SUMMARY
Coordinate control of T cell proliferation, survival, and differentiation are essential for host protection from pathogens and cancer. Long-lived memory cells, whose precursors are formed during the initial immunological insult, provide protection from future encounters, and their generation is the goal of many vaccination strategies. microRNAs (miRNAs) are key nodes in regulatory networks that shape effective T cell responses through the fine-tuning of thousands of genes. Here, using compound conditional mutant mice to eliminate miR-15/16 family miRNAs in T cells, we show that miR-15/16 restrict T cell cycle, survival, and memory T cell differentiation. High throughput sequencing of RNA isolated by cross-linking immuno-precipitation of AGO2 combined with gene expression analysis in miR-15/16-deficient T cells indicates that these effects are mediated through the direct inhibition of an extensive network of target genes within pathways critical to cell cycle, survival, and memory.
In Brief
Coordinate control of T cell proliferation, survival, and differentiation are essential for effective cell-mediated adaptive immunity. Gagnon et al. define roles for the miR-15/16 family of microRNAs in restricting T cell cycle and long-lived memory T cell accumulation through the direct inhibition of a very large network of target mRNAs.
Graphical Abstract

INTRODUCTION
Regulation of T cell proliferation, survival, and differentiation is vital for effective immunity. In response to immunological challenges, naive antigen-specific T cells expand rapidly and undergo massive gene expression changes. As many as 50% of these changes are mediated post-transcriptionally (Cheadle et al., 2005). Within the first division, responding CD8+ T cells acquire sustained gene expression programs that lead to their differentiation into appropriately proportionate populations of terminal effector (TE) and memory precursor (MP) cells, identified by the expression of killer cell lectin-like receptor subfamily G member 1 (KLRG1) and IL-7 receptor alpha (Il7r/CD127), respectively (Kaech et al., 2002, 2003; Kakaradov et al., 2017; Sarkar et al., 2008). Effector T cells are important for pathogen or tumor clearance. MP cells form a long-lived pool of memory cells capable of rapidly responding to subsequent encounters with the same antigen.
microRNAs (miRNAs) are short non-coding RNAs that mediate post-transcriptional regulation, predominantly via Watson-Crick base pairing to 3′ untranslated regions (UTRs) of target mRNAs (Agarwal et al., 2015). miRNAs play key roles in the differentiation and functional characteristics of memory T cells (Ban et al., 2017; Baumjohann and Ansel, 2013; Chen et al., 2017; Guan et al., 2018; Khan et al., 2013; Moffett et al., 2017; Pua et al., 2016; Steiner et al., 2011; Wells et al., 2017). While the effects of miRNAs on the downregulation of individual targets are often less than two-fold, through coordinated tuning of gene networks, their overall biological effects can be profound.
The miR-15/16 family of miRNAs functions as tumor suppressors. Deletions of the Dleu2/Mirc30 locus, which encodes miR-15a and miR-16–1, occur in more than 50% of human chronic lymphocytic leukemia (CLL) cases (Calin et al., 2002), and targeted deletion of these miRNAs in mice induces a CLL-like indolent B lymphocyte proliferative disease (Klein et al., 2010). miR-15/16 restrict the proliferation of B cells through the direct targeting of numerous cell-cycle- and survival-associated genes, including Ccnd1, Ccne1, Cdk6, and Bcl2 (Liu et al., 2008). In addition to Mirc30, T cells strongly express Mirc10 and its two mature miRNA products, miR-15b and miR-16–2. Patients with T cell lymphoblastic lymphoma/leukemia (T-LBL/ALL) exhibiting lower-than-median expression levels of miR-16 exhibit a worse prognosis, suggesting a similar role for miR-15/16 in T cells (Xi et al., 2013). miR-15/16 has also been implicated in T cell anergy, regulatory T cell (Treg) induction, Treg/Th17 balance, and tumor-infiltrating T cell activation (Marcais et al., 2014; Singh et al., 2015; Wu et al., 2016; Yang et al., 2017). However, the requirements for miR-15/16 in T cell development, proliferation, survival, and differentiation remain unknown.
We generated mice with conditional inactivation of both miR-15a/16–1/Mirc30 and miR-15b/16–2/Mirc10 in T cells (miR-15/16Δ/Δ). miR-15/16 restricted T cell proliferation, limited T cell survival ex vivo, and directly targeted numerous cell-cycle- and survival-associated genes. Deletion of miR-15/16 in T cells did not result in overt lymphoproliferative disease. Instead, miR-15/16Δ/Δ mice selectively accumulated memory T cells, and miR-15/16 restricted the differentiation of MP cells in response to the lymphocytic choriomeningitis virus (LCMV). Rather than working through any one critical target, miR-15/16 physically interacted with and repressed the expression of a surprisingly broad network of memory-associated genes.
RESULTS
miR-15/16 Are Dynamically Regulated during T Cell Responses
Activated T cells rapidly reset their mature miRNA repertoire through an increased turnover of the miRNA-induced silencing complex (miRISC) and transcriptional regulation of miRNA precursors (Bronevetsky et al., 2013). Consistent with this prior report, miR-15a, miR-15b, and miR-16 were substantially downregulated over a 4-day course of CD4+ T cell activation in vitro (Figure 1A). miR-155 (upregulated), miR-103/107 (transiently downregulated), and miR-150 (downregulated) also behaved as expected. To assess expression kinetics in a physiologically relevant context, we re-analyzed published data from CD8+ TE and MP cells sorted from LCMV-infected mice (Khan et al., 2013). miR-15/16 were downregulated in both TE and MP cells (Figure 1B). In MP cells, miR-15b and miR-16 downregulation was sustained for at least 30 days post-infection (p.i.), placing these miRNAs among the most downregulated during memory T cell formation. miR-15a expression recovered to naive T cell levels by 30 days p.i. in MP cells (Figure 1B). However, miR-15a accounts for <10% of the total miR-15/16 family miRNAs in resting CD4+ T cells (Figure 1C). These results suggest that limiting the expression of miR-15/16 may be an important component of the gene expression program initiated by T cell activation and sustained among memory CD8+ T cells.
Figure 1. miR-15/16 Are Dynamically Regulated during T Cell Responses.
(A) qPCR of miRNA expression within CD4+ T cells in response to in vitro stimulation with anti-CD3 and anti-CD28 for 3 days followed by 1 day resting v (n = 6 biological replicates from two independent experiments).
(B) Time course miRNA microarray of CD8+ TE and MP cells after infection with LCMV (n = 3 biological replicates from one experiment).
(C) qPCR of miRNAs purified from naive CD4+ T cells of the indicated genotypes (n = 3 biological replicates from one experiment).
See also Figure S1.
T Cell-Specific Inactivation of Both miR-15/16 Gene Loci
miR-15/16 clusters occur in two genomic locations. The miR-15a/16–1 cluster (Mirc30) resides intronic to the long non-coding RNA, Dleu2, on chromosome 14 in mice (13 in humans), and miR-15b/16–2 (Mirc10) resides intronic to Smc4 on chromosome 3 within the genomes of mice and humans. Previous studies of these miRNAs have been restricted to genetic ablation of either one of these clusters independently, overexpression, or transient inhibition. However, cells with CD4-Cre-mediated deletion of either one of these clusters independently (miR-15a/16–1Δ/Δ or miR-15b/16–2Δ/Δ T cells) retained high levels of miR-16 expression, whereas the removal of both clusters (miR-15/16Δ/Δ) effectively abrogated miR-15 and miR-16 expression (Figure 1C).
T cell-specific deletion of both miR-15/16 clusters revealed subtle effects on T cell accumulation in primary and secondary lymphoid tissues without affecting total cellularity (Figures S1A–S1C). These effects were cell intrinsic, as they were also observed among miR-15/16Δ/Δ T cells within mixed bone marrow chimeric mice (Figure S1D). Mice with B cell-specific deletion of miR-15a/16–1 develop a delayed clonal lymphocytosis (Klein et al., 2010). miR-15/16Δ/Δ animals were examined after aging over 1.5 years with no apparent lymphoproliferative disease, lymphoma, or leukemia (data not shown). These findings demonstrate that miR-15/16 restrict the accumulation of T cells in specific-pathogen-free animals without causing disease.
miR-15/16 Bind and Regulate a Large Network of Direct Target RNAs in T Cells
The RNA sequencing (RNA-seq) comparison of miR-15/16fl/fl and miR-15/16Δ/Δ T cells showed that genes predicted to be targeted by miR-15/16 by TargetScan 7.1 (Agarwal et al., 2015) were more likely to be upregulated in miR-15/16Δ/Δ T cells, compared to all expressed genes lacking a miR-15/16 seed match in their 3′UTR (Figure 2A). The full set of genes with 3′UTR seed matches was also enriched for upregulation in miR-15/16Δ/Δ cells. This set contains three times as many putative target genes as the restricted TargetScan subset (3281 versus 944), but also many more false-positive targets.
