Abstract
Molecular network data are increasingly becoming available, necessitating the development of well performing computational tools for their analyses. Such tools enabled conceptually different approaches for exploring human diseases to be undertaken, in particular, those that study the relationship between a multitude of biomolecules within a cell. Hence, a new field of network biology has emerged as part of systems biology, aiming to untangle the complexity of cellular network organization. We survey current network analysis methods that aim to give insight into human disease.
Keywords: biological networks, network topology, human diseasome
INTRODUCTION
Molecular causes of diseases are explored through many different techniques, such as through the examination of their causal genes, the disruption of related pathways, analysis of age factors and various other external influences. Currently, not much is known about the interconnectivity of all these different causes and elucidating the relationship between the malfunctioning of a system and its genomic data would provide insights into disease and set directions for future research. A scientific area that is attempting to address this unification is that of biological networks. It brings together the concepts of the ‘human diseasome’—a combined set of all known disorders and their implicated genetic mutations—and all systems-level molecular data.
Networks, also called ‘graphs’, are defined as sets of nodes (also called vertices) and edges (also called links), where nodes are singular entities and edges represent relations between them. This seemingly simple mathematical concept is a powerful approach for modelling real-world phenomena across various disciplines, including biological data, such as physical interactions between proteins, or synthesis of metabolic compounds. Also, graphs have been used to model relationships between diseases and they are a key part of the rising field of network medicine, which aims to decipher the complex wirings that govern human diseases [1–3]. An important part of this complex cellular wiring is the network of protein–protein interactions (PPIs) [4]. Since proteins interact, a single gene mutation is not confined within the actions of its gene products, but can propagate throughout the system, influencing other gene products that otherwise contain no aberrations. Hence, the final phenotypic effect is a result of a combination of the initial defect along with the influence that it has on other parts of the networked system.
This review is organized into four segments. The first section describes biological data and models used in their network representations. It illustrates the process of reconstructing the human metabolome and the human diseaseome. The second part presents basic methods of linking biological concepts within these networks by virtue of their common features. The third section focuses on more complex graph-theoretical approaches to data analysis and describes key biological insights that such analyses provide. In addition, the section covers scientific controversies that arose in this field and how they led the development of more sophisticated methods and tools. The final section gives an overview of future directions that yield as results of most recent developments in this area.
Data
Advances in biotechnology have led to previously unseen rates of growth in acquisition of biological data, as well as to the increase in understanding how that data can be used to benefit human life [5]. For example, genome-wide association studies are enabling assaying of more than a million of single-nucleotide polymorphisms in thousands of individuals [6,7]. It is to be expected that understanding the functioning of disease-associated genetic variants and elucidating the underlying architecture of diseases will bridge the gap between scientific research and its ultimate application in clinical practice [8]. A decade-long effort to map human disease loci, followed by the positional cloning and genome-wide association studies has produced an impressive database of disease–gene associations. The Online Mendelian Inheritance in Man (OMIM) [9] database contains over 4500 phenotypes for which the molecular basis is known and describes almost 3000 genes with phenotype-causing mutations [10]. We are currently witnessing the shift from a ‘single gene single disease’ paradigm towards the ‘interplay of different disease modules’ [2,3] and ultimately to the notion of a ‘personalized genome/diseasome’ [11–13], but that shift is yet to gain momentum.
Since it is the network of interacting biomolecules, such as proteins, that makes cells work, efforts for gathering these network data are currently under way. The largest available molecular network for a human is that of PPIs. Network data on physical PPIs for many model organisms [14–20], humans [21,22], bacteria [23–25] and viruses [26–28] are obtained using high-throughput screens, such as yeast two-hybrid (Y2H) assays [14–17,21,22,29] and affinity purification with mass spectrometry (AP/MS) [18,19,30,31]. Since techniques for detecting physical interactions between proteins do not work well for membrane proteins, the new technology of membrane Y2H assays is becoming available [32–35]. Discovering membrane-interacting proteins is a key to understanding disease, since integral membrane-interacting proteins have a role in cell signalling and hence, their alterations can produce disorders rooted in disruption of signalling pathways. Membrane proteins account for one-third of the proteome. The difficulty in studying them lies in their hydrophobic nature, which makes conventional biochemical and genetic assays unusable. The above mentioned new technology allows for large-scale screening of membrane proteins’ interactors in a range of organisms by utilizing the split-ubiquitin principle which overcomes this limitation. The effectiveness of this methodology was demonstrated by using the mammalian ErbB3 receptor as a bait to identify previously unknown ErbB3 interactors [32].
Data for physical molecular interaction are publicly available in databases including Human Protein Reference Database (HPRD) [36], the Biological General Repository for Interaction Datasets (BioGRID) [37], IntAct [38], Molecular INTeraction database (MINT) [39], Biomolecular Interaction Network Database (BIND) [40] and the Database of Interacting Proteins (DIP) [41]. Databases such as Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) [42] and iRefIndex [43] aggregate some or all of the above mentioned sources into single datasets.
The entire set of these interactions in humans is termed the human interactome. The complexity of such a network is overwhelming, as humans have approximately 25 000 protein-coding genes and an unknown number of proteins due to many splicing variants [44] and post-translational modifications. Hence, the number of proteins that take part in the interactome is argued to be in the six-digit range [45]. The current state of art datasets are approximated to around 50 000 unique proteins that participate in close to 200 000 interactions [43]. When the quality score of measurements is taken into account, the human PPI dataset is pruned down to around 10 000 proteins participating in some 50 000–60 000 high-confidence interactions. This shows that currently available interactome data are still noisy and incomplete. Numerous biases are introduced by data collecting and data sampling techniques, as well as averaging-out the species population by using universal models of the genome and interactome [46–55]. Nevertheless, even such sparse data are often too large to be efficiently analysed by present day network analysis algorithms. This is due to their large sizes and the fact that many graph theoretic algorithms are computationally intractable (NP-hard or NP-complete) [56]. Hence, new approximate (also called heuristic) methods for analysing network data that can cope with the underlying complexities are being developed [57–60].
Network representations
Constructing the graph model that accurately represents the observed underlying biological process is often not straight-forward. It depends on the way in which the data will be analysed and thus must be tailored for the specific question that is to be answered. For instance, metabolism is a process which keeps the organism in homeostasis. The current representation of metabolism in literature is through metabolic pathways which describe smaller parts of the metabolic system that act upon molecules through a series of reactions. Thus, this description of metabolism can be hierarchically broken down into pathways and further into specific chemical reactions that comprise these pathways. The reactions themselves constitute processes that transform sets of chemical substances. However, modelling this system using a network can be done in a number of different ways, with substantially different outputs [61–74]. Consequently, this dimensionality reduction from the complex intertwined metabolic system down to a graph requires logically splitting up the entire metabolism into two categories: its basic elements and the relations between them, that is, it requires determining which parts of it will be described by nodes and which by edges.
