Abstract
The spatial separation of DNA replication and gene transcription in the nucleus and protein translation in the cytoplasm is a uniform principle of eukaryotic cells. This compartmentalization imposes a requirement for a transport network of macromolecules to shuttle these components in and out of the nucleus. This nucleo‐cytoplasmic transport of macromolecules is critical for both cell physiology and pathology. Consequently, investigating its regulation and disease‐associated alterations can reveal novel therapeutic approaches to fight human diseases, such as cancer or viral infection. The characterization of the nuclear pore complex, the identification of transport signals and transport receptors, as well as the characterization of the Ran system (providing the energy source for efficient cargo transport) has greatly facilitated our understanding of the components, mechanisms and regulation of the nucleo‐cytoplasmic transport of proteins in our cells. Here we review this knowledge with a specific emphasis on the selection of disease‐relevant molecular targets for potential therapeutic intervention.
Keywords: anti‐cancer therapy, anti‐viral therapy, karyopherins, nuclear export, nuclear import, nuclear pore complex, nuclear trafficking
The nucleo‐cytoplasmic transport of macromolecules is critical for cell physiology and pathology. Consequently, investigating its regulation and disease‐associated alterations can reveal novel therapeutic approaches to fight human diseases, such as cancer or viral infection. Here, we review our understanding of the components, mechanisms and regulation of this fundamental process with a specific emphasis on the selection of disease‐relevant molecular targets.
Accompanied by a podcast, http://www.yada-yada.co.uk/Blackwell/PAI/Audio/Wolfgang_Jan2015.mp3. Or http://itunes.apple.com/gb/podcast/febs-journal-podcast/id439631367
Abbreviations
- CRM1
exportin1 or Xpo1, chromosome region maintenance 1
- EM
electron microscopy
- FG
phenylalanineglycine
- FG‐Nups
phenylalanineglycine nucleoporins
- Imp‐α
importin‐α
- Imp‐β
importin‐β
- NE
nuclear envelope
- NES
nuclear export signal
- NF‐κB
nuclear factor‐κB
- NLS
nuclear localization signal
- NPC
nuclear pore complex
- NTR
nuclear transport receptor
- Nups
nucleoporins
- POMs
Pore membrane proteins
- PY‐NLS
proline‐tyrosine NLS
Introduction
The deregulation of the nuclear import and export machinery is a key marker of various diseases 1, 2. For example, many tumor suppressor transcription factors show cytoplasmic sequestration ablating their nuclear functions allowing, for example, uncontrolled cell division 3. The aberrant localization of onco‐proteins can also lead to their inadequate activation. Examples of aberrant nucleo‐cytoplasmic shuttling that correlates with tumor formation, progression or resistance to treatment include p53, FOXO3a, p27, BRCA1, APC, nucleophosmin retinoblastoma, β‐catenin, nuclear factor‐κB (NF‐κB), survivin and cyclin D1 3, 4, 5. Beyond cancer, many viruses including human HIV‐1, influenza A, dengue, respiratory syncytial virus, rabies, Rift Valley fever virus and Venezuelan equine encephalitis virus rely on the transport of specific viral proteins into the host cell nucleus to perturb the anti‐viral response. Therefore, inhibiting the nuclear trafficking of viral proteins has been proposed as a viable therapeutic strategy 6. Over the last decade, the research community has acquired a critical mass of significant knowledge on the constituents of the nuclear transport machinery. The emerging picture, while intriguingly complex, suggests that there is an availability of a broad range of molecular targets enabling specific therapeutic interventions. The transport of cargo proteins is mediated by several distinct types of transport signals that are recognized by specific transport receptors or via a variety of adaptor proteins. These transport receptors can then interact with components of the nuclear pore complex (NPC) and with the Ras family GTPase Ran. Many of the steps within this process have the potential to be therapeutically targeted for innovative anti‐cancer and anti‐viral therapies.
