Figure 2.
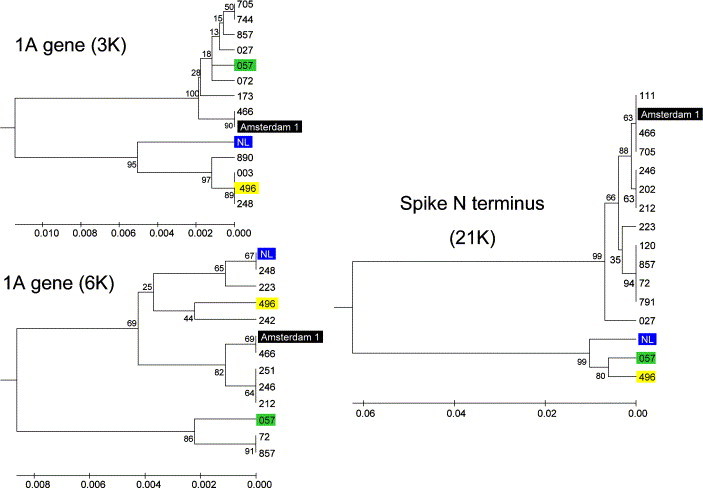
Discordance in phylogenetic clustering of different isolates of HCoV-NL63 at regions 3K, 6K and 21K. Phylogenetic trees were constructed as described in Materials and Methods using an UPGMA algorithm. The scale bar unit represents a 0.002 substitution per site. The trees were rooted with the sequences of it's closest relative: for 3K and 6K the HCoV-229E and for the 21K region the PEDV. Four completely sequenced isolates are marked with colored boxes, illustrating the discordance in clustering in different regions of the genome.