Figure 2. miR-15/16 Bind and Regulate a Large Network of Direct Target RNAs in T Cells.
(A) Cumulative density plot depicting global expression by RNA sequencing as a ratio of the fold change between miR-15/16Δ/Δ (n = 5 biological replicates) and miR-15/16fl/fl (n = 4 biological replicates) resting CD4+ T cells for all genes without a 7-mer or 8-mer miR-15/16 3′UTR seed match (black), genes with a 7-mer or 8-mer miR-15/16 3′UTR seed match (green), genes with a 7-mer or 8-mer miR-15/16 3′UTR seed match and AHC read depth ≥5 (blue), and genes classified as targets of miR-15/16 by TargetScan 7.0 (red) (AHC reads represent the combined depth of n = 10 independent immunoprecipitations).
(B) Relative abundance of miRNAs bound by AGO2 (top) and AHC coverage across transcriptomic locations relative to region length (bottom).
(C) Heatmap of genes with a p value ≤ 0.05 plotted alongside a bar graph of AHC read depth at miR-15/16 seed matches for each gene at which they occur.
(D) Comparison of AHC reads between genes that are downregulated and upregulated (p ≤ 0.05) in miR-15/16Δ/Δ cells among genes with seed matches in the 3′UTR (top) or 5′UTR/CDS (bottom) (Mann-Whitney U test).
See also Figure S2.
To further enrich for genes directly targeted in T cells, we performed AGO2 high-throughput sequencing of RNAs isolated by crosslinking immunoprecipitation (AHC) to globally map AGO2-miRNA complex binding to target RNAs (Chi et al., 2009). Mature miRNAs accounted for 37% of all AHC reads (Figure S2A), and among these, miR-15/16 was the third most abundant family (Figure 2B, top). Non-miRNA AHC relative coverage was highly enriched at TargetScan-predicted miRNA binding sites, and again miR-15/16 seed matches were the third most abundant (Figure 2B, bottom). Indeed, there was a significant correlation between the density of AHC reads mapping to all miRNAs and their corresponding seed matches (Figure S2B).
Restricting the list of putative miR-15/16 targets to the 1280 genes with 3′UTR seed matches corresponding with AHC read depth ≥5 enriched for genes upregulated in miR-15/16Δ/Δ cells equally as well as TargetScan (Figure 2A). Supporting the validity of this approach and the large number of detected direct miR-15/16 target mRNAs expressed in T cells, differentially expressed genes (p ≤ 0.05) upregulated in miR-15/16Δ/Δ cells were frequently occupied by AGO2 at miR-15/16 seed matches, as compared with genes downregulated in miR-15/16Δ/Δ cells (Figure 2C). Furthermore, upregulated genes with 3′UTR seed matches (n = 680) had significantly greater AHC read depth than downregulated genes with 3′UTR seed matches (n = 195) (Figure 2D, top). In contrast, upregulated genes in miR-15/16Δ/Δ cells containing 5′UTR or coding sequence (CDS) but not 3′UTR seed matches had no enrichment for AGO2 binding, compared with downregulated genes (Figure 2D, bottom). Together, these data indicate that miR-15/16 directly binds a large network of target genes in T cells.
miR-15/16 Directly Target Cell-Cycle-Associated Genes and Restrict Accumulation of Antigen-Specific T Cells in Response to LCMV Infection
Gene set enrichment analysis identified KEGG Cell Cycle as a major signature of gene expression in miR-15/16Δ/Δ cells (Figure 3A). Strikingly, all differentially expressed genes (p ≤ 0.05) within the KEGG Cell Cycle gene set containing miR-15/16 seed matches were upregulated in miR-15/16Δ/Δ cells (Figure 3B). All but three of these genes had considerable AHC reads at miR-15/16 seed matches, suggesting that they were directly targeted by miR-15/16.
Figure 3. miR-15/16 Directly Target Cell-Cycle-Associated Genes and Restrict Accumulation of Antigen-Specific T Cells in Response to LCMV Infection.
(A) Gene set enrichment analysis of all expressed genes (normalized read depth ≥5). Plotted are the enrichment curve for and positional location of each target in the KEGG Cell Cycle gene set arrayed in ranked order from most upregulated to most downregulated (left to right) in miR-15/16Δ/Δ (enrichment score (ES) = 0.4354147; nominal p value < 0.001; false discovery rate [FDR] q value = 0.088).
(B) Heatmap of KEGG Cell Cycle genes with miR-15/16 seed matches in their 3′UTRs plotted alongside a bar graph of AHC read depth at miR-15/16 seed matches for each gene at which they occur.
(C) Flow cytometry of EdU-labeled CD8+ and CD4+ T cells 8 days p.i.
(D) Quantification of frequencies (left) and absolute numbers (right) of EdU+ CD8+ and CD4+ T cells 8 days p.i. with LCMV (n = 10 biological replicates representative of two independent experiments, two-tailed t test).
(E) Flow cytometry of antigen-specific T cells.
(F) Quantification of frequencies (left) and absolute numbers (right) of antigen-specific CD8+ T cells (n = 10 biological replicates representative of two independent experiments, two-tailed t test).
(G) Quantification of frequencies of antigen-specific CD8+ T cells in mixed bone marrow chimeras (n ≥ 8 biological replicates, representative of ≥3 independent experiments, two-tailed t test).
*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. See also Figure S3.
To test whether miR-15/16 restrict the T cell response to antigen stimulation, we infected miR-15/16fl/fl and miR-15/16Δ/Δ mice with LCMV. Cycling CD8+ and CD4+ T cells were measured by 5-ethynyl-2’-deoxyuridine (EdU) incorporation 8 days p.i. Compared to miR-15/16fl/fl mice, miR-15/16Δ/Δ mice had significantly more proliferating CD8+ and CD4+ T cells (Figures 3C and 3D). Additionally, miR-15/16Δ/Δ mice had increased frequencies and absolute numbers of cells specific for LCMV immunodominant epitopes (GP33 and GP276) both at the peak of the response and long after viral clearance (Figures 3E and 3F). Accumulation of antigen-specific cells in miR-15/16Δ/Δ mice was cell intrinsic, as it also occurred in mixed bone marrow chimeric mice (Figure 3G). T cells lacking either miR-15a/16–1 or miR-15b/16–2 alone did not exhibit increased proliferation or accumulation of LCMV-specific T cells (Figures S3A–S3D).
These experiments demonstrate that both miR-15/16 clusters act to restrict the accumulation of antigen-specific T cells in response to LCMV infection and suggest that at least part of this effect can be accounted for by decreased proliferation. miR-15/16 effects on the accumulation of CD4+ and CD8+ T cells are likely mediated by direct binding and posttranscriptional regulation of a network of cell-cycle-associated genes.
miR-15/16 Restrict the Formation of CD8+ Memory Cells
We hypothesized that the increased accumulation of antigen-specific T cells in LCMV-infected miR-15/16Δ/Δ mice would be accounted for by increased expansion and survival of virus-specific effector T cells. Surprisingly, however, the absolute number of antigen-specific KLRG1+ effector cells was unchanged in miR-15/16Δ/Δ mice (Figures 4A–4C). Instead, the percentage and absolute number of KLRG1− and CD127+ memory cells increased by more than 50%. This effect was apparent as early as 8 days p.i. and stabilized by 15 days p.i. (Figure 4D). Both miR-15/16 clusters contributed to this effect, since single conditional knockout animals displayed only modest changes in these populations (Figures S4A and S4B).
Figure 4. miR-15/16 Restrict the Formation of CD8+ Memory Cells.
(A) Flow cytometry of tetramer+ CD8+ T cells from spleens harvested 8 days p.i. (top) or 30+ days p.i. (bottom) with LCMV.
(B) Quantification of frequencies of tetramer+ CD8+ T cells from spleens harvested 8 days p.i. (top) or 30+ days p.i. (bottom) with LCMV (n ≥ 10 biological replicates from at least two independent experiments, two-tailed t test).