The nuclear envelope and the nuclear pore complex
The nucleus is surrounded by an envelope composed of two phospholipidic membranes: an outer and an inner nuclear membrane that are 30 nm apart. The nuclear envelope (NE) provides a much stronger physical barrier than the single cordon of the plasma membrane. The outer nuclear membrane is continuous with the endoplasmic reticulum 7, whereas the inner nuclear membrane is associated with a network of intermediate filaments composed of lamin called the nuclear lamina. This acts as a site of attachment for chromosomes and as a shield for the nucleus 8. The NE functions like a selectively permeable barrier allowing macromolecules to move between the nucleus and the cytoplasm via a gatekeeper, the NPC. The NPC is a huge protein complex that fuses the internal and external nuclear membrane to form an aqueous channel (Fig. 1). The NPC is cylindrical measuring 100–150 nm in diameter and 50–70 nm in thickness 9 and is broadly conserved in eukaryotes 10, 11. The molecular mass of the NPC is approximately 125 000 kDa and the number of NPCs per nucleus is highly variable among organisms. The average number of NPCs within a vertebrate cell is between 2000 and 5000 12 and has been extensively studied by electron microscopy (EM). In particular the development of cryo‐EM and cryo‐electron tomography (cryo‐ET) allowed structural preservation to be enhanced and purification steps to be minimized and as a consequence the detailed and artifact‐free analysis of the NPC 12, 13, 14, 15. These EM‐based studies revealed a highly modular and dynamic structure with a doughnut‐shaped central core with an eightfold rotational symmetry 16. A central channel is surrounded by three ring‐like structures, namely the cytoplasmic ring, the central spoke ring and the nuclear ring. Attached to this core structure are eight protein filaments on the cytoplasmic side and eight protein filaments on the nuclear side that converge to a ring‐like structure termed the nuclear basket.
Figure 1.
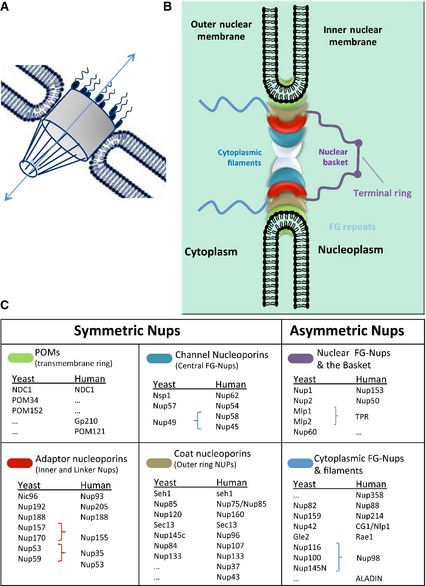
Overall structure and molecular composition of the nuclear pore complexes (NPCs). (A) General structural features of the NPC. (B) A schematic model of the NPC. In this model, the NPC is divided into several groups according to their location and structural characteristics. The symmetrical core is composed of membrane‐anchored POMS (transmembrane ring), channel Nups (central FG‐Nups) and scaffold Nups composed by adaptor Nups (inner and linker Nups) and coat Nups (outer ring). Asymmetric parts of the pore are the nuclear FG‐Nups and the basket plus the cytoplasmic FG‐Nups and filaments. (C) The yeast and vertebrate homolog Nups that are known to constitute each NPC substructure are listed. Symmetric Nups are equally distributed on the cytoplasmic and nucleoplasmic parts of the NPC and form the core region. Asymmetric Nups form the nuclear basket and the cytoplasmic filaments. They serve as docking sites for transport factors and include associated mRNA export factors. See the main text for more information.
The NPC is composed of 30 different proteins termed nucleoporins (Nups) 17 that are organized into several sub‐complexes, each of which is present in multiple copies, resulting in approximately 500–1000 individual proteins in the fully assembled NPC 18. The transport of salts, nucleotides, small molecules/proteins and components required for the syntheses of DNA and RNA occurs passively by diffusion through the NPC. In contrast, proteins larger than 40–65 kDa must be transported into the nucleus through the NPC with the assistance of transport receptors. These receptors recognize the transport signals that are present on cargo proteins allowing their import (reviewed in 19). The key function of the NPC is to form a diffusion barrier between the cytoplasm and the nuclear compartment and to enable the nucleo‐cytoplasmic traffic of macromolecules. In addition, the NPC is also involved in other nuclear processes that include DNA repair 20, the cell cycle 21, chromatin organization 22, transcription regulation 23, 24, epigenetic memory 25 and RNA maturation and quality control 26.
