Abstract
Porcine epidemic diarrhea virus (PEDV) and porcine deltacoronavirus (PDCoV) are epizootic swine viruses. To detect and study the evolution of PEDV and PDCoV in central China (Shanxi, Henan, Hubei province), 70 clinical intestinal and fecal samples from piglets with severe watery diarrhea during August 2015 and June 2016 were collected, tested and analyzed. PEDV was more frequently detected by PCR than PDCoV. Phylogenetic analysis of S genes showed that the 10 PEDV strains from this study clustered into G2a (n = 7) and G2b (n = 3) groups. Additionally, the three G2b strains (PEDV S2△) contained the same specific 3 nt deletion in S2 as other reference strains in G2b. Interestingly, complete genome analysis indicated that CH/hubei/2016 was closer to the US INDEL strain and G2a group. CH/hubei/2016 had one recombination event in S2 gene which may have resulted from AH2012-12 (from G2b group) and CH-ZMDZY-11 (from G2a group). Furthermore, 10 purifying selection sites in S gene indicated an adaptive evolution of PEDV in central China swine herds. These results suggested that Pandemic G2a and G2b are predominant PEDV genotype circulating in central China. In addition, the deletion and recombination identified in S gene suggested PEDV strains of central exhibited an evolutionary variety. However, whether these changes affect the pathogenicity and antigenicity of wild PEDV is unknown and is worth for further investigation.
Keywords: Porcine epidemic diarrhea virus, Porcine deltacoronavirus, Spike gene, Diversity, China
Highlights
-
•
PEDV (84.2%) infection could be more commonly detected than PDCoV (2.9%) in central China.
-
•
A specific 3 nt-deletion in S2 gene was firstly reported in PEDV strains of central China.
-
•
The further analyses provided evidence of the relationship between PEDV S2△ and previous PEDV stains (3-deletion in S2).
1. Introduction
Porcine epidemic diarrhea (PED) is an acute, highly contagious swine disease caused by porcine epidemic diarrhea virus (PEDV), which leads to severe watery vomiting, diarrhea, and high mortality in newborn piglets (Pensaert and de Bouck, 1978; Song and Park, 2012). PEDV is an enveloped single-stranded RNA Coronavirus that belongs to the family Coronaviridae, genus Alphacoronavirus. PEDV genome (28 kb in length) has seven open reading frames (ORFs) and encodes four structural proteins: spike (S), envelope (E), membrane (M) and nucleocapsid (N) as well as three non-structural proteins: replicates 1a, 1b and ORF3 (Song and Park, 2012). Among these proteins, PEDV S glycoprotein shows an important role on host cell entry and viral virulence and could be divided structurally into the S1 and S2 regions (Song and Park, 2012), which is also a key protein exhibiting major genetic variations among different stains (Sun et al., 2015).
PEDV was first reported in UK in 1971 and then was detected in other European countries during the following 6 years (Chasey and Cartwright, 1978; Wood, 1977). Since 1980s, it has been widespread in Asia (Song and Park, 2012). A massive PED outbreaks were reported in China in 2010, which associated with a novel PEDV strain (Pandemic strains) that was genetically distant from the Prototype PEDV strain CV777 (Chen et al., 2010). The first case of PED (Pandemic strains) in North America was reported in April 2013. In 2014, the new variants, INDEL strains, containing insertions and deletions in the N terminal region of the S1 gene was found in American (Vlasova et al., 2014). Genetic and phylogenetic analyses indicated PEDV strains fell clearly into two distinct groups, G1 (Prototype strain) and G2 (Pandemic strains) based on neighbor-joining method; G2 further separated into two subgroups:G2a and G2b (Huang et al., 2013).
Porcine deltacoronavirus (PDCoV) is a proposed member of family Coronaviridae, genus deltacoronavirus, which also lead to severe watery vomiting, diarrhea (Woo et al., 2012). PDCoV has been firstly identified in Hong Kong 2012, and then successively detected in China mainland and USA (Song et al., 2015; Woo et al., 2012). Like PEDV S gene, the phylogenetic relationship of PDCoV is also constructed in terms of S genes (Ma et al., 2015; Zhai et al., 2016).