(C) Quantification of absolute numbers of tetramer+ CD8+ T cells from spleens harvested 8 days p.i. (top) or 30+ days p.i. (bottom) with LCMV (n = 10 biological replicates from two independent experiments, two-tailed t test).
(D) Quantification off requencies of tetramer+ CD8+ T cells from peripheral blood over the course of 30+ days p.i. with LCMV (n ≥ 9 biological replicates from at least two independent experiments, two-tailed t test).
(E and F) Quantification of flow cytometry to identify CXCR3+ or CD27+ (E) and CXCR3− or CD27− (F) populations of LCMV-specific CD8+ T cells 30+ days p.i. (n ≥ 5 biological replicates from ≥2 independent experiments, two-tailed t test).
(G) Quantification of frequencies of tetramer+ CD8+ T cells from spleens of mixed bone marrow chimeric mice 30+ days p.i. (n ≥ 8 biological replicates, representative of R3 independent experiments, two-tailed t test).
**p < 0.01; ***p < 0.001; ****p < 0.0001. See also Figures S4 and S5.
Responding T cell subsets can be distinguished using a variety of markers (Baars et al., 2005; Hikono et al., 2007; Hu et al., 2011; Kaech et al., 2002; Olson et al., 2013; Wherry et al., 2003). miR-15/16Δ/Δ mice generated higher frequencies and absolute numbers of antigen-specific CD8+ T cells with long-lived memory surface phenotypes, including high expression of CD127, CXCR3 and CD27, and low or absent KLRG1 (Figures 4A–4E). Furthermore, even among CD27− and CXCR3− populations, miR-15/16-deficient T cells were enriched for an increased expression of long-lived memory surface marker CD127 (Figure S5). In contrast, effector cells marked as KLRG1+, CD127−, CXCR3−, or CD27− were present at a reduced frequency but equivalent absolute numbers (Figure 4F). Mixed bone marrow chimera experiments demonstrated that these effects were cell intrinsic (Figure 4G).
miR-15/16 deficiencies preferentially affected memory T cell accumulation. The spleens and inguinal lymph nodes (iLNs) of unchallenged miR-15/16Δ/Δ mice contained increased numbers of both CD8+CD44hi and CD4+CD44hi memory T cells (Figures S4C and S4D). These cells were not acutely activated, as the frequencies of CD25+ T cells were not increased among CD44hi populations (Figure S4E). Memory cells accounted for most of the accumulated T cells. CD44loCD62L+ naive T cell numbers were relatively stable, with differences between the control and miR-15/16Δ/Δ mice reaching significance only for CD8+ cells in the spleen (Figure S4F). Taken together, these findings suggest that miR-15/16 specifically restrict the accumulation of long-lived memory cells in unchallenged animals and in response to infection with LCMV, but not at the expense of the naive or effector cell compartment.
miR-15/16 Restrict Memory CD8+ T Cell Differentiation
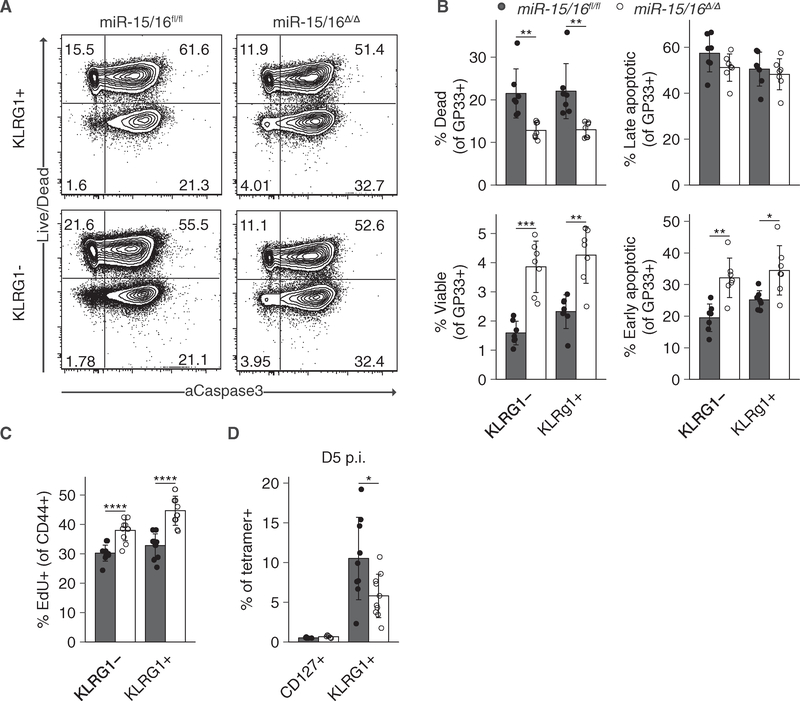
We investigated the possibility that memory CD8+ T cells preferentially accumulate in miR-15/16-deficient mice due to MP-specific effects on cell survival or proliferation. Following antigen clearance, LCMV-specific TE CD8+ T cells undergo a massive contraction phase, leaving behind a population of memory cells that provide protection from future encounters with the same virus. One possible explanation for the increase in MP CD8+ T cells in miR-15/16-deficient animals is that they may have a selective survival advantage over miR-15/16-deficient TE cells. To determine whether miR-15/16Δ/Δ CD8+ T cells were exhibiting an MP-specific survival advantage compared to miR-15/16-sufficient cells, splenocytes from mice 8 days p.i. with LCMV were cultured ex vivo overnight in the absence of exogenous cytokines. These conditions revealed a significant increase in viable and early apoptotic cells, with a corresponding decrease in dead cells among miR-15/16-deficient, virus-specific CD8+ T cells within both MP and TE populations (Figures 5A and 5B). The cell cycle effects of miR-15/16 were also apparent in both MP and TE cells, as measured by in vivo EdU incorporation 8 days p.i. with LCMV. Either miR-15b/16–2 or miR-15a/16–1 was sufficient to restrict cell survival and the proliferation of both MP and TE populations (Figures S6A and S6B). miR-15/16Δ/Δ CD8+ T cells were less likely to express the TE marker KLRG1 as early as 5 days p.i. with LCMV (Figure 5D). These results support a model wherein miR-15/16 preferentially restrict memory CD8+ T cell accumulation through effects on differentiation. Reciprocal reduction of the alternate effector T cell differentiation is likely balanced by increased survival and proliferation.
Figure 5. miR-15/16 Restrict Memory CD8+ T Cell Differentiation.
(A) Flow cytometry of GP33+ CD8+ T cells from spleens harvested 8 days p.i. with LCMV cultured overnight in vitro (n = 7 biological replicates from two Independent experiments).
(B) Quantification of frequencies of Live/Dead− activated caspase-3− GP33+ CD8+ T cells from spleens harvested 8 days p.i. with LCMV cultured overnight in vitro (n = 7 biological replicates from two independent experiments, two-tailed t test).
(C) Quantification of frequencies of EdU+ CD44+ CD8+ T cells from spleens harvested 8 days p.i. with LCMV (n = 7 biological replicates from two independent experiments, two-tailed t test).
(D) Quantification of frequencies of MP and TE populations among tetramer+ CD8+ T cells from spleens harvested 5 days p.i. with LCMV (n = 8 biological replicates from two independent experiments).
*p < 0.05; **p < 0.01; ***p < 0.001. See also Figure S6.
miR-15/16Δ/Δ Memory CD8+ T Cells Exhibit Functional Hallmarks of Long-Lived Memory Cells
We further characterized the quality of miR-15/16Δ/Δ memory CD8+ T cell responses to an antigen re-challenge and secondary infection. First, we pulsed splenocytes from LCMV-infected mice with GP33 or GP276 ex vivo. Consistent with the increases in antigen-specific cells detected by tetramer staining, greater frequencies and absolute numbers of CD8+ T cells from miR-15/16Δ/Δ mice produced the effector cytokine’s tumor necrosis factor (TNF) and IFNγ 8 days p.i. (Figures 6A and 6B), with a non-significant trend toward an increase at day 30+ p.i. (Figure 6C). Furthermore, greater frequencies and absolute numbers of CD8+ T cells from miR-15/16Δ/Δ mice exhibited a polyfunctional phenotype, characterized by production of TNF, IFNγ, and IL2 8 days p.i. (Figures 6D and 6E).
Figure 6. miR-15/16Δ/Δ Memory CD8+ T Cells Exhibit Functional Hallmarks of Long-Lived Memory Cells.
(A) Flow cytometry of CD8+ T cells 8 days p.i. with LCMV responding to splenocytes pulsed with either GP33 (top) or GP276 (bottom).