Nucleoporin proteins
Despite its enormous dimensions, the NPC is built from a surprisingly small number of proteins called nucleoporins (Nups) 18. The NPC displays a high degree of internal symmetry and can be divided into a symmetric part, enclosed in the nuclear membrane, and an asymmetric part, with extensions into the nucleus or cytoplasm (Fig. 1). Nups from the symmetric part of the NPC are generally classified into three categories: membrane‐anchored (POMs, part of the nuclear envelope), scaffold (coat Nups and adaptor Nups) and channel (barrier Nups). Each category has unique structural features that are essential to execute specific functions. The Nups that form the asymmetric part of the NPC are called nuclear basket Nups and cytoplasmic filament Nups.
Membrane Nups
Membrane Nups (POMs) anchor the symmetric part of the NPC to the pore membrane where the inner and outer nuclear membranes fuse to form the nuclear pore binding the assembly complex to the NE. Membrane Nups contain transmembrane α‐helices that allow the protein to anchor onto the membrane while large regions extend toward the luminal and pore sides of the membrane 27, 28.
Scaffold Nups
The scaffold Nups (coat Nups and adaptor Nups) form the skeleton of the NPC connecting the membrane Nups to the barrier Nups. The scaffold Nups contain mainly α‐solenoid and β‐propeller folds and are classified into outer ring Nups, inner ring Nups and linker Nups according to their location and function. The high flexibility of the outer ring Nup components allows for conformational changes of the scaffold that enable large cargo to pass through the central channel 29.
Barrier Nups
Barrier Nups (channel Nups) are phenylalanineglycine Nups (FG‐Nups) and form the innermost cylindrical layer that acts as a selective gatekeeper for nuclear transport regulation. FG‐Nups contain multiple stretches of FG sequences that form intrinsically disordered regions. These motifs are present in about one‐third of Nups forming an unstructured meshwork lining the central channel. The tentacle‐like structures provide several low affinity, high specificity interactions with transport receptors that escort cargo proteins through the nuclear pore. Collectively, FG‐rich regions build the diffusion barrier of the NPC.
Asymmetric Nups
Asymmetric Nups (formed by nuclear basket Nups and cytoplasmic filament Nups) are key components in establishing the directionality of the nucleo‐cytoplasmic transport process. These structures mediate specific interactions with transport complexes and several asymmetric Nups that contain FG repeats serving as binding sites with important roles in cargo–NPC interactions.
The transport signals
For nuclear import and export, macromolecules generally require specific transport signals, namely a nuclear localization signal (NLS) or a nuclear export signal (NES) (Fig. 2). Soluble transport receptors of the karyopherin family of proteins (known as importins and exportins) recognize these sequences within macromolecules. NLS and NES can be defined as sequences within proteins that are necessary and sufficient for their import/export (summarized in Table 1). These sequences bind transport receptors either directly or via adaptor molecules and enable the release of the transport complex at the end of the translocation process 30. The transport signals that interact with importin‐α (Imp‐α), importin‐β (Imp‐β), CRM1 (chromosome region maintenance 1, also known as exportin1 or Xpo1) and transportin‐1 (also known as karyopherin‐β2) are well described but they remain to be determined for the other 16 human karyopherin‐βs 31, 32, 33, 34, 35. The known NLSs can be classified into either classical NLSs or non‐classical NLSs. The classical NLSs can be further divided into monopartite or bipartite NLSs. The non‐classical NLSs include proline‐tyrosine NLSs (PY‐NLSs) that allow the cargo protein to bind to karyopherin‐β2 that mediates the direct interaction with Imp‐β. For nuclear export, the leucine‐rich NES (recognized by CRM1) is the most extensively characterized export signal while the structure of the CRM1‐snurportin1 and RanGTP complex has been elucidated 36, 37.