One way of making this simplification is to represent elementary metabolic compounds as nodes and reactions between them as edges of a network. This is known as the ‘metabolite-centric’ mapping [66–69], and is the most common one in use for general-purpose extraction of knowledge about metabolism from these networks, as it closely mirrors the real-world structure of metabolic reactions. The second way is ‘enzyme-centric’ [70,71]: nodes represent enzymes, and a pair of nodes is connected by an edge if the corresponding enzymes catalyse at least one common reaction. This representation is used for understanding molecular wirings around enzymes. The third model, a reaction-centric map [72–74], is obtained by having nodes represent individual reactions and placing links between them if they are commonly catalysed by at least one enzyme, or if they act upon the same chemical compounds. However, even when we choose the nodes in the network representation of metabolism, it is still not clear how the edges between them should be drawn (details given in Figure 1).
Figure 1:
If we consider a metabolite-centric map of the following irreversible metabolic reaction from substrates A and B to products C and D, A + B→C + D, we can chose (a) to link only the main substrate–product pair (say, A and C) while leaving out the transitive elements, such as energy or water (say, B and D). However, it is not always the case that a reaction has transitive elements. If there are no transitive elements in this reaction, (b) the metabolite-centric network map would usually link A to both C and D, as well as B to both C and D, even though this might not be completely biologically accurate, since for the production of C (and D) both A and B are needed together, and this subtlety is lost in this type of network representation. However, the issue could be solved by using more involved mathematical concepts, such as hypergraphs instead of graphs, as edges in hypergraphs consist of any sub-sets of nodes and not just node pairs. By using hypergraphs (c), {A, B, C} and {A, B, D} would be hyperedges, which would better describe the real-world product–substrate relationships. However, algorithms for analysing hypergraphs are far more mathematically and conceptually involved than those for graphs.
Similarly, there are analysis-dependent options in representing the seemingly simple ‘diseaseome bipartite graph’, which is a bipartite graph linking diseases to genes known to be causing them [1]; a ‘bipartite graph’ is a graph whose nodes can be divided into two separate sets, U and V, such that every edge in the graph connects a node from U to one node in V. From this bipartite graph, two projections are made as follows: that of diseases, in which nodes are disorders and a pair of disorders is linked if they share at least one gene whose mutation is known to be involved in both disorders, and that of genes, where nodes are genes and a pair of genes is linked if they are both involved in at least one same disorder. When the former projection network is clustered, major disease classes are discovered, such as the cancer cluster, which is densely connected due to the fact that many genes are associated with multiple types of cancers [1]. Similarly, neurological disorders cluster together, but metabolic disorders do not, and are dispersed throughout the network [1]. Subsequent studies have shown that metabolic disorders are better modelled using adjacency via metabolic pathways, rather than via sharing of disease-related genes [75] (see below).
GUILT BY ASSOCIATION
In this section we outline some commonly used approaches that are based on shared features for extracting disease information out of networks.
Shared genes
Linking several diseases with the same gene points to the possibility of their common genetic origin. Goh et al. [1] used data from OMIM to construct such a network. Their human disease network contained 1284 diseases, out of which 867 were linked to one or more other diseases. It is expected that linked diseases would exhibit congruent phenotypes. Indeed, Park et al. [76] showed co-morbidity between linked pairs of diseases: they found that patients with a primary disease are twice as likely to develop a secondary (co-morbid) disease if the secondary disease shares genes with the primary one. On the other hand, many linked disease pairs in the network representation did not exhibit these co-morbidity effects, which was attributed to different contextual scenarios of their genetic mutations.
Recently, a number of substantially different findings regarding predicting biological function based on shared gene features were presented in works of Gillis and Pavlidis [77–79]. In these studies they showed that multi-functionality of a gene, rather than its association, is a primary cause of high efficiency in gene function prediction [79]. Also, they found that it was possible for a small number of edges to accounts for all prediction performance in the biological networks and even that high quality predictions on gene function can be made regardless whether information on which gene interacts with which is available or not [77].
Shared metabolic processes
A metabolic reaction can be affected by a disruption of the enzymes that catalyse it, which then potentially disrupts all downstream metabolic reactions, ultimately leading to a metabolically-induced disease phenotype. To model this, Lee et al. [80] constructed the Metabolic Disease Network (MDN) in which two diseases are linked by an edge if the enzymes associated with them catalyse adjacent metabolic reactions. Co-morbidity analysis of MDN showed a 1.8-fold co-morbidity increase in diseases linked in this network when compared to the ones that are not. A substantially different representation of metabolism, termed Network of Interacting Pathways (NIP), was used to show that the complexity of an organism’s lifestyle determines how large, dense and efficiently organized its metabolism is, quantifying the changes in evolution of metabolism across archaea, bacteria and eukarya [75]. In a NIP model, pathways are represented by nodes and nodes are connected by an edge if the corresponding pathways overlap.
Shared microRNAs
microRNAs (miRNAs) are responsible for post-transcriptional regulation of protein-coding genes by means of inhibiting, destabilizing or degrading target mRNAs. A single miRNA down-regulates hundreds of target mRNA, thus having a key role in cellular functions such as development, differentiation, proliferation, apoptosis and metabolism. Recently, miRNA-based network reconstruction was implemented by Lu et al. [81], where they connected disease pairs whose associated genes are targeted by one or more shared miRNAs. A network constructed in such a way had clusters associated with diseases, such as cancer or cardiovascular diseases. Also, a negative correlation was found between tissue-specificity of a miRNA and the number of diseases associated with it. Another study has shedded additional light on the biological meaning of miRNA-based network reconstruction by strengthening the fact that a set of mRNA targets which are regulated by a single miRNA generally consist of functionally-associated molecules in human cells, rather than a random set of functionally-independent genes [82].
EXAMINING NETWORK TOPOLOGY
Regardless of data representation, networks of interconnected biomolecules in the cell and the concept of the human diseasome offer an opportunity for data analyses that aim to increase our understanding of why particular diseases co-manifest their phenotypes more often than others. The hope is that variants of network representation of these data and the new tools for their analyses will lead to novel approaches for disease diagnosis and treatment and aid drug discovery [83]. Even though only ∼10% of human genes have a known association to diseases [84], such limited knowledge about disease genes has still yielded insight into the link between network topology around a gene and its involvement in disease.