(B) Quantification of cytokine-producing CD8+ T cells re-stimulated in vitro with peptide-pulsed splenocytes 8 days p.i. (n = 5–7 biological replicates representative of at least two independent experiments, two-tailed t test).
(C) Quantification of cytokine-producing CD8+ T cells re-stimulated in vitro with peptide-pulsed splenocytes 30+ days p.i. (n = 5 biological replicates representative of at least two independent experiments, two-tailed t test).
(D) Flow cytometry of CD8+ T cells 8 days p.i. with LCMV responding to splenocytes pulsed GP33 and analyzed for multi-functional cytokine production.
(E) Quantification of multi-functional cytokine-producing CD8+ T cells re-stimulated in vitro with peptide-pulsed splenocytes 8 days p.i. (n = 5–7 biological replicates from a single experiment, two-tailed t test).
(F) Experimental outline of direct re-challenge model.
(G) LM-GP33 colony forming units (c.f.u.) from the spleen and liver of mice infected with LM-GP33 30+ days p.i. with LCMV and harvested 48 h later normalized to the naive group of each respective experiment (n ≥ 9 biological replicates from two independent experiments, Mann-Whitney U test).
*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
To assess the effectiveness of miR-15/16-deficient antigen-specific memory CD8+ T cells in vivo, we re-challenged animals 30 days p.i. with LCMV with Listeria monocytogenes expressing GP33 (LM-GP33) (Figure 6F). CD27low memory CD8+ T cells provide superior protection to Listeria, compared to CD27+ cells (Olson et al., 2013). miR-15/16Δ/Δ and miR-15/16fl/fl animals had equivalent numbers of this subset of memory T cells (Figure 4F) and exerted equivalent control of the LM-GP33 bacterial burden (Figure 6G). Thus, miR-15/16 are dispensable for the establishment of functionally protective CD8+ memory T cells, but is required for the generation of a normally proportioned memory cell pool. In other contexts, the imbalanced increase in CD27+ T cells in miR-15/16Δ/Δ animals may generate improved protection or hyperactive secondary immune responses (Hikono et al., 2007; Roberts et al., 2010).
A Network of Memory-Associated Genes Is Regulated by miR-15/16
In order to shed light on the mechanistic targets of miR-15/16 that contribute to memory CD8+ T cell differentiation, we coupled our RNA-seq and AHC data with existing data that defined a set of genes upregulated in memory T cells following LCMV infection (Kaech et al., 2002). Since long-lived memory CD4+ and CD8+ T cells share more than 95% similarity in gene expression (Chang et al., 2014), we reasoned that experiments conducted using CD4+ T cells could provide valuable insights into miR-15/16 regulation of CD8+ T cells. Twenty-two memory-associated genes contained a 3′UTR seed match and were differentially expressed in miR-15/16Δ/Δ T cells (p ≤ 0.1). Of these, 19 were upregulated in miR-15/16Δ/Δ cells (Figure 7A, left). AHC detected AGO2 binding at miR-15/16 seed-match sites in most of these genes, further supporting direct regulation by miR-15/16 (Figure 7A, right). Among the genes upregulated in miR-15/16Δ/Δ T cells and containing AGO2 binding at miR-15/16 seed-match sites, Adrb2 (Slota et al., 2015), Pim1 (Knudson et al., 2017), Il7r (Nanjappa et al., 2008), Cd28, and Bcl2 (Kurtulus et al., 2011) have been extensively studied in the context of CD8+ memory.
Figure 7. A Network of miR-15/16 Targets Is Upregulated in Memory Cells.
(A) Row z-score normalized expression of genes found to be upregulated in memory cells compared to day 8 p.i. effector cells, differentially expressed in miR-15/16Δ/Δ cells (p value ≤ 0.1), and containing at least one miR-15/16 seed match in their 3′UTR. Black bars indicate AHC reads at miR-15/16 seed matches.
(B) Flow cytometry of CD127 on naive CD4+ and CD8+ T cells (n ≥ 14 biological replicates from three independent experiments, two-tailed t test).
(C) Flow cytometry of CD127 on naive CD4+ and CD8+ T cells from mixed bone marrow chimeric mice (n ≥ 9 biological replicates from two independent experiments, two-tailed t test).
(D) RNA-seq (top) and AHC (bottom) for the 3′UTRs of Ccnd2, Il7r, Cd28, and Adrb2 with red-shaded regions indicating the locations of miR-15/16 seed matches.
(E) Relative luciferase activity between empty vector, Il7r 3′UTR, Cd28 3′UTR, Adrb2 3′UTR, and all three 3′UTRs with their respective miR-15/16 seed matches scrambled (n R 3 biological replicates from at least two independent experiments, two-tailed t test).
*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. See also Figure S7.
This approach identified numerous putative targets of miR-15/16 that may contribute to memory cell differentiation. Il7r was of particular interest, as it has been previously reported that the addition of exogenous recombinant IL-7 can boost memory formation (Nanjappa et al., 2008). Surface CD127 protein abundance was elevated in naive miR-15/16Δ/Δ CD8+ and CD4+ T cells (Figure 7B). This effect was cell intrinsic, as it was also observed in mixed bone marrow chimeric mice (Figure 7C). Similar to the well-characterized target of miR-15/16, Ccnd2, the Il7r 3′UTR contains multiple major AHC peaks corresponding to miR-15/16 seed matches (Figure 7D). Dual luciferase assays in miR-15/16Δ/Δ cells transfected with control (CM) or miR-16 mimic validated the capacity of miR-16 to directly target the Il7r 3′UTR (Figure 7E). Site-directed mutagenesis of both miR-15/16 seed-match sites completely rescued luciferase expression to CM-treated levels, proving that the effects of miR-15/16 on the Il7r 3′UTR are dependent upon these seed matches (Figure 7E). These results provide strong evidence that Il7r is a bona fide target of miR-15/16.
AHC peaks also sharply coincided with miR-15/16 3′UTR seed matches in many of these putative memory-associated targets (Figure S7), especially Cd28 and Adrb2 (Figure 7D). Dual luciferase assays confirmed these two additional targets; mutating the miR-15/16 seed matches within their 3′UTRs completely rescued luciferase expression to CM-treated levels (Figure 7E). While it is unlikely that any one of these targets could account for the effects of miR-15/16 on CD8+ memory cell differentiation, regulation of this network of memory-associated genes provides a plausible mechanism by which miR-15/16 may regulate memory differentiation.
DISCUSSION
The experiments in this study demonstrated roles for miR-15/16 in the restriction of T cell cycle, survival, and CD8+ memory cell differentiation through the direct targeting of networks of genes associated with cell cycle, signaling, metabolism, and survival. Both the miR-15a/16–1 and miR-15b/16–2 clusters were sufficient to restrict cell cycle, survival, and CD8+ memory cell differentiation during the response to LCMV infection. Moreover, all four mature miRNAs from these clusters are downregulated during the response, and both miR-15b and miR-16 remain low long after viral clearance. Given the importance of the quantity and quality of cells generated in response to an immunologic insult such as infection or cancer, our findings mark miR-15/16 as critical tuners of the cell-mediated adaptive immune response.
miRNAs exert their biological effects through multiple target genes (Pua et al., 2016), and our data indicate that miR-15/16 regulate a remarkably extensive target network. We combined biochemical and bioinformatic approaches to map AGO2 binding at thousands of miR-15/16 seed-match sequences in mRNAs and linked this with gene expression analysis in miR-15/16Δ/Δ T cells to identify a large network of high-confidence direct miR-15/16 target genes that restrict cell cycle and CD8+ long-lived memory T cell differentiation. This approach is best suited to highly expressed miRNAs like miR-15/16 since seed-match occupancy correlated robustly with miRNA AGO2 occupancy. Deletion of both the endogenous Mirc30 (miR-15a/16–1) and Mirc10 (miR-15b/16–2) clusters was also critical to unveiling this large set of target genes.