Figure 2.
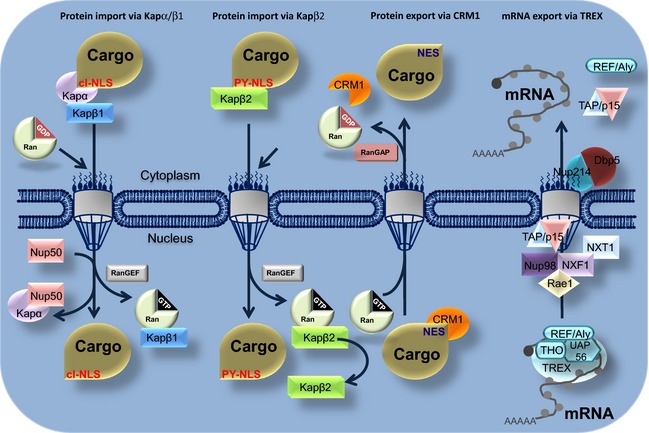
Schematic overview of Ran‐dependent nucleo‐cytoplasmic transport. Nuclear export. CRM1 exports a great part of NES‐containing protein. Nuclear import. Importin‐α (Imp‐α)/importin‐β (Imp‐β) heterodimer (designated as α and β) and karyopherin‐β2 mediate the import of NLS‐containing proteins. See the main text for details.
Table 1.
Examples of different types of nuclear transport signals
| Amino acid sequence | Protein | Type of signal | References |
|---|---|---|---|
| PKKKRKV | SV40 T antigen | Classical NLS | 33 |
| KRx{10}KKKL | Nucleoplasmin | Bipartite NLS | 31, 33 |
| VRILESWFAKNIENPYLDT | Matα2 | Polar/nonpolar residues NLS | 196 |
| PAAKRVKLD | c‐Myc | cMyc‐NLS | 197 |
| YNDFGNYNNSSNFGPMKGGNFGGRSSGPYGGGGQY | hnRNP A1 (M9 sequence hydrophobic subclasses) | hPY‐NLS | 198 |
| KVSRRG‐GHQNSYKPY | hnRNP D (basic enriched) | bPY‐NLS | 45 |
| RQARRNRRRRWR | VIH Rev protein | Arginine‐rich NLS | 199 |
| DNSQRFTQRGGGAVGKNRRGGRGGNRGGRNNNSTRFNPLAK | Nab2p | Arginine/glycine‐rich NLS | 200 |
| KTPGKKKKGK | Parathyroid hormone‐related protein (PTHrP) | Lysine‐rich‐NLS | 201 |
| QDLNSTAAPHPRLSQYKSKYSSLEQSERRRRL | Snurportin1 | UsnRNPs‐NLS | 202 |
| LPPLERLTL | HIV Rev | Hydrophobic‐NES | 32 |
| LALKLAGLKI | PKI | NES | 35, 203 |
| LCQAFSDVIL | Cyclin B1 | NES | |
| LQKKLEELEL | MAPKK | NES | 204 |
| LAEMLEDLHI | NMD3 | NES | 77 |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Classical NLSs
The first nuclear transport signals were described in the SV40 large T antigen and nucleoplasmin in the early 1980s comprising a short lysine‐rich sequence classified as classical NLSs 31, 38. These bind the armadillo (ARM) domain in the C terminus of Imp‐α. The adaptor protein Imp‐α also binds the transport receptor Imp‐β through its N‐terminal αIBB domain forming a ternary complex 39. The small size and relatively simple sequence patterns of these basic NLSs have facilitated identification of similar signals in many proteins. The classical NLSs contain one or two clusters of positively charged amino acids, typically lysine or arginine, and are divided into two classes, monopartite and bipartite classical NLSs 40. The monopartite classical NLS is a short and highly basic signal. The two stretches of basic amino acids within a bipartite classical NLS are separated by a linker region that is usually 10–12 residues long, although longer linker sequences have been reported 41. In addition, several atypical NLSs that bind to classical NLS binding sites in Imp‐α have been characterized, including the hydrophobic NLS from phospholipid scramblase 1 40.