Since proteins are the main workhorses of the cell and they aggregate to perform a function, PPI networks have been analysed to elucidate cellular processes and disease. Early studies tried to use very simple methods to analyse PPI networks and link their topology to biological function [85–92]. For example, it was noticed that essential genes in early PPI networks of baker’s yeast obtained by Y2H methods tend to code for hub proteins [85], which was later refuted on more recent PPI data [93]. Also, it was postulated that there is a negative correlation between the connectivity of a gene and its rate of evolution and concluded that hub-coding genes are older and tend to evolve more slowly than non-hub-coding genes [86–88]. The number of phenotypic outcomes upon the removal of hub-coding genes has also been examined [55,89]. Similarly, 346 cancer-related proteins have been shown to have, on average, two times more direct interacting partners than non-cancer-related ones [91].
However, using such simple analysis methods on noisy data that are obtained by biased data collection and sampling may lead to questionable conclusions [46–55].
Hence, more sophisticated methods that give consistent results even in the presence of noise in the data have been designed.
Rather than looking at the network as a whole on a global, ‘macroscopic’ level, the local, ‘microscopic’ topology of networks has been examined. ‘Network motifs’, small sub-graphs that occur in real networks much more often than is expected at random, have been introduced to examine the structure of complex networks [94]. They enabled systematic detection of repeated appearances of topological substructures in Escherichia coli transcriptional regulation network and related them to specific biological responses to external signals [95]. However, since by definition motifs need to be over-represented in the data when compared to random graph models, the question is what models are the best fitting to real biological networks. This problem cannot be answered exactly due to NP-completeness (i.e. provable computational intractability) of the underlying sub-graph isomorphism problem that test whether one network exist as a copy in another network [56]. Hence, various methods for approximately comparing [96–98] and aligning [99–107] networks have been proposed.
‘Graphlets’ have been designed to further strengthen the bond between network topology and biological function and disease: they are small sub-graphs of large networks that do not need to be over-represented in the data (Figure 2) [96,98]. The statistics of frequencies of appearance of graphlets in entire networks, or around nodes in network have been used to classify networks into models [96,98], as well as to link the topology around a node in a network with the node’s biological function and involvement in disease: proteins with similar wiring up to a four-deep neighbourhood (Figure 3) were shown to belong to the same protein complexes, perform the same biological functions, are localized in the same sub-cellular compartments, and are co-expressed in tissues [108–113]. Furthermore, clustering of nodes based solely on graphlet-based topology of human PPI networks was used to successfully predict RNAi targets as novel components of melanogenesis regulatory pathways that could not have been identified by other existing approaches [111]. Similarly, involvement of genes in cancer was successfully predicted and validated both through literature curation and experimentally, thus providing evidence that topological wiring around cancer genes differs from wiring around non-cancer genes [113].
Figure 2:
Graphlets with up to five nodes. There are 30 of them, G0, G1, G2, … G29, and they contain 73 topologically unique node types, which are called ‘automorphism orbits’. Nodes belonging to the same orbit are of the same shade [98].
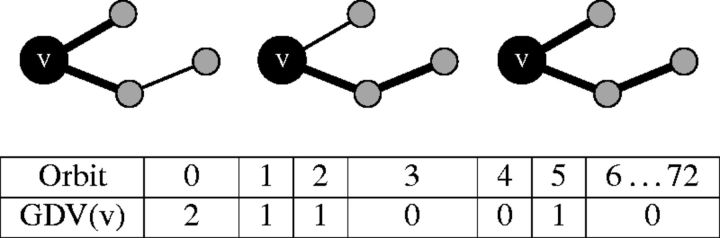
Figure 3:
An illustration of the graphlet degree vector (GDV) of node v. GDV represents one way in which graphlets can be used to describe topology around a node. GDV(v) = (2,1,1,0,0,1,0 … ,0), meaning that v is touched by two edges (Orbit 0, illustrated in the left panel), an end-node of one graphlet G1 (Orbit 1, illustrated in the middle panel), the middle node of one graphlet G1 (Orbit 2, illustrated in the left panel again), no nodes of a triangle (Orbit 3 in graphlet G2), no end-node of graphlet G3 (Orbit 4), one middle node of graphlet G3 (Orbit 5, illustrated in the right panel), and no other orbits. In this way, GDV essentially ‘quantifies’ the four-level-deep topological environment of a node.
The role of ‘topologically central’ proteins has also been examined. Several measures of topological centrality have been proposed, including degree centrality (i.e. hub nodes described above), betweenness centrality, closeness centrality, sub-graph centrality and graphlet centrality [114]. A general consensus is that topologically central proteins in PPI networks are involved in disease and aging. The concept of a dominating set (DS), that is, the set of nodes in a network such that all nodes are either in it or adjacent to it, has also been explored. Finding a DS of minimum size in a network is another computationally intractable problem, so heuristics are being sought. It was shown that proteins in a DS of the human PPI network constitute the ‘spine’ of the PPI network: this DS is statistically significantly enriched with disease, aging, and proteins participating in signalling pathways [114].
Predicting new disease proteins has also been based on examining disease gene DNA neighbourhoods and cellular co-localization of proteins [112,115–121]. Despite their simplicity, these approaches provided new insight: for example, Oti et al. [121] searched 432 loci for candidate disease genes achieving a 10-fold enrichment in success rate of valid disease–gene predictions. In the same study, additional consideration of cellular co-localization led to 1000-fold enrichment. Other methods base their predictions on overlaying network topology with additional information, such as the knowledge of functional or disease modules in the network [122–124]. They do this by, for instance, deriving a phenotype similarity score to identify new protein complexes associated with disease.
Recently, ‘driver’ genes have been proposed to be those whose mutations trigger genetic instability and cancer formation [125–128]. Also, genetic interactions, that is, pairs of genes whose joint mutation produces a distinct phenotype, have been indicated as potential targets for therapeutic intervention [125]. A very small number of driver genes are currently known and it is believed that the complete set of driver genes is not very large [125]. Thus far, there does not exist a standard method for finding driver genes. Hence, a new method for analysing PPI network topology has been proposed aiming to give insight into network parts that are rich in driver genes [129]. This method, which effectively captures a large portion of known driver and other disease-related genes, is based on an iterative pruning of the human PPI network: it uses k-core decomposition, a method that first removes nodes of degree one (k = 1), then from the remaining network it removes nodes of degree at most two (k ≤ 2) etc., until it reaches the value of k where removing nodes would result in an empty graph [130–133]. The largest value of k that leaves the graph in a non-empty state is called kmax. In this way, the part of the human PPI network that remains after kmax-core decomposition is obtained and examined for structural (i.e. topological) and functional uniqueness: this central, tightly-knit sub-network of the human PPI network is statistically significantly enriched with disease genes, driver genes and genetic interactions currently targeted by many drugs. Hence, it is termed ‘The Core Diseasome’ [129].