In present study, a total of 70 samples from central China during 2015 to 2016 were simultaneously tested for PEDV and PDCoV by PCR. S genes of PEDVs and complete gene of PEDV from positive samples were sequenced, respectively. Phylogenetic, sequence, recombination and selection pressures analysis of PEDV were performed to construct evolution relationships between the strains from central China and other country, whereas one PDCoV S gene was also sequenced and analyzed. This work may provide new insights in the epidemiology of PEDV in central China.
2. Methods
2.1. Sample collection
A total of 70 clinical intestinal and fecal samples were collected from different regions of central China (Henan, Hubei, Shanxi province) between 2015 and 2016. These samples came from pigs with a variety of clinical signs including vomiting, anorexia, and severe watery diarrhea. All clinical samples were homogenized for RNA extraction and virus isolation.
2.2. Total RNA extraction and RT-PCR
Total RNA was extracted from samples using Trizol reagent (TaKaRa, Japan) according to the manufacturer's instructions. Reverse transcription was performed as previously described (Kim et al., 2001). Briefly, using 20 ul of the reaction mixture containing 11 ul of the total RNA extraction solution, 1 ul 10 mM OligodT primer, 4 ul 5 × RTbuffer, 2 ul 10 Mm dNTP, 1 ul (40 U) RNase inhibitor, 1 ul M-MLV (100 U) Reverse Transcriptase. After 1 h kept under 42 °C, the resulting cDNA was stored at −20 °C. All sample's cDNA was performed by PCR method as previously described (Kim et al., 2001; Song et al., 2015). In this study, to amplify the S gene of PEDV and PDCoV primers were designed based on PEDV CV777 strain (AF353511) and PDCoV HKU15 strain (KJ568769) (Table 1 ). The reaction mixture consisted of 1 ul of cDNA, 10 ul ExTaq (TAKARA), 1 ul F-primer (10 pmol), 1 ul R-primer (10 poml), and RNase-free water in a total volume of 20 ul. And the amplification was carried out as follows: 95 °C for 5 min, followed by 30 cycles of 95°Cfor 1 min, 53 °C for 1 min and 72 °Cfor 1 min, and then final elongation for 10 min at 72 °C·The complete genome of PEDV was amplified as previously described (Wang et al., 2013). Products were visualized using 1% agarose gel under ultraviolet light.
Table 1.
Primers used in this study.
| Primer name | Nucleotide sequence, 5′-3′ | Size(bp) | aPrimer location |
|---|---|---|---|
| PEDV S1-F | GGTAAGTTGCTAGTGCGTAA | 1689 | 20570–22258 |
| PEDV S1-R | CACAGAAAGAACTAAACCC | ||
| PEDV S2-F | TTTGGTGGTCTTAGTAGTGCC | 1420 | 22189–23608 |
| PEDV S2-R | GCTGTAGAACATCCGTCTGTA | ||
| PEDV S3-F | GGGCGAGACTCAATTATCTTGC | 1312 | 23564–24875 |
| PEDV S3-R | CTGGACAGCATCCAAAGACAAG | ||
| PDCoV S1-F | TTGGCGGAACTCACACACTT | 1799 | 18210–20009 |
| PDCoV S1-R | TGACCCCGATACAACCTAACA | ||
| PDCoV S2-F | GTGAGCAGTTTAACTACACCACT | 1710 | 19796–21506 |
| PDCoV S2-R | TTCTCAGCATCAACAACACCA | ||
| PDCoV S3-F | AGCAGCATACTAACCACCAGA | 1744 | 21310–23054 |
| PDCoV S3-R | ACTAGGGTGAAGGGTTGGAGCA |
2.3. Cloning and sequencing of PEDV and PDCoV gene
Three pairs of primers were used to determine the PEDV S gene or PDCoV S gene (Table 1). The amplicons were purified and cloned into pMD-18 T, then sequenced by Sanger method. Each fragment was independently sequenced at least three times. The sequenced products were splicing to obtain the PEDV S gene. After analysis of the S gene, the positive samples were selected and sequenced for the complete genomes (Wang et al., 2013).