Non‐classical NLSs
Some cargo proteins bypass the requirement for an adaptor protein and bind directly to transport receptors through non‐classical NLSs. Proteins that are directly recognized by Imp‐β include ribosomal proteins, CREB, the human immunodeficiency virus (HIV) Rev and Tat, SREBP‐2, the human T‐cell leukemia virus type 1 (HTLV‐1) protein Rex, PTHrP, cyclin B1, Smad3, TRF and SRY 40. In contrast to the interaction between classical NLS and Imp‐α, proteins that bind to Imp‐β directly do not obey strict rules and the NLSs that confer Imp‐β recognition vary significantly in both size and charge 40.
Many proteins that are imported into the nucleus can bind directly to the transport receptor transportin‐1. Common characteristics between the apparently disparate signals recognized by transportin‐1 were described unifying them into a new class of NLS termed PY‐NLS 34. The consensus motif of PY‐NLS consists of a loose N‐terminal hydrophobic motif, a central arginine residue and a C‐terminal PY sequence 42. The physical rules that describe the PY‐NLSs as structurally disordered in free cargoes, positively charged and with weak consensus motifs predicted approximately 100 candidate transportin‐1 cargoes 34. Several of these have been confirmed experimentally 34, 43, 44. Interestingly, arginine–glycine‐rich NLSs known as RG‐NLSs in the yeast proteins Hrp1 and Nab2 and the 38 amino acid long NLS of the hnRNP A1 protein, designated M9, were also shown to have the same characteristics as the PY‐NLS 42, 45. Recently other non‐classical NLSs such as the extensive coiled‐coil domain of STAT5a 46 have been characterized. Furthermore, a number of proteins have been identified that contain both classical and non‐classical NLS motifs that can interact directly with both Imp‐α and Imp‐β family members.
Leucine‐rich NESs containing cargoes
The first signals that direct the nuclear export of a protein were identified in HIV Rev and protein kinase inhibitor A 32, 35. The consensus sequence for NES is ϕ1‐X(2–3)‐ϕ2‐X(2–3)‐ϕ3‐X‐ϕ4 (where ϕ represents one of the hydrophobic residues L, V, I, F or M, and X can be any amino acid but preferentially is charged, polar or a small amino acid) 47, 48, 49.
More complex export signatures incorporate the three‐dimensional features of the whole protein that are recognized and targeted for export 50, 51. Proteins containing these NESs are recognized and exported by CRM1 52, 53, 54, 55 (Fig. 2). Güttler et al. showed that CRM1 contains five pockets for binding the conserved hydrophobic residues of NESs. Accordingly, a structure‐based NES consensus with an additional hydrophobic position (five instead of four) has been proposed 56. Kosugi et al. generated a large number of NES peptides in a random peptide library screen and delineated multiple distinct consensus sequences that described more than 80% of known functional NESs 57. A predictor, NESsential, that uses sequence derived meta‐features, such as predicted disorder and solvent accessibility, in addition to a primary sequence that can identify promising NES‐containing candidate proteins 58, 59, http://validness.ym.edu.tw/index.php. Other recently developed NES predictor tools include wregex, which uses position‐specific scoring matrices for motif prediction 60, and nesmapper (http://sourceforge.net/projects/nesmapper), which has been developed based on the activity profile of all classes of NESs 61.