FUTURE DIRECTIONS
Despite noise and incompleteness in PPI and other systems-level biological network data, the structural properties of these networks have already given insight into biological function and involvement in disease of individual proteins. As we gather more network data and as the network data matures and becomes more reliable, we need to ensure that our models keep representing the data well and that our methods can cope with increased data complexity. Also, systems-level biological networks are currently only static representations of all interactions that we have ever observed under any condition and in any tissue, while cells are in fact dynamic, time- and condition-dependent systems. Hence, our data and methods should be extended to capture this systems-wide dynamics of biological processes [134–139]. Analysing the Human Diseasome in such a systems-level dynamic network framework has a potential to fully explain molecular, and environmental causes for onset and progression of disease and substantially change therapeutic practices.
Putting into the context of biological network data other approaches for analysing molecular causes of disease may to lead further insight. For example, it has been demonstrated that a significant number of diseases with early-life onset result from defective enzyme-coding genes, whereas adulthood onset diseases are caused by alterations in receptors and modifiers of protein function [140]. Also, it has been indicated that age-related diseases are a consequence of accumulation of mitochondrial dysfunction over the life of an individual [141,142]. Mitochondria perform oxidative phosphorylation that produces adenosine triphosphate (ATP) by utilizing energy released from oxidation of nutrients. In this process, toxic side-products, reactive oxygen species (ROS), are generated, which are molecules such as oxygen ions and peroxides. Increased ROS levels may lead to significant aberration of cell structure, specifically to DNA damage. Hence, many studies examine the role of energy and in particular, energy deficiency in human disease [141–143]. A direct link between mitochondrial dysfunction and disease was established and it was shown that mutations in mitochondrial DNA (mtDNA) alone are sufficient to generate major clinical phenotypes [144]. In particular, mtDNA is present in thousands of copies in a cell and it mediates effects of the environment onto genes by accumulating somatic mutations in post-mitotic tissue and resulting in delayed-onset of age-related diseases [141]. Analysing these processes in the context of systems-level biological networks may yield further insight. Another way in which the disruption of one gene was shown to trigger the onset of a seemingly unrelated disease is through what is known as the neighbouring gene effect (NGE) [145], which is also termed the ‘uncertainty principle of genetics’ [146]; It posits that the deletion of a genomic locus may affect the function of one or more neighbouring loci [146], effectively disrupting events downstream of the unintentionally affected loci. Hence, NGE may lead to erroneous gene annotation: it is estimated that NGE erroneously affects the annotation of 10% of the human interactome [147]. Due to these effects, global changes in the currently available interaction maps may soon be necessary.
Key points.
Various biological network data are being analysed for elucidating mechanisms of human disease.
Network topology is beginning to yield insight into biological function and disease.
Data and methods for analysing the dynamics of systems-level molecular networks are needed for better understanding of biology and disease.
Funding
The work was supported by ERC Starting Independent Researcher Grant 278212, NSF CDI OIA–1028394 grant, and the Serbian Ministry of Education and Science Project III44006.
References
- 1.Goh K-I, Cusick ME, Valle D, et al. The human disease network. Proc Natl Acad Sci USA. 2007;104(21):8685–90. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barabási A-L. Network medicine–from obesity to the ‘diseasome’. N Eng J Med. 2007;357(4):404–7. doi: 10.1056/NEJMe078114. [DOI] [PubMed] [Google Scholar]
- 3.Barabási A-L, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat Rev Genet. 2011;12(1):56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ideker T, Sharan R. Protein networks in disease. Genome Res. 2008;18:644–52. doi: 10.1101/gr.071852.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daly AK. Genome-wide association studies in pharmacogenomics. Nat Rev Genet. 2010;11(4):241–6. doi: 10.1038/nrg2751. [DOI] [PubMed] [Google Scholar]
- 6.Hindorff LA, Sethupathy P, Junkins HA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA. 2009;106(23):9362–7. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freedman ML, Monteiro ANA, Gayther SA, et al. Principles for the post-gwas functional characterization of cancer risk loci. Nat Genet. 2011;43(6):513–8. doi: 10.1038/ng.840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Urbach D, Moore JH. Mining the diseasome. BioData Mining. 2011;4(1):25. doi: 10.1186/1756-0381-4-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rashbass J. Online mendelian inheritance in man ‘omim’. Ind J Dermat Venereol Leprol. 1995;69(7):291–2. [PubMed] [Google Scholar]
- 10.Hamosh A, Scott AF, Amberger J, et al. Online mendelian inheritance in man. Hum Mutat. 2000;15(3):811–2. doi: 10.1002/(SICI)1098-1004(200001)15:1<57::AID-HUMU12>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 11.Wheeler DA, Srinivasan M, Egholm M, et al. The complete genome of an individual by massively parallel dna sequencing. Nature. 2008;452(7189):872–6. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 12.Ng PC, Murray SS, Levy S, et al. An agenda for personalized medicine. Nature. 2009;461(7265):724–6. doi: 10.1038/461724a. [DOI] [PubMed] [Google Scholar]
- 13.Roukos DH. Personal genomics and genome-wide association studies: novel discoveries but limitations for practical personalized medicine. Ann Surg Oncol. 2009;16(3):772–3. doi: 10.1245/s10434-008-0109-6. [DOI] [PubMed] [Google Scholar]