2.4. Phylogenetic and sequence analysis of PEDV and PDCOV
Reference PEDV and PDCoV strains are shown in Table 2 (Supplemental Material 1). PEDVs and PDCoV isolates from this study are shown in Table 3 (Supplemental Material 2)
The PEDV S genes, PEDV complete genomes, and PDCoV S genes were aligned using ClustalW in DNASTAR7.1. Phylogenetic analysis for each dataset was performed using neighbor-joining method with 1000 bootstrap replicates in MEGA7.
2.5. Recombination analysis
ClustalW program was used to align the full-length genome sequences of CH/hubei/2016 and reference sequence from GenBank (Table 2) (Supplemental Material 1). RDP3 recombination detection program was used to detect the potential recombination events. RDP, BootScan, MaxChi, Chimaera, SiScan, and 3Seq methods embedded in the RDP3 software package (p < 0.01) were utilized (Chen et al., 2013), only those recombination events supported by >4 programs were considered to avoid dependence on a single methodology (Li et al., 2016). Confirmation of the potential recombination events was carried out by BootScan analysis and fast neighbor-joining tree(Sun et al., 2016).
2.6. Selection pressures analysis of S gene
PEDV S gene deduced amino acid sequences were aligned by ClustalW program in DNASTAR7.1 software, and selection pressures for this gene at individual codon sites were estimated by the ratio of non-synonymous (dN) and synonymous (dS) mutations, which was calculated by the Single Likelihood Ancestor Counting method and Fixed Effects Likelihood method available at the Datamonkey online version of the Hy-Phy package, using a significance level of 0.1 (Mu et al., 2013).
3. Results
3.1. PEDV and PDCoV of positive samples among diagnostic samples
All the samples were simultaneously tested for PEDV and PDCoV by PCR. 84.2% (59/70samples) were positive for PEDV, but only 2.9% (2/70samples) samples were PDCoV positive, which were also PEDV positive. (These data suggested PEDV infection could be more commonly detected than PDCoV in central China.) These data suggested that PEDV could be more commonly detected than PDCoV in central China.
3.2. Sequence analyses of PEDV S gene
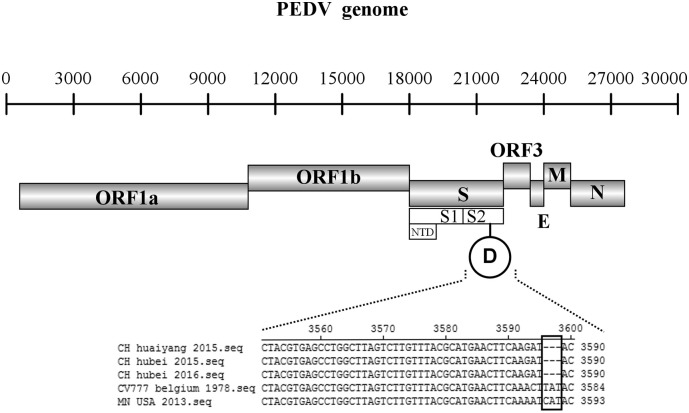
To reveal the characteristics of PEDV strains currently circulating in central China, 10 PEDV positive samples (Table 3 Supplemental Material 2) were selected and sequenced in terms of S gene. Compared to the S gene of CV777 (4152), sequencing of the field strains revealed a range of lengths: three were 6 nt longer (4158 nt), seven were 9 nt longer (4161 nt). The analysis of PEDV S gene showed that at N-terminal domain (NTD) of S gene, these 10 strains included two notable insertions at 166 nt to 177 nt and 337 nt to 339 nt, respectively, and one deletion between 477 nt to 483 nt at S gene compared with CV777. And same deletions and insertions segments were also observed in the same position on global Pandemic strains as previous study indicated (Huang et al., 2013). Besides, a specific deletion from 3596 nt to 3598 nt at S2 gene was firstly found in three PEDV strains (CH/hubei/2015, CH/hubei/2016, CH/huaiyang/2015) (Fig. 1 ), respectively, which was named as PEDV S2△.