At the peak of infection with LCMV, antigen-specific CD8+ T cells divide as rapidly as once every 2 h (Yoon et al., 2010) and complete >14 divisions in 7 days p.i. (Blattman et al., 2002). Even with this remarkably rapid rate of division, removal of miR-15/16 increased the accumulation of virus-specific cells, emphasizing the degree of control exerted by miR-15/16. Previous studies in CLL as well as malignant pleural mesothelioma demonstrated that miR-15/16 are frequently downregulated or deleted and directly target several cell cycle genes (Klein et al., 2010; Reid et al., 2013). Our findings expand this cell cycle target network and extend the influence of miR-15/16 to the control of antigen-driven T cell clonal selection.
miR-150, which is also downregulated in response to T cell activation (Figures 1A and 1B), also restricts memory cell development (Ban et al., 2017; Chen et al., 2017). However, unlike miR-150, miR-15/16 expression is maintained at reduced levels during the memory phase (Figure 1B), suggesting that targets of miR-15/16 may be critical to the maintenance of memory cells. In fact, a number of miR-15/16 targets, including Bcl2 (Kurtulus et al., 2011), Pim1 (Knudson et al., 2017), Il7r (Kaech et al.,2003), and Cd28 (Klein Geltink et al., 2017), have been linked with the maintenance and functional capacity of the memory CD8+ T cell pool. Interestingly, Il7r/CD127 may represent not only a target of miR-15/16, but also part of a feed-forward negative regulator of miR-15/16 expression through STAT5, which is activated downstream of CD127 signaling, and a direct transcriptional repressor of miR-15b/16–2 (Li et al., 2010; Lindner et al., 2017). This feed-forward circuit may play important roles in other lymphocyte cell fate decisions. For example, miR-15 family activity was inversely correlated with IL-7 receptor surface expression in pre-B cells (Lindner et al., 2017).
We conclude that miR-15/16 represents an important node in the coordinate regulation of cell proliferation, survival, and early differentiation of memory CD8+ T cells. High precursor frequencies of long-lived memory cells reduce the proliferative burden required for a response to secondary challenge, thereby preserving mitochondrial integrity and allowing for a more productive secondary response (Sheridan et al., 2014). Therefore, reducing miR-15/16 expression might be beneficial in the context of vaccination and cancer immunotherapies where the capacity of memory CD8+ T cells to expand and respond is critical for protection. Future studies investigating the regulation of miR-15/16 in response to T cell stimulation, as well as the maintenance of its suppression in memory CD8+ T cells, could identify up-stream drivers of memory formation and maintenance and provide potential therapeutic targets to modulate T cell memory.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, K. Mark Ansel (mark.ansel@ucsf.edu). Mirc10tm1Mtm ES cells and mice generated in this study have been deposited to the Mutant Mouse Resource & Research Center (MMRRC:037643-JAX).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice generated from Mirc10tm1Mtm ES cells (Park et al., 2012)} were crossed with Rosa26-Flp mice (GT(ROSA)26Sortm1(FLP1)Dym; 009086, Jackson Laboratory) to delete the selection cassette, yielding a loxP-flanked allele. “miR-15/16fl/fl” animals were generated by crossing the resulting miR-15b/16–2fl/fl animals with miR-15a/16–1fl/fl (Mirc30tm1.1Rdf) animals (Klein et al., 2010). “miR-15/16Δ/Δ” animals were generated by further crossing miR-15/16fl/fl animals with CD4-Cre mice (Tg(CD4-cre)1Cwi; 4196, Taconic) (Sawada et al., 1994). For hematopoietic chimeras, B6-Ly5.1/Cr (CD45.1+; Charles River) mice were lethally irradiated (2×550 rad), reconstituted with 5×106 bone marrow cells, and analyzed 8–10 weeks later. Male and female age and sex matched mice were used between 5 to 12 weeks of age. All mice were housed and bred in specific pathogen-free conditions in the Animal Barrier Facility at the University of California, San Francisco. Animal experiments were approved by the Institutional Animal Care and Use Committee of the University of California, San Francisco.
LCMV model
Animals were infected intraperitoneally with 2×105 plaque forming units (p.f.u.) LCMV Armstrong. Mice were bled retroorbitally with heparinized 75mm hematocrit tubes (Drummond) or sacrificed for spleen harvesting. Splenocytes were re-stimulated in vitro with 0.2μg/ml GP33–41 (KAVYNFATM) or GP276–286 (SGVENPGGYCL) for 5 hours in the presence of brefeldin A. For EdU labeling, 1mg EdU was injected retroorbitally 16 hours before sacrifice and detected using the Click-iT EdU flow cytometry assay kit (Thermo Fisher Scientific).
Listeria monocytogenes model
Animals 30+ days post-infection (p.i.) with LCMV Armstrong and naive control mice (no prior infection with LCMV) were infected with 1–2×105 colony forming units (c.f.u.) of LM-GP33. Two days later, spleen and liver samples were harvested, and bacterial content was analyzed as described (Shehata et al., 2018). Spleens and livers were isolated, weighed, placed in 1% FBS in PBS, and homogenized. Single-cell suspensions were made from the spleens and livers of recipient mice in PBS containing 0.5% Triton X-100. Serial dilutions of the supernatants were inoculated on brain-heart-infusion agar plates and were incubated for 24h at 37°C. Bacterial colonies were enumerated, and c.f.u. were normalized per gram of spleen or liver plated. Data were normalized to the mean c.f.u of naive control mice.
METHOD DETAILS
in Vitro Cultures and qPCR
CD4+ T cells from the spleen and lymph nodes of mice were enriched by Dynabead-positive selection (Invitrogen, L3T4. Cells were stimulated with biotinylated anti-CD3 (clone 2C11, 1μg/ml) and anti-CD28 (clone 37.51, 0.5μg/ml) for 3 days on plates pre-coated with NeutrAvidin (5μg/ml, Thermo Fischer Scientific), then rested with 20 units/mL recombinant IL-2 (National Cancer Institute) for an additional 2 days. Cells were cultured in DMEM high glucose media supplemented with 10% FCS, pyruvate, nonessential amino acids, MEM vitamins, L-arginine, L-asparagine, L-glutamine, folic acid, beta mercaptoethanol, penicillin, and streptomycin. For miRNA qPCR analysis, cells were lysed in Trizol LS (Life Technologies), total RNA isolated, and RNA quantified on an ND1000 Spectrophotometer (NanoDrop). Reverse transcription of miRNA was performed with the NCode miRNA First-Strand cDNA Synthesis Kit (Life Technologies). Forward primers were the mature miRNA sequences (miRbase.org) and a universal reverse primer was used from the kit. Expression values were normalized to the 5.8S ribosomal RNA (F: ATCGTAGGCACCGCTACGCCTGTCTG). qPCR was performed in technical duplicates using FastStart Universal SYBR Green Master mix (Roche) on a Realplex2 instrument (Eppendorf).
Flow Cytometry
Cells were harvested from thymus, spleen, and lymph nodes by mashing through 70μM filters. Splenic and blood RBCs were lysed with ACK buffer for 5 min at room temperature. Single cell suspensions were prepared in PBS 2% FCS and stained for analysis on an LSRFortessa (Becton Dickenson). Tetramer reagents were generated mixing biotinylated peptide-MHC monomers with fluorophore-conjugated Streptavidin. ForGP33 (H-2Db GP33–41 KAVYNFATM), 200ug peptide-MHC monomer was incubated with 318uLStrep-tavidin-PE (Life Sciences). For GP276 (H-2Db GP276–286 SGVENPGGYCL), 200ug peptide-MHC monomer was incubated with 176uL Streptavidin-APC (Life Sciences). 1/10 total volume of Streptavidin was added every 15 minutes on ice in the dark and gently mixed. For EdU assays, cells were stained with anti-CD3 PE-dazzle 594, anti-CD4 PerCP-Cy5.5, anti-CD8 BV785. Cells were then fixed and permeabilized and labeled with EdU AF647 (Thermo-Fisher) according to the manufacturer’s instructions. For transcription factor stains, cells were fixed and permeabilized using the Foxp3 Transcription Factor Staining Buffer Set (eBioscience). For cytokine stains, cells were fixed for 8 min at room temperature with 4% paraformaldehyde, and permeabilized with 0.5% saponin buffer. Normalized MFI is geometric mean fluorescence intensity normalized to the average for the miR-15/16fl/fl samples in each independent experiment. Data were analyzed with FlowJo.
AGO2 HITS-CLIP
Libraries were constructed as previously described (Loeb et al., 2012) using CD4+ T cells, with the following modifications. To eliminate nuclei, SDS was excluded from the lysis buffer and physical lysis was not performed with a needle. SDS was included before adding the lysate to anti-AGO2 coated beads. AGO2 immunoprecipitation was performed using a monoclonal antibody (Wako; clone 2D4). To increase resolution and precision, we used a 10% TBE gel (BioRad) for extracting final PCR products. 10% PEG was added to 3′ and 5′ linker ligation steps to improve ligation efficiency. A randomized dinucleotide was added to the 3′ linker to reduce ligation bias. Maximum read depths across mature miRNAs and miRNA targets were generated using the samtools package (Li et al., 2009). Bedfiles used for mapping to transcriptomic regions were acquired from the UCSC table browser using the mm10 reference genome.