Nuclear transport receptors
Proteins larger than 40 kDa are transported through the NPC by soluble nuclear transport receptors (NTRs). NTRs continuously shuttle between the nucleus and the cytoplasm, bind their cargo on one side of the nucleus and release it on the other side. The majority of the nuclear‐cytoplasmic transport receptors are members of the β‐karyopherin protein family with each member recognizing a unique group of cargo proteins or RNAs (summarized in Table 2) 62. There are 22 putative members of the karyopherin family in humans 63 that share only modest sequence homology with the greatest homology noted within their Ran‐binding domain 64. A specific architecture within the karyopherin family is the tandem HEAT repeat fold formed by anti‐parallel helices that are linked by a short intra‐repeat loop 65. These repetitive structures are inherently flexible and contribute substantially to Ran‐controlled cargo recognition and cargo release 66. Karyopherin‐β proteins can either directly or indirectly, through adaptors, interact with their cargo. In addition there are six homologs of adaptor proteins that belong to the Imp‐α protein family that have been identified in humans which adds a further level of complexity to this process 67. For example, Imp‐β mediated nuclear import of the uridine‐rich small nuclear RNPs involves a different adaptor protein called snuportin‐1 and, as in the case of Imp‐α, snuportin‐1 binds Imp‐β through an IBB domain. The karyopherin‐β proteins are multi‐domain transport factors that contain a cargo binding domain, an NPC binding domain 68 and a binding domain for the small Ras‐like GTPase Ran 69, 70. Family members contain both import and export receptors; however, only a few of them have been functionally characterized in higher eukaryotes (indicated in Table 2).
Table 2.
Transport receptors and cargoes
| Vertebrate karyopherins | Saccharomyces cerevisiae karyopherins | Cargoes (selection) vertebrate (V) yeast (Y) |
|---|---|---|
| Imp‐β/Kapβ1 | Kap95 | Classical NLS via Imp‐α; cyclin B1; snurportin1; SRY; PTHrP; CREB, AP‐1, TRF1, Smad3; SREBP‐2; HTLV‐1 Rex, HIV‐1 Tat, HIV‐1 Rev; NF‐YA; adenovirus core protein pVII; aristaless/arx; cJun; H1; HPV16E6; H1; H2A; H2B; H3; H4; rPL23a, rPS7, rPL5, rPL18a, rPL6, rPL4; PP2A (PR65) |
| Kapβ2 (Transportin/Transportin‐2) | Kap104 | (V) PY‐NLS cargoes; PQBP‐1, YBP1, PABP2, EWS, FUS, SAM68, hnRNP M, hnRNP A1, hnRNP A0, hnRNP A2, hnRNP A3, hnRNP D, hnRNP F, JKTP‐1, TAP (NXF1), HuR, HEXIM1, RB15B, Clk3, WBS16, Cyclin T1, TAFII68, CPSF6, HCC1, ETLE; tfg2p non PY‐NLS cargoes: TAFI48; NPM‐ALK, SRP19; H2A; H2B; H3; H4; c‐Jun; rPL23a, rPS7, rPL5; adenovirus core protein pVII; HIV‐1 Rev; HPV16 E6, HPV16 L2, HPV18 L2 (Y) Nab2p; Hrp1p; Tfg2p |
| Importin‐5 (Kapβ3 or RanBP5) | Kap121 (Pse1) | (V) p60TRP;Rag‐2; PGC7/Stella; apolipoprotein A‐I; influenza A PB1‐PA; HPV18 L2; HPV16 L2; CDK5 activator p35; TAFI48, c‐Jun; HIV‐1 Rev; rPL23a, rPS7, rPL5, rPS3a; H2A; H2B; H3; H4 (Y) Aft1p; Asr1p; Egd1p; Nop1p, Nup53p; Pdr1p; Pho4p; Sas2p; Sof1p; Spo12p; Ste12p; Yap1p; Yra1p; secondary pathway for histones, ribosomal proteins, Ho, SRP, TBP |