Fig. 1.
S gene nucleotide alignments of CH/hubei/2016, CH/huaiyang/2015, CH/hubei/2016, and two reference PEDV strains: MN/USA/2013 (Pandemic strain) and CV777/Belgium/1978 (Prototype strain). NTD, N-terminal domain of the spike gene is hypervariable region of PEDV S gene. Black rectangle indicates the position of 3-nt deletions sites on PEDV genome.
From nucleotide identity analysis, the nucleotide of these 3 PEDV S2△strains from our study had 99.3%–99.8% identity to one another, while the identity of the remaining 7 strains was 98.3%–99.8% to one another. However, the identity between these 3 PEDV S2△strains and the remaining 7 strains were at 97.4%–98.5%. The 10 PEDV strains shared 96.9%–99.2% nucleotide identities with reference Pandemic strains, 95.2%–96.8% with reference INDEL strains and 93.5% -94.8% with reference Prototype strains, respectively. Moreover, the 3 PEDV S2△strains were closely related to reference Pandemic G2b strains with 97.6%–98.6% nucleotide identity. However, the remaining 7 strains from our study were closely related to reference Pandemic G2a strains with 97.6%–99.2% nucleotide identified.
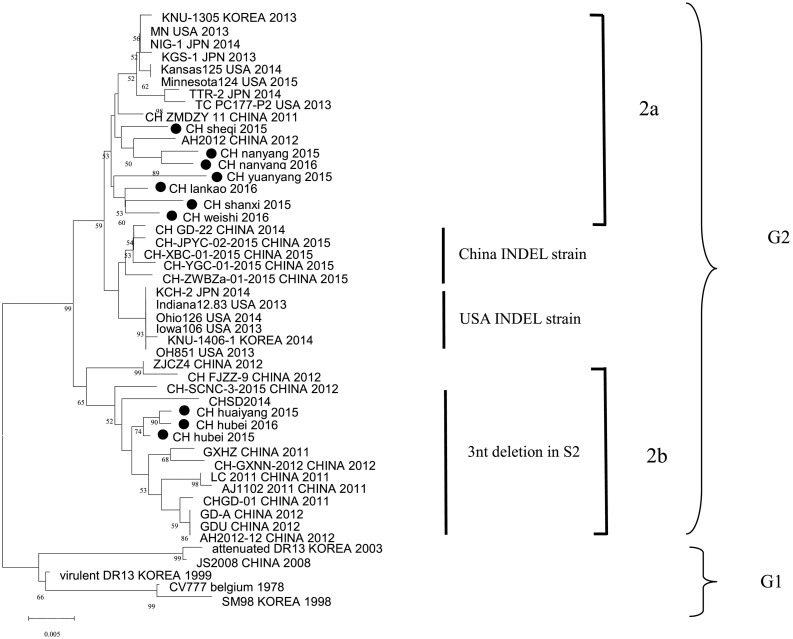
3.3. Phylogenetic analysis of PEDV S gene
The Fig. 2 phylogenetic tree was established based upon multiple sequences alignment of S gene according to (Huang et al., 2013). All PEDV strains fell clearly into two genetic groups, designated as G2 (Pandemic strains) and G1 (Prototype PEDV).
Fig. 2.
Phylogenetic tree based on the spike gene (S) sequence of PEDV strains were constructed using the neighbor-joining method in MEGA7 software. Numbers above branches indicate bootstrap p values calculated from 1000 bootstrap replicates; only values >50 are shown. ‘●’indicates the 10 isolates from this study.
Obviously, PEDV Pandemic strains (G2) consisted of two groups. In current study, these 3 PEDV S2△strains and other reference PEDV strains with 3 nt-deletion in S2 gene were grouped into G2b. The remaining 7 PEDV strains in this study were grouped into G2a. None of the strains isolated in this study were included in G1 (Prototype strain) and PEDV INDEL strain.