- 14.Ito T, Tashiro K, Muta S, et al. Toward a protein-protein interaction map of the budding yeast: a comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins. Proc Natl Acad Sci USA. 2000;97(3):1143–7. doi: 10.1073/pnas.97.3.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uetz P, Giot L, Cagney G, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–7. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 16.Giot L, Bader J, Brouwer C, et al. A protein interaction map of drosophila melanogaster. Science. 2003;302(5651):1727–36. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 17.Li S, Armstrong C, Bertin N, et al. A map of the interactome network of the metazoan c. elegans. Science. 2004;303:540–3. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gavin AC, Aloy P, Grandi P, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440(7084):631–6. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 19.Krogan N, Cagney G, Yu H, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–43. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 20.Tong AHY, Lesage G, Bader GD, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–13. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 21.Stelzl U, Worm U, Lalowski M, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–68. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 22.Rual J, Venkatesan K, Hao T, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–8. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 23.Rain J-D, Selig L, De Reuse H, et al. The protein-protein interaction map of helicobacter pylori. Nature. 2001;409:211–5. doi: 10.1038/35051615. [DOI] [PubMed] [Google Scholar]
- 24.Parrish JR, Yu J, Liu G, et al. A proteome-wide protein interaction map for Campylobacter jejuni. Genome Biol. 2007;8:R130. doi: 10.1186/gb-2007-8-7-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.LaCount DJ, Vignali M, Chettier R, et al. A protein interaction network of the malaria parasite plasmodium falciparum. Nature. 2005;438:103–7. doi: 10.1038/nature04104. [DOI] [PubMed] [Google Scholar]
- 26.Uetz P, Dong Y-A, Zeretzke C, et al. Herpesviral protein networks and their interaction with the human proteome. Science. 2006;311:239–42. doi: 10.1126/science.1116804. [DOI] [PubMed] [Google Scholar]
- 27.von Brunn1 A, Teepe C, Simpson JC, et al. Analysis of intraviral protein-protein interactions of the sars coronavirus orfeome. PLoS One. 2007;2:e459. doi: 10.1371/journal.pone.0000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chatr-aryamontri A, Ceol A, Peluso D, et al. Virusmint: a viral protein interaction database. Nucleic Acids Res. 2009;37:D669–73. doi: 10.1093/nar/gkn739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simonis N, Rual J, Carvunis A, et al. Empirically controlled mapping of the Caenorhabditis elegans protein–protein interactome network. Nat. Methods. 2009;6(1):47–54. doi: 10.1038/nmeth.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gavin AC, Bosche M, Krause R, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415(6868):141–7. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 31.Ho Y, Gruhler A, Heilbut A, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415(6868):180–3. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 32.Thaminy S, Auerbach D, Arnoldo A, et al. Identification of novel erbb3-interacting factors using the split-ubiquitin membrane yeast two-hybrid system. Genome Res. 2003;13(7):1744–53. doi: 10.1101/gr.1276503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snider J, Kittanakom S, Damjanovic D, et al. Detecting interactions with membrane proteins using a membrane two-hybrid assay in yeast. Nat Protocols. 2010;5(7):1281–93. doi: 10.1038/nprot.2010.83. [DOI] [PubMed] [Google Scholar]
- 34.Petschnigg J, Moe OW, Stagljar I. Using yeast as a model to study membrane proteins. Curr Opin Nephrol Hypertension. 2011;20(4):425–32. doi: 10.1097/MNH.0b013e3283478611. [DOI] [PubMed] [Google Scholar]
- 35.Iyer K, Būrkle L, Auerbach D, et al. Utilizing the split-ubiquitin membrane yeast two-hybrid system to identify protein-protein recipes. Sciences STKE. 2009;2005:pl3. doi: 10.1126/stke.2752005pl3. [DOI] [PubMed] [Google Scholar]
- 36.Peri S, Navarro JD, Kristiansen TZ, et al. Human protein reference database as a discovery resource for proteomics. Nucleic Acids Res. 2004;32:D497–501. doi: 10.1093/nar/gkh070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Breitkreutz BJ, Stark C, Reguly T, et al. The BioGRID interaction database: 2008 update. Nucleic Acids Res. 2008;36:D637–40. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kerrien S, Aranda B, Breuza L, et al. The intact molecular interaction database in 2012. Nucleic Acids Res. 2012;40:D841–6. doi: 10.1093/nar/gkr1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chatr-aryamontri A, Ceol A, Palazzi LM, et al. Mint: the molecular interaction database. Nucleic Acids Res. 2007;35:D572–4. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bader GD, Betel D, Hogue CWV. Bind: the biomolecular interaction network database. Nucleic Acids Res. 2003;31(1):248–50. doi: 10.1093/nar/gkg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salwinski L, Miller CS, Smith AJ, et al. The database of interacting proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–51. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jensen LJ, Kuhn M, Stark M, et al. STRING: a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–6. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Razick S, Magklaras G, Donaldson IM. irefindex: a consolidated protein interaction database with provenance. BMC Bioinformatics. 2008;9(1):405. doi: 10.1186/1471-2105-9-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao Y, Jensen ON. Modification-specific proteomics: strategies for characterization of post-translational modifications using enrichment techniques. Proteomics. 2009;9(20):4632–41. doi: 10.1002/pmic.200900398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Venkatesan K, Rual J-F, Vazquez A, et al. An empirical framework for binary interactome mapping. Nat Methods. 2009;6(1):83–90. doi: 10.1038/nmeth.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hakes L, Pinney JW, Robertson DL, et al. Protein-protein interaction networks and biology–whats the connection? Nat Biotechnol. 2008;26(1):69–72. doi: 10.1038/nbt0108-69. [DOI] [PubMed] [Google Scholar]
- 47.Wodak SJ, Pu S, Vlasblom J, et al. Challenges and rewards of interaction proteomics. Mol Cell Proteomics. 2009;8(1):3–18. doi: 10.1074/mcp.R800014-MCP200. [DOI] [PubMed] [Google Scholar]
- 48.De Silva E, Stumpf MPH. Complex networks and simple models in biology. J Roy Soc Inter Roy Soc. 2005;2(5):419–30. doi: 10.1098/rsif.2005.0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stumpf MPH, Wiuf C, May RM. Subnets of scale-free networks are not scale-free: sampling properties of networks. Proc Natl Acad Sci USA. 2005;102(12):4221–4. doi: 10.1073/pnas.0501179102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Han J-DJ, Dupuy D, Bertin N, et al. Effect of sampling on topology predictions of protein-protein interaction networks. Nat Biotechnol. 2005;23(7):839–44. doi: 10.1038/nbt1116. [DOI] [PubMed] [Google Scholar]