RNA sequencing
1×106 CD4+ T cells cultured as described above were lysed in Trizol (Life Technologies) and RNA isolated using miRNeasy Micro Kit (QIAGEN) with on-column DNase digestion. cDNA was synthesized using the TruSeq PolyA library kit. Single-end 50 base-pair RNA sequencing was performed on the Illumina HiSeq 4000. Alignment was performed using STAR_2.4.2a (Dobin et al., 2013) against the Ensembl Human GRCm38.78 alignment genome. Differential expression was tested using DESeq2 v1.14.0 (Love et al., 2014). Analysis was performed on datasets filtered on genes with an average of at least 5 reads-per-million across all samples. Expression values were analyzed as log2(fold change) of miR-15/16 deficient T cells compared with miR-15/16 sufficient T cells. Raw data and processed datasets have been deposited in GEO.
The cumulative distribution function plot was generated in R (www.r-project.org) to display the log2(miR-15/16Δ/Δ/miR-15/16fl/fl) against the cumulative distribution of genes. The expression values from all genes lacking a miR-15/16 3′UTR seed match were plotted along with all genes predicted to be targeted by miR-15/16 by TargetScan (www.targetscan.org), all genes containing a miR-15/16 3′UTR seed match, and the subset of all genes containing a miR-15/16 3′UTR seed match with above 5 AHC reads mapping to the seed match. Gene Set Enrichment Analysis (http://software.broadinstitute.org/gsea/index.jsp) was performed using the set of genes with an average of at least 5 reads-per-million across all samples. Plotted were the enrichment curve and rank order location of each gene contained within the KEGG Cell Cycle gene set from most upregulated to most downregulated (left to right).
Luciferase Assays
CD4+ T cells were transfected on day 1 of culture with luciferase reporter constructs and/or 500nM miRIDIAN miRNA mimics (Dharmacon) using the Neon Transfection System (Thermo Fisher Scientific). Luciferase activity was measured 24 h after transfection with the Dual Luciferase Reporter Assay System (Promega) and a FLUOstar Optima plate-reader (BMG Labtech). The near full-length 3′UTRs of Il7r, Cd28, and Adrb2 were cloned into the psiCHECK-2 luciferase reporter construct (Promega). Primers for cloning and site-directed-mutagenesis (SDM) were: Il7r F: CAGGCTCGAGATTATAAGAAAACCCTTCCATCGACAACC, Il7r R: GATGCGGCCGCTTCCAGAAA ATAGCGCATGCTTGTATTTG; Cd28 F: TAGCTCGAGCAGGGACCCCTATCCAGAAG, Cd28 R: CTAGCGGCCGCAGTCAATGAAT AAATATTTATTGCAGGCTAAGC; Adrb2 F: CAGCTCGAGAGGCTTTCTACTCTCTAAGACCC, Adrb2 R: CAGGCGGCCGCCCAC TCATCGGTCACGACAC. SDM was performed using QuickChange Lightning Site-Directed Mutagenesis Kit (Agilent) using primers: Il7r SDM F: GATAACCACAACAGTCCTGAATGCTTGATTATATTCTCAGG, Il7r SDM R: CCTGAGAATATAATCAAGCATTCAGGA CTGTTGTGGTTATC; Cd28 SDM F: GAAAACTATGTCACTTGTCCTGATTATTGTAAGAGTCTAAGAAC, Cd28 SDM R: GAAAACTATG TCACTTGTCCTGATTATTGTAAGAGTCTAAGAAC; Adrb2 SDM F: GAATGATATATATTGTCCTGGGAAATCCATATCTAAAGGAG AGAG, Adrb2 SDM R: CTCTCTCCTTTAGATATGGATTTCCCAGGACAATATATATCATTC.
QUANTIFICATION AND STATISTICAL ANALYSIS
Excel (Windows) and R were used for data analysis. Some plots were generated using the Bioconductor package, plotGrouper (Gagnon, 2018). For all figures, bar graphs display mean ± s.d. and each point represents an individual mouse unless otherwise stated. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 for significance. Parametric statistics were employed except in cases of non-normally distributed data. In those cases, differences between groups were assessed using a Mann-Whitney U test.
DATA AND CODE AVAILABILITY
The RNA-seq data reported in this paper are archived at the NCBI Gene Expression Omnibus database (accession number GEO: GSE111568 and GEO: GSE130655).
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE Antibodies | SOURCE | IDENTIFIER |
|---|---|---|
| Anti-CD3 PE-dazzle 594 (145-2C11) | BioLegend | Cat# 100348; RRID:AB_2564029 |
| Anti-CD8a BV785 (53-6.7) | BioLegend | Cat# 100749; RRID:AB_11218801 |
| Anti-CD8a BUV805 (53-6.7) | BD Biosciences | Cat# 564920; RRID:AB_2716856 |
| Anti-CD4 PerCP-Cy5.5 (RM4-5) | Thermo Fisher Scientific | Cat# 45-0042-80; RRID:AB_906231 |
| Anti-Foxp3 APC (FJK-16 s) | Thermo Fisher Scientific | Cat# 17-5773-82; RRID:AB_469457 |
| Anti-CXCR3 FITC (CXCR3-173) | Thermo Fisher Scientific | Cat# 11-1831-82; RRID:AB_11040010 |
| Anti-CD127 BV421 (A7R34) | BioLegend | Cat# 135024; RRID:AB_11218800 |
| Anti-CD62L BV605 (MEL-14) | BioLegend | Cat# 104438; RRID:AB_2563058 |
| Anti-CD45.1 BV785 (A20) | BioLegend | Cat# 110743; RRID:AB_2563379 |
| Anti-CD45.2 BUV395 (104) | BD Biosciences | Cat# 564616; RRID:AB_2738867 |
| Anti-CD43 PerCP-Cy5.5 (1B11) | BioLegend | Cat# 121223; RRID:AB_893336 |
| Anti-KLRG1 PE-Cy7 (2F1/KLRG1) | BioLegend | Cat# 138416; RRID:AB_2561736) |
| Anti-CD44 Af700 (IM7) | Thermo Fisher Scientific | Cat# 56-0441-82; RRID:AB_494011 |
| Anti-CD27 FITC (LG.7F9) | Thermo Fisher Scientific | Cat# 11-0271-82; RRID:AB_465001 |
| Anti-GR1 APC-Cy7 (RB6-8C5) | BioLegend | Cat# 108423; RRID:AB_2137486 |
| Anti-CD11b APC-Cy7 (M1/70) | BD Biosciences | Cat# 557657; RRID:AB_396772 |
| Anti-active caspase 3 (C92-605) | BD Biosciences | Cat# 559565; RRID:AB_397274 |
| Anti-cd19 APC-H7 (1D3) | BD Biosciences | Cat# 560245; RRID:AB_1645233 |
| Anti-TNF AF700 (MP6-XT22) | BD Biosciences | Cat# 558000; RRID:AB_396980 |
| Anti-IFNγ FITC (XMG1.2) | BioLegend | Cat# 505806; RRID:AB_315400 |
| Anti-IL2 APC (JES6-5H4) | Thermo Fisher Scientific | Cat# 17-7021-81; RRID:AB_469489 |
| Anti-CD16/32 (2.4G2) | Bio X Cell | Cat# BE0307; RRID:AB_2736987 |
| Anti-Rabbit A647 (Donkey polyclonal) | BioLegend | Cat# 406414; RRID:AB_2563202 |
| Anti-CD3 (2C11) | Bio X Cell | Cat# BP0001-1; RRID:AB_1107634 |
| Anti-CD28 (37.51) | Bio X Cell | Cat# BE0015-1; RRID:AB_1107624 |
| Anti-AGO2 (2D4) | Wako | Cat# 018-22021; RRID:AB_1106838 |
| Bacterial and Virus Strains | ||
| LCMV (Armstrong) | in house, M. Matloubian lab | NA |
| Listeria monocytogenes | in house, S. Sanjabi lab | NA |
| Chemicals, Peptides, and Recombinant Proteins | ||
| GP33 | NIH | NA |
| GP276 | NIH | NA |
| Streptavidin PE | Life Sciences | Cat# S-866 |
| Streptavidin APC | Life Sciences | Cat# S-868 |
| Recombinant IL-2 | NCI | NA |
| Critical Commercial Assays | ||
| Click-iT EdU Alexa Fluor 647 Flow Cytometry Assay Kit | Thermo-Fisher | Cat# C10419 |
| Foxp3/Transcription Factor Staining Buffer Set | eBioscience | Cat# 00-5523-00 |
| Deposited Data | ||
| RNA sequencing | This study | GEO: GSE111568 |
| Ago2 HITS-CLIP | This study | GEO: GSE130655 |
| Experimental Models: Organisms/Strains | ||
| Cd4-cre Transgenic (B6.Cg-Tg(CD4-cre)1Cwi | Taconic | Cd4-cre Transgenic (Model #: 4196) |
| miR15a/16-1 fl Mirc30tm11Rdf | The Jackson Laboratory | JAX: 017641 |
| miR15b/16-2 fl Mirc10tm1Mtm | This study; now available via The Jackson Laboratory | MMRRC Stock No: 37643-JAX |
| NCI B6-Ly5.1/Cr (B6.SJL-PtprcaPepcb/BoyCrCrl) | Charles River | NCI B6-Ly5.1/Cr (Strain code: 564) |
| Oligonucleotides | ||
| miR15a-5p miRIDIAN miRNA mimic | Dharmacon | C-310510-05-0002 |
| miR16 miRIDIAN miRNA mimic | Dharmacon | C-310511-05-0002 |
| miRIDIAN control mimic | Dharmacon | CN-001000-01-05 |
| miR15b-5p miRIDIAN miRNA mimic | Dharmacon | C-310378-05-0002 |
| Recombinant DNA | ||
| Plasmid: psiCheck-2 | Promega | C8021 |
| Plasmid: psiCHECK-2-IL7r | In this study | NA |
| Plasmid: psiCHECK-2-Cd28 | In this study | NA |
| Plasmid: siCHECK-2-Adrb2 | In this study | NA |
| Software and Algorithms | ||
| Flow jo V9 | FlowJo, LLC | NA |
| Plot Grouper | Bioconductor | https://doi.org/10.18129/B9.bioc.plotGrouper |
| microRNA.TEvMP | This study, available at GitHub | https://github.com/jdgagnon/microRNA.TEvMP |
| DE.AHC | This study, available at GitHub | https://github.com/jdgagnon/DE.AHC |
Highlights.