3.4. Sequence analyses of CH/hubei/2016 complete genome
After culturing on vero cell, only CH/hubei/2016 was successfully isolated, and its genome was future amplified and sequenced (Wang et al., 2013). CH/hubei/2016 of complete genome had 28063 nt. Like PEDV Pandemic strains, CH/hubei/2016 strain contained a notable U insertion at 48 nt, and two notable nucleotide deletion between 72 nt and 73 nt (an A deletion) and between 84 nt and 85 nt (a 4-nt UUCC deletion) at the 5’untranslated region (UTR) in comparison with the prototype CV777 strain (Huang et al., 2013).
From nucleotide identity analysis, CH/hubei/2016 strain had an identity at 98.2% with Pandemic strains G2b, 97.9%–98.0% with Pandemic strains G2a, 97.6%–97.7% with US INDEL strains, and 96.4%–97.2% with Prototype strains respectively.
3.5. Phylogenetic analysis of PEDV complete genome
As shown in Fig. 3 , all PEDV strains were clearly grouped into G1 and G2 group. PEDV Pandemic strains (G2) consisted of two groups. CH/hubei/2016 belonged to an independent subgroup which was more closely related to the US S INEDL and G2a strains than the reference strains with 3 nt deletion in S2 gene in the other subgroup.
Fig. 3.
Phylogenetic tree based on the complete genome of PEDV strains were constructed using the neighbor-joining method in MEGA7 software. Numbers above branches indicate bootstrap p values calculated from 1000 bootstrap replicates; only values >50 are shown. ‘●’indicates the one isolates from this study.
3.6. Recombination analysis
The analysis revealed that four putatively recombinant breakpoints in CH/hubei/2016 strain were detected. One recombination event of putative breakpoint was located at position 23310 to 24012 nt on S2 gene, which was supported by 6 programs (RDP, p = 1.961E-6; GENECONV, p = 1.400E-3; BootScan, p = 7.920E-5; MaxChi, p = 1.037E-3; Chimaera, p = 7.286E-5; SiScan, p = 3.351E-4; 3Seq, p = 4.564E-2). Additionally, phylogenetic trees based on the recombination region of each potential recombinant event and the non-recombinant region of each potential recombinant event displayed potential minor parent (CH-ZMDZY-11), potential major parent (AH2012-12) which had 3 nt-delection in S2 and potential recombinant strain evolutionary relationships (Fig. 4 ).
Fig. 4.
Detection of potential recombination events in CH hubei 2016 by RDP4. a One recombination breakpoints were located at 23310 to 24,12 nt. The analysis was performed with an RDP distance model, a window size of 1000 base pairs and a step size of 200 base pair; b Fast neighbor-joining (NJ) tree constructed using the recombinant region of each potential recombinant event; c Fast neighbor-joining (NJ) tree constructed using the non-recombinant region of each potential recombinant event.
3.7. Selection pressures in PEDV S gene
A ratio of dN/dS > 0 or dN/dS < 0 indicates positive and purify in selection, respectively, and a valued dN/dS = 0 indicates neutral evolution. Significant selection is p <0.1.The analysis revealed that PEDV strains isolated from central China from 2015 to 2016 had 7 purifying selection sites in S1; and there were 3 purifying selection sites in S2 (Fig. 5 ). No significant positive selected positions were found in S gene.
Fig. 5.
Identifying signatures of selection pressures in S gene of PEDV central China isolated. Imaginary lines indicate sites under significant selection (p < 0.1) and ‘●’shows the dN/dS estimates by site position. The x-axis refers to the amino acid position of S gene.
3.8. Sequence and phylogenetic analysis of PDCoV S gene
The S of PDCoV CH/HN/2016 had 3480 nt which was similar with reference strains (3480 nt or 3483 nt). The S gene homology of PDCoV CH/HN/2016 strain was measured by aligning with the other S genes of PDCoV strains from USA, Korea, and Thailand. The S gene of CH/HN/2016 had an identity 98.3%–98.9 with most of Chinese strains, 98.2%–98.4% with USA strains, 98.0% with the CHN-AH-2004, 97.5% with Asian leopard cat coronavirus and Chinese ferret badger coronavirus and 96% with all the stains from Thailand, respectively.