- 51.De Silva E, Thorne T, Ingram P, et al. The effects of incomplete protein interaction data on structural and evolutionary inferences. BMC Biol. 2006;4(1):39. doi: 10.1186/1741-7007-4-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Von Mering C, Krause R, Snel B, et al. Comparative assessment of large-scale data sets of protein-protein interactions. Nature. 2002;417(6887):399–403. doi: 10.1038/nature750. [DOI] [PubMed] [Google Scholar]
- 53.Collins SR, Kemmeren P, Zhao X-C, et al. Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol Cell Proteomics. 2007;6(3):439–50. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- 54.Schwartz AS, Yu J, Gardenour KR, et al. Cost-effective strategies for completing the interactome. Nat Methods. 2009;6(1):55–61. doi: 10.1038/nmeth.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu H, Braun P, Yildirim MA, et al. High-quality binary protein interaction map of the yeast interactome network. Science. 2008;322(5898):104–10. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cook SA. The complexity of theorem-proving procedures. Proc 3rd Annu ACM Symp Theor Comput STOC 71. 1971;22(1):151–8. [Google Scholar]
- 57.Pržulj N. Protein-protein interactions: making sense of networks via graph-theoretic modeling. BioEssays. 2011;33(2):115–23. doi: 10.1002/bies.201000044. [DOI] [PubMed] [Google Scholar]
- 58.Barabási A-l, Oltvai ZN. Network biology: understanding the cells functional organization. Genetics. 2004;5:101–13. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 59.Zhu X, Gerstein M, Snyder M. Getting connected: analysis and principles of biological networks. Genes Dev. 2007;21(9):1010–24. doi: 10.1101/gad.1528707. [DOI] [PubMed] [Google Scholar]
- 60.Barabási A-L, Bonabeau E. Scale-free networks. Sci Am. 2003;288(5):60–9. doi: 10.1038/scientificamerican0503-60. [DOI] [PubMed] [Google Scholar]
- 61.Nikoloski Z, Grimbs S, May P, et al. Metabolic networks are np-hard to reconstruct. J Theor Biol. 2008;254(4):807–16. doi: 10.1016/j.jtbi.2008.07.015. [DOI] [PubMed] [Google Scholar]
- 62.Radrich K, Tsuruoka Y, Dobson P, et al. Integration of metabolic databases for the reconstruction of genome-scale metabolic networks. BMC Sys Biol. 2010;4(1):114. doi: 10.1186/1752-0509-4-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Terzer M, Maynard ND, Covert MW, et al. Genome-scale metabolic networks. Wiley Interdisciplinary Rev Sys Biol Med. 2009;1(3):285–97. doi: 10.1002/wsbm.37. [DOI] [PubMed] [Google Scholar]
- 64.Janga SC, Babu MM. Network-based approaches for linking metabolism with environment. Genome Biol. 2008;9(11):239. doi: 10.1186/gb-2008-9-11-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ōsterlund T, Nookaew I, Nielsen J. Fifteen years of large scale metabolic modeling of yeast: developments and impacts. Biotechnol Adv. 2011;3:1–10. doi: 10.1016/j.biotechadv.2011.07.021. [DOI] [PubMed] [Google Scholar]
- 66.Kim TY, Kim HU, Lee SY. Metabolite-centric approaches for the discovery of antibacterials using genome-scale metabolic networks. Metab Eng. 2010;12(2):105–11. doi: 10.1016/j.ymben.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 67.Chung BKS, Lee D-Y. Flux-sum analysis: a metabolite-centric approach for understanding the metabolic network. BMC Sys Biol. 2009;3:117. doi: 10.1186/1752-0509-3-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Imieliński M, Belta C, Halász A, et al. Investigating metabolite essentiality through genome-scale analysis of Escherichia coli production capabilities. Bioinformatics. 2005;21(9):2008–16. doi: 10.1093/bioinformatics/bti245. [DOI] [PubMed] [Google Scholar]
- 69.Kim P-J, Lee D-Y, Kim TY, et al. Metabolite essentiality elucidates robustness of Escherichia coli metabolism. Proc Natl Acad Sci USA. 2007;104(34):13638–42. doi: 10.1073/pnas.0703262104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Horne AB, Hodgman TC, Spence HD, et al. Constructing an enzyme-centric view of metabolism. Bioinformatics. 2004;20(13):2050–5. doi: 10.1093/bioinformatics/bth199. [DOI] [PubMed] [Google Scholar]
- 71.Yang C-R. An enzyme-centric approach for modelling non-linear biological complexity. BMC Sys Biol. 2008;2(1):70. doi: 10.1186/1752-0509-2-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pharkya P, Maranas CD. An optimization framework for identifying reaction activation/inhibition or elimination candidates for overproduction in microbial systems. Metab Engineer. 2006;8(1):1–13. doi: 10.1016/j.ymben.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 73.Oberhardt MA, Chavali AK, Papin JA. Flux balance analysis: interrogating genome-scale metabolic networks. Methods Mol Biol (Clifton, NJ) 2009;500:61–80. doi: 10.1007/978-1-59745-525-1_3. [DOI] [PubMed] [Google Scholar]
- 74.Thiele I, Vo TD, Price ND, et al. Expanded metabolic reconstruction of helicobacter pylori (iit341 gsm/gpr): an in silico genome-scale characterization of single- and double-deletion mutants. Society. 2005;187(16):5818–30. doi: 10.1128/JB.187.16.5818-5830.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mazurie A, Bonchev D, Schwikowski B, et al. Evolution of metabolic network organization. BMC Sys Biol. 2010;4(1):59. doi: 10.1186/1752-0509-4-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Park J, Lee D-S, Christakis NA, et al. The impact of cellular networks on disease comorbidity. Mol Sys Biol. 2009;5(262):262. doi: 10.1038/msb.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gillis J, Pavlidis P. ‘guilt by association’ is the exception rather than the rule in gene networks. PLoS Comput Biol. 2012;8(3):e1002444. doi: 10.1371/journal.pcbi.1002444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gillis J, Pavlidis P. The role of indirect connections in gene networks in predicting function. Bioinformatics. 2011:1–8. doi: 10.1093/bioinformatics/btr288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gillis J, Pavlidis P. The impact of multifunctional genes on ‘guilt by association’ analysis. PLoS One. 2011;6(2):16. doi: 10.1371/journal.pone.0017258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee DS, Park J, Kay KA, et al. The implications of human metabolic network topology for disease comorbidity. Proc Natl Acad Sci USA. 2008;105(29):9880–5. doi: 10.1073/pnas.0802208105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lu M, Zhang Q, Deng M, et al. An analysis of human microrna and disease associations. PLoS One. 2008;3(10):5. doi: 10.1371/journal.pone.0003420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Satoh J-i, Tabunoki H. Comprehensive analysis of human microrna target networks. BioData Mining. 2011;4(1):17. doi: 10.1186/1756-0381-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim HU, Sohn SB, Lee SY. Metabolic network modeling and simulation for drug targeting and discovery. Biotechnology J. 2012;7(3):330–42. doi: 10.1002/biot.201100159. [DOI] [PubMed] [Google Scholar]