miR-15/16 and their target mRNAs are abundant in Ago2 complexes in T cells
miR-15/16 regulate the number and type of T cells generated during viral infection
miR-15/16 target acts in pathways critical for T cell expansion and differentiation
ACKNOWLEDGMENTS
We thank Andrea Barczak, Matthew Aber, Walter Eckalbar, and David Erle of the UCSF SABRE Functional Genomics Core Facility for help with RNA sequencing; Chong Park and the Gladstone Transgenic Gene Targeting Core for help with generating mice; Priti Singh for help with establishing and maintaining mutant mouse colonies; and the Single-Cell Analysis Core (SCAC) for use of their instruments.
The SCAC was supported by the NIH (NIH P30 DK063720 and NIH S10 1S10OD021822–01). This work was supported by the NIH (HL107202 and HL109102), the Sandler Asthma Basic Research Center, a Scholar Award from The Leukemia & Lymphoma Society (K.M.A.), the W.M. Keck Foundation (M.T.M.), and the UCSF Diabetes Center (NIH P30 DK063720). A.J.L. was supported by a Cancer Research Institute Irvington Fellowship and the UCSF Immunology Program (T32AI007334). L.T.J. was supported by the Swiss Foundation for Grants in Biology and Medicine (SFGBM)/SNSF (PASMP3–124274/1). A.M. holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund, is an investigator at the Chan Zuckerberg Biohub, has received funding from the Innovative Genomics Institute (IGI), and is a member of the Parker Institute for Cancer Immunotherapy (PICI). This material is based upon work supported by the National Science Foundation Graduate Research Fellowship Program under grant 2015202541 (J.D.G.). Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.
Footnotes
DECLARATION OF INTERESTS
A.M. is a co-founder of Arsenal Biosciences and Spotlight Therapeutics. A.M. serves on the scientific advisory board of PACT Pharma, is an advisor to Trizell, and was a former advisor to Juno Therapeutics. The Marson Laboratory has received sponsored research support from Juno Therapeutics, Epinomics, Sanofi, and a gift from Gilead. J.D.G. and A.J.L. are now employees of PACT Pharma. S.S. is now an employee of Genentech and holds stock in Roche, Genentech’s parent company. K.M.A. holds stock in Roche. P.O. is now an employee of Vir Biotechnology. H.M.S. is now an employee of Gilead Sciences and holds stock in Gilead Sciences.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.07.064.
REFERENCES
- Agarwal V, Bell GW, Nam J-W, and Bartel DP (2015). Predicting effective microRNA target sites in mammalian mRNAs. eLife 4, 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baars PA, Sierro S, Arens R, Tesselaar K, Hooibrink B, Klenerman P, and van Lier RAW (2005). Properties of murine (CD8+)CD27- T cells. Eur. J. Immunol 35, 3131–3141. [DOI] [PubMed] [Google Scholar]
- Ban YH, Oh S-C, Seo S-H, Kim S-M, Choi I-P, Greenberg PD, Chang J, Kim T-D, and Ha S-J (2017). miR-150-Mediated Foxo1 Regulation Programs CD8+ T Cell Differentiation. Cell Rep. 20, 2598–2611. [DOI] [PubMed] [Google Scholar]
- Baumjohann D, and Ansel KM (2013). MicroRNA-mediated regulation of T helper cell differentiation and plasticity. Nat. Rev. Immunol 13, 666–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattman JN, Antia R, Sourdive DJD, Wang X, Kaech SM, Murali-Krishna K, Altman JD, and Ahmed R (2002). Estimating the precursor frequency of naive antigen-specific CD8 T cells. J. Exp. Med 195, 657–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronevetsky Y, Villarino AV, Eisley CJ, Barbeau R, Barczak AJ, Heinz GA, Kremmer E, Heissmeyer V, McManus MT, Erle DJ, et al. (2013). T cell activation induces proteasomal degradation of Argonaute and rapid remodeling of the microRNA repertoire. J. Exp. Med 210, 417–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al. (2002). Frequent deletions and down-regulation of micro- RNAgenes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 99, 15524–15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JT, Wherry EJ, and Goldrath AW (2014). Molecular regulation of effector and memory T cell differentiation. Nat. Immunol 15, 1104–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheadle C, Fan J, Cho-Chung YS, Werner T, Ray J, Do L, Gorospe M, and Becker KG (2005). Control of gene expression during T cell activation: alternate regulation of mRNA transcription and mRNA stability. BMC Genomics 6, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Stelekati E, Kurachi M, Yu S, Cai Z, Manne S, Khan O, Yang X, and Wherry EJ (2017). miR-150 Regulates Memory CD8 T Cell Differentiation via c-Myb. Cell Rep. 20, 2584–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, and Darnell RB (2009). Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 460, 479–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon JD (2018). plotGrouper: Shiny app GUI wrapper for ggplot with built-in statistical analysis (, San Francisco: University of California; ). https://jdgagnon.github.io/plotGrouper. [Google Scholar]
- Guan T, Dominguez CX, Amezquita RA, Laidlaw BJ, Cheng J, Henao-Mejia J, Williams A, Flavell RA, Lu J, and Kaech SM (2018). ZEB1, ZEB2, and the miR-200 family form a counterregulatory network to regulate CD8+ T cell fates. J. Exp. Med 215, 1153–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikono H, Kohlmeier JE, Takamura S, Wittmer ST, Roberts AD, and Woodland DL (2007). Activation phenotype, rather than central- or effector-memory phenotype, predicts the recall efficacy of memory CD8+ T cells. J. Exp. Med 204, 1625–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JK, Kagari T, Clingan JM, and Matloubian M (2011). Expression of chemokine receptor CXCR3 on T cells affects the balance between effector and memory CD8 T-cell generation. Proc. Natl. Acad. Sci. USA 108, E118–E127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech SM, Hemby S, Kersh E, and Ahmed R (2002). Molecular and functional profiling of memory CD8 T cell differentiation. Cell 111, 837–851. [DOI] [PubMed] [Google Scholar]
- Kaech SM, Tan JT, Wherry EJ, Konieczny BT, Surh CD, and Ahmed R (2003). Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat. Immunol 4, 1191–1198. [DOI] [PubMed] [Google Scholar]
- Kakaradov B, Arsenio J, Widjaja CE, He Z, Aigner S, Metz PJ, Yu B, Wehrens EJ, Lopez J, Kim SH, et al. (2017). Early transcriptional and epigenetic regulation of CD8+ T cell differentiation revealed by single-cell RNA sequencing. Nat. Immunol 18, 422–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AA, Penny LA, Yuzefpolskiy Y, Sarkar S, and Kalia V (2013). MicroRNA-17~92 regulates effector and memory CD8 T-cell fates by modulating proliferation in response to infections. Blood 121, 4473–4483. [DOI] [PubMed] [Google Scholar]
- Klein U, Lia M, Crespo M, Siegel R, Shen Q, Mo T, Ambesi-Impiombato A, Califano A, Migliazza A, Bhagat G, and Dalla-Favera R (2010). The DLEU2/miR-15a/16–1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell 17, 28–40. [DOI] [PubMed] [Google Scholar]