The phylogenetic analysis show that all PDCoV strains fell into G1 except Thailand strains belonged to G2 (Fig. 6 ). The PDCoV strains of China, USA and Korea had a closer relationship with Asian leopard cat and Chinese ferret-badger coronavirus than Thailand strains. In addition, PDCoV CH/HN/2016 from our study had a closer relationship with other PDCoV strains of China than CH/AH/2004. Further sequence analysis showed CHN/AH/2004 strain has 3-nt insertions at positions 153–155 (Wang et al., 2016).
Fig. 6.
Phylogenetic tree based on the spike gene (S) sequence of PDCoV strains were constructed using the neighbor-joining method in MEGA 7 software. Numbers above branches indicate bootstrap p values calculated from 1000 bootstrap replicates; only values >50 are shown. ‘●’indicates the one isolates from this study.
4. Discussion
Since 2012, PEDV has re-emerged and rapidly disseminated all over the world(Huang et al., 2013). In this study, the data indicates that PEDV was more frequently found than PDCoV among the tested samples. The positive rate of PEDV in all diarrhea samples tested was 84.2%, which was similar with previous study (Li et al., 2014). But, the positive rate of PDCoV was only 2.9% and lower than PDCoV in the south of China (Song et al., 2015).
From sequence and phylogenetic analyses of PEDV S gene, the 10 PEDV strains in our study from central China were clustered into G2a and G2b. The 3 PEDV S2△strains were grouped into G2b Pandemic strains, and the remaining 7 PEDV strains were grouped into G2a Pandemic strains (Fig. 2). In G2b, PEDV S2△strains (CH/hubei/2015, CH/hubei/2016, CH/huaiyang/2015) with a part of reference PEDVs strains possessed a same specific 3-nt deletion in S2 gene (Fig. 1). Interestingly, phylogenetic analysis of PEDV complete genomes showed that the PEDV S2△strain (CH/hubei/2016) was not grouped into same subgroup with the other reference PEDV strains possessed S2 3 nt deletion (Fig. 3). This phenomenon maybe explained by the fact that CH/hubei/2016 may result from the recombination between AH2012-12 (G2b) which had 3 nt-delection in S2 and CH-ZMDZY-11(G2a) (Fig. 4). Previous study revealed that recombination events in S1 gene between different PEDV groups may contribute to the genetic diversity of PEDV (Jarvis et al., 2016; Li et al., 2016; Sun et al., 2016). Furthermore, Taiwan New PEDV variants have two potential recombination breakpoints in S1 and S2 (Chiou et al., 2017). All these fact indicted that recombination, insertion and deletion in the S gene may contribute to the genetic diversity of PEDV. However, because of only one complete PEDV S2△strain genomes was included into the phylogenetic analysis, further study need to explore the evolution position of PEDV S2△strains.
Previous studies demonstrated that spike gene of Coronavirus is a determinant of virulence, and a single amino acid substitution can influence virulence (Macnamara et al., 2005; Sánchez et al., 1999). According to previous study, the deletion and insertion in PEDV S1 gene may affect the virulence of PEDV (Lin et al., 2015). Therefore, the specific 3 nt-deletion in PEDV S2 gene (Fig. 1) firstly reported in current study could also affect the pathogenicity and antigenicity of wild virus.
More effective purifying selections on RNA viruses than DNA viruses was proved and purifying selection is an important mechanism for adaption of RNA viruses (Hughes and Hughes, 2007). Our study demonstrated 10 purifying selection sites were in central China of PEDV S gene which indicated an adaptive evolution of PEDV in central China swine herd.
In current study, the strains with specific genetic signatures-3 nt deletion in S2 gene was firstly identified and reported (PEDV S2△strains), and one of strain (CH/hubei/2016) was clustered into different groups based on PEDV S gene or complete genomes. This fact may result from the recombination between AH2012-12 (G2b) which had 3 nt-delection in S2 and CH-ZMDZY-11(G2a). In contrast with central China of PEDV strains, PDCoV had low positive rates and had a close relationship with other PDCoV strains of China in 2012 to 2015. Future work should continue to trace the variability of PEDV strains and PDCoV strains.