- 84.Amberger J, Bocchini CA, Scott AF, et al. Mckusicks online mendelian inheritance in man (omim) Nucleic Acids Res. 2009;37:D793–6. doi: 10.1093/nar/gkn665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jeong H, Mason SP, Barabási A-L, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411(6833):41–2. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 86.Fraser HB, Hirsh AE, Steinmetz LM, et al. Evolutionary rate in the protein interaction network. Science. 2002;296(5568):750–2. doi: 10.1126/science.1068696. [DOI] [PubMed] [Google Scholar]
- 87.Eisenberg E, Levanon EY. Preferential attachment in the protein network evolution. Phys Rev Lett. 2003;91(13):138701. doi: 10.1103/PhysRevLett.91.138701. [DOI] [PubMed] [Google Scholar]
- 88.Saeed R, Deane CM. Protein–protein interactions, evolutionary rate, abundance and age. BMC Bioinformatics. 2006;7(2003):128. doi: 10.1186/1471-2105-7-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jordan IK, Wolf YI, Koonin EV. No simple dependence between protein evolution rate and the number of protein-protein interactions: only the most prolific interactors tend to evolve slowly. BMC Evol Biol. 2003;3(1):1. doi: 10.1186/1471-2148-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wachi S, Yoneda K, Wu R. Interactome-transcriptome analysis reveals the high centrality of genes differentially expressed in lung cancer tissues. Bioinformatics. 2005;21(23):4205–8. doi: 10.1093/bioinformatics/bti688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jonsson PF, Bates PA. Global topological features of cancer proteins in the human interactome. Bioinformatics. 2006;22(18):2291–7. doi: 10.1093/bioinformatics/btl390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xu J, Li Y. Discovering disease-genes by topological features in human protein-protein interaction network. Bioinformatics. 2006;22(22):2800–5. doi: 10.1093/bioinformatics/btl467. [DOI] [PubMed] [Google Scholar]
- 93.Coulomb S, Bauer M, Bernard D, et al. Gene essentiality and the topology of protein interaction networks. Proc Roy Soc B. 2005;272:1721–5. doi: 10.1098/rspb.2005.3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Milo R, Shen-Orr SS, Itzkovitz S, et al. Network motifs: simple building blocks of complex networks. Science. 2002;298:824–7. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- 95.Shen-Orr SS, Milo R, Mangan S, Alon U. Network motifs in the transcriptional regulation network of Escherichia coli. Nat Genet. 2002;31(1):64–8. doi: 10.1038/ng881. [DOI] [PubMed] [Google Scholar]
- 96.Pržulj N, Corneil DG, Jurisica I. Modeling interactome: scale-free or geometric? Bioinformatics. 2004;20(18):3508–15. doi: 10.1093/bioinformatics/bth436. [DOI] [PubMed] [Google Scholar]
- 97.Sharan R, Ideker T. Modeling cellular machinery through biological network comparison. Nat Biotechnol. 2006;24:427–33. doi: 10.1038/nbt1196. [DOI] [PubMed] [Google Scholar]
- 98.Pržulj N. Biological network comparison using graphlet degree distribution. Bioinformatics. 2007;23(2):e177–83. doi: 10.1093/bioinformatics/btl301. [DOI] [PubMed] [Google Scholar]
- 99.Kelley BP, Bingbing Y, Lewitter F, et al. PathBLAST: a tool for alignment of protein interaction networks. Nucleic Acids Res. 2004;32:83–8. doi: 10.1093/nar/gkh411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Flannick J, Novak A, Balaji S, et al. Graemlin general and robust alignment of multiple large interaction networks. Genome Res. 2006;16(9):1169–81. doi: 10.1101/gr.5235706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Koyuturk M, Kim Y, Topkara U, et al. Pairwise alignment of protein interaction networks. J Comput Biol. 2006;13(2):182–89. doi: 10.1089/cmb.2006.13.182. [DOI] [PubMed] [Google Scholar]
- 102.Singh R, Xu J, Berger B. Pairwise global alignment of protein interaction networks by matching neighborhood topology. In: Terence PS, Haiyan Huang, editors. Research in Computational Molecular Biology. Oakland, CA, USA: Springer; 2007. pp. 16–31. [Google Scholar]
- 103.Liao C-S, Lu K, Baym M, et al. IsoRankN: spectral methods for global alignment of multiple protein networks. Bioinformatics. 2009;25(12):i253–58. doi: 10.1093/bioinformatics/btp203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kuchaiev O, Milenković T, Memišević V, et al. Topological network alignment uncovers biological function and phylogeny. J R Soc Interface. 2010;7:1341–54. doi: 10.1098/rsif.2010.0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Milenković T, Leong Ng W, Hayes W, et al. Optimal network alignment with graphlet degree vectors. Cancer Inform. 2010;9:121–37. doi: 10.4137/cin.s4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kuchaiev O, Pržulj N. Integrative network alignment reveals large regions of global network similarity in yeast and human. Bioinformatics. 2011;27(10):1390–6. doi: 10.1093/bioinformatics/btr127. [DOI] [PubMed] [Google Scholar]
- 107.Memišević V, Pržulj N. C-GRAAL: common-neighbors-based global graph alignment of biological networks. Integr Biol. 2012;4:10. doi: 10.1039/c2ib00140c. [DOI] [PubMed] [Google Scholar]
- 108.Milenkovič T, Pržulj N. Uncovering biological network function via graphlet degree signatures. Cancer Inform. 2008;6:257–73. [PMC free article] [PubMed] [Google Scholar]
- 109.Guerrero C, Milenković T, Pržulj N, et al. Characterization of the yeast proteasome interaction network by qtax-based tag-team mass spectrometry and protein interaction network analysis. Proc Natl Acad Sci USA. 2008;105(36):13333–8. doi: 10.1073/pnas.0801870105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Memišević V, Milenković T, Pržulj N. Complementarity of network and sequence structure in homologous proteins. J Integr Bioinform. 2010;7(3):135. doi: 10.2390/biecoll-jib-2010-135. [DOI] [PubMed] [Google Scholar]
- 111.Ho H, Milenković T, Memišević V, et al. Protein interaction network topology uncovers melanogenesis regulatory network components within functional genomics datasets. BMC Syst Biol. 2010;4(1):84. doi: 10.1186/1752-0509-4-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Milenković T, Pržulj N. Uncovering biological network function via graphlet degree signatures. Cancer Inform. 2008;4:257–73. [PMC free article] [PubMed] [Google Scholar]