- Klein Geltink RI, O’Sullivan D, Corrado M, Bremser A, Buck MD, Buescher JM, Firat E, Zhu X, Niedermann G, Caputa G, et al. (2017). Mitochondrial Priming by CD28. Cell 171, 385–397.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudson KM, Pritzl CJ, Saxena V, Altman A, Daniels MA, and Teixeiro E (2017). NFκB-Pim-1-Eomesodermin axis is critical for maintaining CD8 T-cell memory quality. Proc. Natl. Acad. Sci. USA 114, E1659–E1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtulus S, Tripathi P, Moreno-Fernandez ME, Sholl A, Katz JD, Grimes HL, and Hildeman DA (2011). Bcl-2 allows effector and memory CD8+ T cells to tolerate higher expression of Bim. J. Immunol 186, 5729–5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, and Durbin R; 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Miskimen KL, Wang Z, Xie XY, Brenzovich J, Ryan JJ, Tse W, Moriggl R, and Bunting KD (2010). STAT5 requires the N-domain for suppression of miR15/16, induction of bcl-2, and survival signaling in myeloproliferative disease. Blood 115, 1416–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner SE, Lohmüller M, Kotkamp B, Schuler F, Knust Z, Villunger A, and Herzog S (2017). The miR-15 family reinforces the transition from proliferation to differentiation in pre-B cells. EMBO Rep. 18, 1604–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Fu H, Sun F, Zhang H, Tie Y, Zhu J, Xing R, Sun Z, and Zheng X (2008). miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res. 36, 5391–5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb GB, Khan AA, Canner D, Hiatt JB, Shendure J, Darnell RB, Leslie CS, and Rudensky AY (2012). Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Mol. Cell 48, 760–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcais A, Blevins R, Graumann J, Feytout A, Dharmalingam G, Carroll T, Amado IF, Bruno L, Lee K, Walzer T, et al. (2014). microRNA-mediated regulation of mTOR complex components facilitates discrimination between activation and anergy in CD4 T cells. J. Exp. Med 211, 2281–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffett HF, Cartwright ANR, Kim H-J, Godec J, Pyrdol J, Äijö T, Martinez GJ, Rao A, Lu J, Golub TR, et al. (2017). The microRNA miR-31 in-hibitsCD8+ T cell function in chronic viral infection. Nat. Immunol 18, 791–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanjappa SG, Walent JH, Morre M, and Suresh M (2008). Effects of IL-7 on memory CD8 T cell homeostasis are influenced by the timing of therapy in mice. J. Clin. Invest 118, 1027–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson JA, McDonald-Hyman C, Jameson SC, and Hamilton SE (2013). Effector-like CD8+ T cells in the memory population mediate potent protective immunity. Immunity 38, 1250–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CY, Jeker LT, Carver-Moore K, Oh A, Liu HJ, Cameron R, Richards H, Li Z, Adler D, Yoshinaga Y, et al. (2012). A resource for the conditional ablation of microRNAs in the mouse. Cell Rep. 1, 385–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pua HH, Steiner DF, Patel S, Gonzalez JR, Ortiz-Carpena JF, Kageyama R, Chiou N-T, Gallman A, de Kouchkovsky D, Jeker LT, et al. (2016). MicroRNAs 24 and 27 Suppress Allergic Inflammation and Target a Network of Regulators of T Helper 2 Cell-Associated Cytokine Production. Immunity 44, 821–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid G, Pel ME, Kirschner MB, Cheng YY, Mugridge N, Weiss J, Williams M, Wright C, Edelman JJ, Vallely MP, et al. (2013). Restoring expression of miR-16: a novel approach to therapy for malignant pleural mesothelioma. Ann. Oncol 24, 3128–3135. [DOI] [PubMed] [Google Scholar]
- Roberts DJ, Franklin NA, Kingeter LM, Yagita H, Tutt AL, Glennie MJ, and Bullock TNJ (2010). Control of established melanoma by CD27 stimulation is associated with enhanced effector function and persistence, and reduced PD-1 expression of tumor infiltrating CD8(+) T cells. J. Immunother 33, 769–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Kalia V, Haining WN, Konieczny BT, Subramaniam S, and Ahmed R (2008). Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J. Exp. Med 205, 625–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada S, Scarborough JD, Killeen N, and Littman DR (1994). A line-age-specific transcriptional silencer regulates CD4 gene expression during T lymphocyte development. Cell 77, 917–929. [DOI] [PubMed] [Google Scholar]
- Shehata HM, Khan S, Chen E, Fields PE, Flavell RA, and Sanjabi S (2018). Lack of Sprouty 1 and 2 enhances survival of effector CD8+ T cells and yields more protective memory cells. Proc. Natl. Acad. Sci. USA 115, E8939–E8947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan BS, Pham Q-M, Lee Y-T, Cauley LS, Puddington L, and Lefrançois L (2014). Oral infection drives a distinct population of intestinal resident memory CD8(+) T cells with enhanced protective function. Immunity 40, 747–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh Y, Garden OA, Lang F, and Cobb BS (2015). MicroRNA-15b/16 Enhances the Induction of Regulatory T Cells by Regulating the Expression of Rictor and mTOR. J. Immunol 195, 5667–5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slota C, Shi A, Chen G, Bevans M, and Weng N-P (2015). Norepinephrine preferentially modulates memory CD8 T cell function inducing inflammatory cytokine production and reducing proliferation in response to activation. Brain Behav. Immun 46, 168–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner DF, Thomas MF, Hu JK, Yang Z, Babiarz JE, Allen CDC, Matloubian M, Blelloch R, and Ansel KM (2011). MicroRNA-29 regulates T-box transcription factors and interferon-γ production in helper T cells. Immunity 35, 169–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells AC, Daniels KA, Angelou CC, Fagerberg E, Burnside AS, Markstein M, Alfandari D, Welsh RM, Pobezinskaya EL, and Pobezinsky LA (2017). Modulation of let-7 miRNAs controls the differentiation of effector CD8 T cells. eLife 6, 403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry EJ, Teichgräber V, Becker TC, Masopust D, Kaech SM, Antia R, von Andrian UH, and Ahmed R (2003). Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat. Immunol 4, 225–234. [DOI] [PubMed] [Google Scholar]
- Wu Y-H, Liu W, Xue B, Zhang L, Liu X-Y, Liu B, Wang Y, Cai Y, and Duan R (2016). Upregulated Expression of microRNA-16 Correlates with Th17/Treg Cell Imbalance in Patients with Rheumatoid Arthritis. DNA Cell Biol. 35, 853–860. [DOI] [PubMed] [Google Scholar]
- Xi Y, Li J, Zan L, Wang J, Wang G, and Ning Y (2013). Micro-RNA-16 expression in paraffin-embedded specimen correlates with overall survival of T-lymphoblastic lymphoma/leukemia. Hum. Pathol 44, 1011–1016. [DOI] [PubMed] [Google Scholar]
- Yang J, Liu R, Deng Y, Qian J, Lu Z, Wang Y, Zhang D, Luo F, and Chu Y (2017). MiR-15a/16 deficiency enhances anti-tumor immunity of glioma-infiltrating CD8+ T cells through targeting mTOR. Int. J. Cancer 141, 2082–2092. [DOI] [PubMed] [Google Scholar]
- Yoon H, Kim TS, and Braciale TJ (2010). The cell cycle time of CD8+ T cells responding in vivo is controlled by the type of antigenic stimulus. PLoS ONE 5,e15423. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data reported in this paper are archived at the NCBI Gene Expression Omnibus database (accession number GEO: GSE111568 and GEO: GSE130655).