, 574–579. [DOI] [PubMed] [Google Scholar]
- Heemskerk J, DiNardo S, Kostriken R, O’Farrell PH, 1991. Multiple modes of engrailed regulation in the progression towards cell fate determination. Nature 352, 404–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo A, Ingham P, 1990. Cell patterning in the Drosophila segment: spatial regulation of the segment polarity gene patched. Development 110, 291–301. [DOI] [PubMed] [Google Scholar]
- Hooper JE, Scott MP, 1992. The molecular genetic basis of positional information in insect segments In: Hennig W (Ed.), Early Embryonic Development of Animals. Springer, Berlin, pp. 1–49. [DOI] [PubMed] [Google Scholar]
- Ingham PW, 1998. Transducing hedgehog: the story so far. EMBO J. 17, 3505–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham PW, 2000. How cholesterol modulates the signal. Curr. Biol 10, R180–R183. [DOI] [PubMed] [Google Scholar]
- Ingham PW, McMahon AP, 2001. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 15, 3059–3087. [DOI] [PubMed] [Google Scholar]
- Ingham PW, Taylor AM, Nakano Y, 1991. Role of the Drosophila patched gene in positional signaling. Nature 353, 184–187. [DOI] [PubMed] [Google Scholar]
- Kauffman SA, 1969. Metabolic stability and epigenesis in randomly constructed genetic netw. J. theor. Biol 22, 437–467. [DOI] [PubMed] [Google Scholar]
- Kauffman SA, 1993. The Origins of Order. Oxford University Press, New York. [Google Scholar]
- Lecourtois M, Alexandre C, Dubois L, Vincent J-P, 2001. Wingless capture by frizzled and frizzled2 in Drosophila embryos. Dev. Biol 235, 467–475. [DOI] [PubMed] [Google Scholar]
- Lefers MA, Wang QT, Holmgren R, 2001. Genetic dissection of the Drosophila cubitus interruptus signaling complex. Dev. Biol 236, 411–420. [DOI] [PubMed] [Google Scholar]
- Martinez-Arias A, Baker N, Ingham PW, 1988. Role of segment polarity genes in the definition and maintenance of cell states in the Drosophila embryo. Development 103, 157–170. [DOI] [PubMed] [Google Scholar]
- Mendoza L, Thieffry D, Alvarez-Buylla ER, 1999. Genetic control of flower morphogenesis in Arabidopsis thaliana: a logical analysis. Bioinformatics 15, 593–606. [DOI] [PubMed] [Google Scholar]
- Méthot N, Basler K, 1999. Hedgehog controls limb development by regulating the activities of distinct transcriptional activator and repressor forms of Cubitus interruptus. Cell 96, 819–831. [DOI] [PubMed] [Google Scholar]
- Ohlmeyer JT, Kalderon D, 1998. Hedgehog stimulates maturation of Cubitus interruptus into a labile transcriptional activator. Nature 396, 749–753. [DOI] [PubMed] [Google Scholar]
- Pfeiffer S, Vincent J-P, 1999. Signaling at a distance: transport of wingless in the embryonic epidermis of Drosophila. Cell Dev. Biol 10, 303–309. [DOI] [PubMed] [Google Scholar]
- Reinitz J, Sharp DH, 1995. Mechanism of eve stripe formation. Mech. Dev 49, 133–158. [DOI] [PubMed] [Google Scholar]
- Sánchez L, Thieffry D, 2001. A logical analysis of the Drosophila gap-gene system. J. theor. Biol 211, 115–141. [DOI] [PubMed] [Google Scholar]
- Sanson B, 2001. Generating patterns from fields of cells. EMBO Rep. 2, 1083–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz C, Locke J, Nishida C, Kornberg TB, 1995. Analysis of cubitus interruptus regulation in Drosophila embryos and imaginal disks. Development 121, 1625–1635. [DOI] [PubMed] [Google Scholar]
- Tabata T, Eaton S, Kornberg TB, 1992. The Drosophila hedgehog gene is expressed specifically in posterior compartment cells and is a target of engrailed regulation. Genes Dev. 6, 2635–2645. [DOI] [PubMed] [Google Scholar]
- Taylor AM, Nakano Y, Mohler J, Ingham PW, 1993. Contrasting distributions of patched and hedgehog proteins in the Drosophila embryo. Mech. Dev 42, 89–96. [DOI] [PubMed] [Google Scholar]
- The I, Bellaiche Y, Perrimon N, 1999. Hedgehog movement is regulated through tout velu-dependent synthesis of a heparan sulfate proteoglycan. Mol. Cell 4, 633–639. [DOI] [PubMed] [Google Scholar]
- Thomas R, 1973. Boolean formalization of genetic control circuits. J. theor. Biol 42, 563–585. [DOI] [PubMed] [Google Scholar]
- Thomas R, D’Ari R, 1990. Biological Feedback. CRC Press, Boca Raton, Ann Arbor, Boston. [Google Scholar]
- van den Heuvel M, Ingham PW, 1996. smoothened encodes a receptor-like serpentine protein required for hedgehog signaling. Nature 382, 547–551. [DOI] [PubMed] [Google Scholar]
- Vincent J-P, Lawrence PA, 1994. Drosophila wingless sustains engrailed expression only in adjoining cells: evidence from mosaic embryos. Cell 77, 909–915. [DOI] [PubMed] [Google Scholar]
- von Dassow G, Meir E, Munro EM, Odell GM, 2000. The segment polarity network is a robust developmental module. Nature 406, 188–192. [DOI] [PubMed] [Google Scholar]
- von Dassow G, Odell GM, 2002. Design and constraints of the Drosophila segment polarity module: robust spatial patterning emerges from intertwined cell state switches. J. Exp. Zool 294, 179–215. [DOI] [PubMed] [Google Scholar]
- von Ohlen T, Hooper JE, 1997. Hedgehog signaling regulates transcription through Gli/Ci binding sites in the wingless enhancer. Mech. Dev 68, 149–156. [DOI] [PubMed] [Google Scholar]
- Wolpert L, Beddington R, Brockes J, Jessell T, Lawrence P, Meyerowitz E, 1998. Principles of Development. Current Biology Ltd., London. [Google Scholar]
- Wuensche A, 1997. Attractor basins of discrete networks. D. Phil. Thesis, University of Sussex. [Google Scholar]
- Yuh C-H, Bolouri H, Davidson EH, 2001. Cis-regulatory logic in the endo16 gene: switching from a specification to a differentiation mode of control. Development 128, 617–629. [DOI] [PubMed] [Google Scholar]