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Acknowledgements
This work was supported by grants from National Key R&D Program (2016YFD0500704 and 2017YFD0501103) and Program of Henan finance (201776-21).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.rvsc.2018.06.001.
Appendix A. Supplementary data
Supplementary material: Information about PEDV strains used in this study.
References
- Chasey D., Cartwright S.F. Virus like particles associated with porcine epidemic diarrhoea. Res. Vet. Sci. 1978;25:255–256. doi: 10.1016/S0034-5288(18)32994-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Wang C., Shi H., Qiu H., Liu S., Chen X., Zhang Z., Feng L. Molecular epidemiology of porcine epidemic diarrhea virus in China. Arch. Virol. 2010;155:1471–1476. doi: 10.1007/s00705-010-0720-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Yu X., Wang L., Wu J., Zhou Z., Ni J., Li X., Zhai X., Tian K. Two natural recombinant highly pathogenic porcine reproductive and respiratory syndrome viruses with different pathogenicities. Virus Genes. 2013;46:473–478. doi: 10.1007/s11262-013-0892-4. [DOI] [PubMed] [Google Scholar]
- Chiou H.Y., Huang Y.L., Deng M.C., Chang C.Y., Jeng C.R., Tsai P.S., Yang C., Pang V.F., Chang H.W. Phylogenetic analysis of the spike (S) gene of the new variants of porcine epidemic Diarrhoea virus in Taiwan. Transbound. Emerg. Dis. 2017;64:157–166. doi: 10.1111/tbed.12357. [DOI] [PubMed] [Google Scholar]
- Huang Y.W., Dickerman A.W., Pineyro P., Li L., Fang L., Kiehne R., Opriessnig T., Meng X.J. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. MBio. 2013;4:e00713–e00737. doi: 10.1128/mBio.00737-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes A.L., Hughes M.A. More effective purifying selection on RNA viruses than in DNA viruses. Gene. 2007;404:117–125. doi: 10.1016/j.gene.2007.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis M.C., Lam H.C., Zhang Y., Wang L., Hesse R.A., Hause B.M., Vlasova A., Wang Q., Zhang J., Nelson M.I., Murtaugh M.P., Marthaler D. Genomic and evolutionary inferences between American and global strains of porcine epidemic diarrhea virus. Prev. Vet. Med. 2016;123:175–184. doi: 10.1016/j.prevetmed.2015.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.Y., Song D.S., Park B.K. Differential detection of transmissible gastroenteritis virus and porcine epidemic diarrhea virus by duplex RT-PCR. J. Vet. Diagn. Investig. 2001;13:516–520. doi: 10.1177/104063870101300611. [DOI] [PubMed] [Google Scholar]
- Li R., Qiao S., Yang Y., Su Y., Zhao P., Zhou E., Zhang G. Phylogenetic analysis of porcine epidemic diarrhea virus (PEDV) field strains in Central China based on the ORF3 gene and the main neutralization epitopes. Arch. Virol. 2014;159:1057–1065. doi: 10.1007/s00705-013-1929-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Qiao S., Yang Y., Guo J., Xie S., Zhou E., Zhang G. Genome sequencing and analysis of a novel recombinant porcine epidemic diarrhea virus strain from Henan, China. Virus Genes. 2016;52:91–98. doi: 10.1007/s11262-015-1254-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.M., Annamalai T., Liu X., Gao X., Lu Z., El-Tholoth M., Hu H., Saif L.J., Wang Q. Experimental infection of a US spike-insertion deletion porcine epidemic diarrhea virus in conventional nursing piglets and cross-protection to the original US PEDV infection. Vet. Res. 2015;46:134. doi: 10.1186/s13567-015-0278-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Zhang Y., Liang X., Lou F., Oglesbee M., Krakowka S., Li J. Origin, evolution, and virulence of porcine deltacoronaviruses in the United States. MBio. 2015;6 doi: 10.1128/mBio.00064-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnamara K.C., Chua M.M., Phillips J.J., Weiss S.R. Contributions of the viral genetic background and a single amino acid substitution in an Immunodominant CD8(+) T-cell epitope to murine coronavirus Neurovirulence. J. Virol. 2005;79:9108–9118. doi: 10.1128/JVI.79.14.9108-9118.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu C., Lu X., Duan E., Chen J., Li W., Zhang F., Martin D.P., Yang M., Xia P., Cui B. Molecular evolution of porcine reproductive and respiratory syndrome virus isolates from Central China. Res. Vet. Sci. 2013;95:908–912. doi: 10.1016/j.rvsc.2013.07.029. [DOI] [PubMed] [Google Scholar]