- 113.Milenković T, Memišević V, Ganesan A, et al. Systems-level cancer gene identification from protein interaction network topology applied to melanogenesis-related functional genomics data. J R Soc Interface. 2010;44(7):353–0. doi: 10.1098/rsif.2009.0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Milenković T, Memišević V, Bonato A, et al. Dominating biological networks. PLoS One. 2011;6(8):12. doi: 10.1371/journal.pone.0023016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sharan R, Ulitsky I, Ideker T. Network-based prediction of protein function. Mol Syst Biol. 2007;3(88):88. doi: 10.1038/msb4100129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Schwikowski B, Fields S. A network of protein-protein interactions in yeast. Nat Biotechnol. 2000;18:1257–61. doi: 10.1038/82360. [DOI] [PubMed] [Google Scholar]
- 117.Radivojac P, Peng K, Clark WT, et al. An integrated approach to inferring gene-disease associations in humans. Proteins. 2008;72(3):1030–7. doi: 10.1002/prot.21989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Goh K, Cusick ME, Valle D, et al. The human disease network. Proc Natl Acad Sci USA. 2007;104(21):8685–90. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yidirim MA, Goh K-I, Cusick ME, et al. Drug-target network. Nat Biotechnol. 2007;25(10):1119–26. doi: 10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]
- 120.Vanunu O, Magger O, Ruppin E, et al. Associating genes and protein complexes with disease via network propagation. PLoS Comput Biol. 2010;6:e1000641. doi: 10.1371/journal.pcbi.1000641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Oti M, Snel B, Huynen MA, et al. Predicting disease genes using protein-protein interactions. J Med Genet. 2006;43(8):691–8. doi: 10.1136/jmg.2006.041376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Navlakha S, Kingsford C. The power of protein interaction networks for associating genes with diseases. Bioinformatics. 2010;26(8):1057–63. doi: 10.1093/bioinformatics/btq076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lage K, Karlberg EO, Storling ZM, et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat Biotechnol. 2007;25(3):309–16. doi: 10.1038/nbt1295. [DOI] [PubMed] [Google Scholar]
- 124.Wu X, Jiang R, Zhang MQ, et al. Network-based global inference of human disease genes. Mol Syst Biol. 2008;4(189):189. doi: 10.1038/msb.2008.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ashworth A, Lord CJ, Reis-Filho JS. Genetic interactions in cancer progression and treatment. Cell. 2011;145(1):30–8. doi: 10.1016/j.cell.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 126.Ji X, Tang J, Halberg R, et al. Distinguishing between cancer driver and passenger gene alteration candidates via cross-species comparison: a pilot study. BMC Cancer. 2010;10:426. doi: 10.1186/1471-2407-10-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Youn A, Simon R. Identifying cancer driver genes in tumor genome sequencing studies. Bioinformatics. 2011;27(2):175–81. doi: 10.1093/bioinformatics/btq630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Akavia UD, Litvin O, Kim J, et al. An integrated approach to uncover drivers of cancer. Cell. 2010;143(6):1005–17. doi: 10.1016/j.cell.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Janjić V, Pržulj N. The core diseasome. Mol. BisSyst. 2012 doi: 10.1039/c2mb25230a. doi:10.1039/C2MB25230A. [DOI] [PubMed] [Google Scholar]
- 130.Miorandi D, Pellegrini FD. K-shell decomposition for dynamic complex networks. Modeling and Optimization in Mobile Ad Hoc and Wireless Networks WiOpt 2010 Proceedings of the 8th International Symposium on, Avignon. 2010:488–96. [Google Scholar]
- 131.Carmi S, Havlin S, Kirkpatrick S, et al. A model of internet topology using k-shell decomposition. Proc Natl Acad Sci USA. 2007;104(27):11150–4. doi: 10.1073/pnas.0701175104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Batagelj V, Zaversnik M. An o(m) algorithm for cores decomposition of networks. Symp Quart J Modern Foreign Literat. 2003 cs.DS/0310(m):1–10. [Google Scholar]
- 133.Leskovec J, Lang KJ, Dasgupta A, et al. Community structure in large networks: natural cluster sizes and the absence of large well-defined clusters. Inter Math. 2008;6(1):66. [Google Scholar]
- 134.Piston DW, Kremers G-J. Fluorescent protein fret: the good, the bad and the ugly. Trends Biochem Sci. 2007;32(9):407–14. doi: 10.1016/j.tibs.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 135.Kentner D, Sourjik V. Dynamic map of protein interactions in the Escherichia coli chemotaxis pathway. Mol Syst Biol. 2009;5(238):238. doi: 10.1038/msb.2008.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Cristea IM, Carroll J-WN, Rout MP, et al. Tracking and elucidating alphavirus-host protein interactions. J Biol Chem. 2006;281(40):30269–78. doi: 10.1074/jbc.M603980200. [DOI] [PubMed] [Google Scholar]
- 137.Tarassov K, Messier V, Landry CR, et al. An in vivo map of the yeast protein interactome. Science. 2008;320(5882):1465–70. doi: 10.1126/science.1153878. [DOI] [PubMed] [Google Scholar]
- 138.Przytycka TM, Singh M, Slonim DK. Toward the dynamic interactome: it’s about time. Brief Bioinform. 2010;11(1):15–29. doi: 10.1093/bib/bbp057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Siegal ML, Promislow DEL, Bergman A. Functional and evolutionary inference in gene networks: does topology matter? Genetica. 2007;129(1):83–103. doi: 10.1007/s10709-006-0035-0. [DOI] [PubMed] [Google Scholar]
- 140.Jimenez-Sanchez G, Childs B, Valle D. Human disease genes. Nature. 2001;409(6822):853–5. doi: 10.1038/35057050. [DOI] [PubMed] [Google Scholar]
- 141.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wallace DC. The mitochondrial genome in human adaptive radiation and disease: on the road to therapeutics and performance enhancement. Gene. 2005;354:169–80. doi: 10.1016/j.gene.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 143.Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283(5407):1482–8. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 144.Wallace DC, Fan W. The pathophysiology of mitochondrial disease as modeled in the mouse. Genes Dev. 2009;23(15):1714–36. doi: 10.1101/gad.1784909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ben-Shitrit T, Yosef N, Shemesh K, et al. Systematic identification of gene annotation errors in the widely used yeast mutation collections. Nat Meth. 2012;9(4):373–8. doi: 10.1038/nmeth.1890. [DOI] [PubMed] [Google Scholar]
- 146.Baryshnikova A, Andrews B. Neighboring-gene effect: a genetic uncertainty principle. Nat Meth. 2012;9(4):341–3. doi: 10.1038/nmeth.1936. [DOI] [PubMed] [Google Scholar]
- 147.Costanzo M, Baryshnikova A, Bellay J, et al. The genetic landscape of a cell. Science. 2010;327(5964):425–31. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]