- Pensaert M.B., de Bouck P. A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 1978;58:243–247. doi: 10.1007/BF01317606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez C.M., Izeta A., Sánchez-Morgado J.M., Alonso S., Sola I., Balasch M., Plana-Durán J., Enjuanes L. Targeted recombination demonstrates that the spike gene of transmissible gastroenteritis coronavirus is a determinant of its enteric tropism and virulence. J. Virol. 1999;73:7607–7618. doi: 10.1128/jvi.73.9.7607-7618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D., Park B. Porcine epidemic diarrhoea virus: a comprehensive review of molecular epidemiology, diagnosis, and vaccines. Virus Genes. 2012;44:167–175. doi: 10.1007/s11262-012-0713-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D., Zhou X., Peng Q., Chen Y., Zhang F., Huang T., Zhang T., Li A., Huang D., Wu Q., He H., Tang Y. Newly emerged porcine Deltacoronavirus associated with Diarrhoea in swine in China: identification, prevalence and full-length genome sequence analysis. Transbound. Emerg. Dis. 2015;62:575–580. doi: 10.1111/tbed.12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M., Ma J., Wang Y., Wang M., Song W., Zhang W., Lu C., Yao H. Genomic and epidemiological characteristics provide new insights into the phylogeographical and spatiotemporal spread of porcine epidemic diarrhea virus in Asia. J. Clin. Microbiol. 2015;53:1484–1492. doi: 10.1128/JCM.02898-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D., Wang X., Wei S., Chen J., Feng L. Epidemiology and vaccine of porcine epidemic diarrhea virus in China: a mini-review. J. Vet. Med. Sci. 2016;78:355–363. doi: 10.1292/jvms.15-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasova A.N., Marthaler D., Wang Q., Culhane M.R., Rossow K.D., Rovira A., Collins J., Saif L.J. Distinct characteristics and complex evolution of PEDV strains, North America, may 2013-February 2014. Emerg. Infect. Dis. 2014;20:1620–1628. doi: 10.3201/eid2010.140491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.M., Niu B.B., Yan H., Gao D.S., Yang X., Chen L., Chang H.T., Zhao J., Wang C.Q. Genetic properties of endemic Chinese porcine epidemic diarrhea virus strains isolated since 2010. Arch. Virol. 2013;158:2487–2494. doi: 10.1007/s00705-013-1767-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Hayes J., Sarver C., Byrum B., Zhang Y. Porcine deltacoronavirus: histological lesions and genetic characterization. Arch. Virol. 2016;161:171–175. doi: 10.1007/s00705-015-2627-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C.Y., Lau S.K.P., Lam C.S.F., Lau C.C.Y., Tsang A.K.L., Lau J.H.N., Bai R., Teng J.L.L., Tsang C.C.C., Wang M., Zheng B.-J., Chan K.-H., Yuen K.-Y. Discovery of seven novel mammalian and avian coronaviruses in the genus Deltacoronavirus supports bat coronaviruses as the gene source of Alphacoronavirus and Betacoronavirus and avian coronaviruses as the gene source of Gammacoronavirus and Deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood E.N. An apparently new syndrome of porcine epidemic diarrhoea. Vet. Rec. 1977;100:243–244. doi: 10.1136/vr.100.12.243. [DOI] [PubMed] [Google Scholar]
- Zhai S.L., Wei W.K., Li X.P., Wen X.H., Zhou X., Zhang H., Lv D.H., Li F., Wang D. Occurrence and sequence analysis of porcine deltacoronaviruses in southern China. Virol. J. 2016;13:136. doi: 10.1186/s12985-016-0591-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material: Information about PEDV strains used in this study.