| Importin‐4 (RanBP4) | Kap123 | (V) Vitamin D receptor, TP2, HIF1‐α, rPS3a (Y) Egd1p; H3 (Hht2p, H4 (Hhf2p); Sas2p; SRP; Rpl25p, Rp10a, Rps1p, Rpl4p, Rpl15p, Rpl16p, Rpl18p,Rpl25p, Rpl41p; secondary pathway for Asr1p, Asf1p, H2A, H2B, Htz1p, Yap1p, Yra1p, TBP |
| Importin‐9 | Kap114 | (V) Hepatocellular carcinoma associated protein, HSP27, rPS3, rPS9, rPL19, rPL18a, rPS7, rPL6, rPL4; c‐Jun; H2A, H2B, H3, H4; aristaless (Arx); PP2A (PR65); (Y) H2A (Hta1p), H2B (Htb1p, Nap1p, TBP (Spt15p); TFIIB Sua7p); rfp1; secondary pathway for Asr1p |
| Importin‐7 | Kap119 (Nmd5) | (V) Proline‐rich homeodomain; EZI; RK‐2, MEK1, Smad3; HIV‐1 integrase; CDK5 activator p35; HIF1‐α; c‐Jun; glucocorticoid receptor;HIV‐1 Rev; rPL23a; rPS7; rPL5; H2A, H2B, H3, H4; Imp‐β/‐7 heterodimer: H1; HIV‐1 integrase, adenovirus core protein pVII; rPL6, rPL4, rPS3a; (Y) Crz1p; Gal4p; Hog1p; Ssa4p; TFIIS (Dst1p), Rpf1p; secondary pathway for histones H3 and H4 |
| Importin‐8 (RanBP8) | Kap108 (Sxm1) | (V) Ago2; Smad4, Smad1; NPM‐ALK; SRP19; (Y) Lhp1p, Pab1p, Rpl16p, Rpl25p, Rpl34p; secondary pathway for Ho, histones H3 and H4 |
| Transportin‐SR (‐SR2/‐3/TNPO3) | Kap111 (Mtr10) | (V) ASF/SF2, SC35, TRA2α, TRA2β; HPV E2, RBM4, ALEX3, BAB71287, BAP1, MLF2, ODF2; dASF, dSC35, d9G8, Rbp1, B52, RSF1; HIV1 IN; (Y) Gbp2p; Hrb1p; Npl3p; tRNAs |
| – | Kap122 (Pdr6p) | (Y) Imports sc‐cargo – Toa1 and Toa2, TFIIA |
| Importin‐13 | – | NF‐YB/NF‐YC; NC2α/NC2β; Myopodin, hUBC9, elF1A Y14‐Mago; glucocorticoid receptor CHRAC‐15/CHRAC‐17, p12/CHRAC‐17; PAX6, Pax3, Crx; Aristaless (Arx); rPL5; histone fold heterodimers |
| CRM1 (Exportin‐1) | CRM1 (Xpo1/Kap124p) | Leu‐rich NES cargoes; HIV genomic RNA; m7G‐ capped UsnRNAs; 40S and 60S pre‐ribosomal subunits via NMD3 adaptor; snurportin1 (SPN1) |
| CAS (Exportin‐2) | Kap109 (Cse1) | Imp‐αs (Y) Kap60/Srp1 |
| Exportin‐4/Bidirectional NTRs | – | Sox‐2, SRY; eIF5A, Smad3 |
| Exportin‐5 | Kap142 (Msn5) | tRNA, eEF1A (via aa‐tRNA); dsRNA‐binding proteins (via dsRNA); pre‐miRNAs; 60S pre‐ribosomal subunits |
| Exportin‐6 | – | Actin‐profilin complexes |
| Exportin‐7 (RanBP16) | – | p50‐RhoGAP, 14‐3‐3‐σ |
| Exportin‐t (Xpo‐t) | Kap127 (Los1) | tRNA |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Import receptors
A protein that contains a classical NLS is bound by the NLS binding pocket formed by the armadillo repeats within the Imp‐α adaptor protein. The transport receptor Imp‐β binds to Imp‐α and then targets the NLS‐containing protein into and through the NPC. Intriguingly, Imp‐β has been shown to contain two binding sites for FG‐rich motifs that are located away from the cargo binding site 71. Most karyopherin‐β proteins do not rely on adaptors to bind to cargoes as they can directly interact with basic NLSs of core histones and ribosomal proteins or arginine–glycine‐rich NLSs of some RNA binding proteins.
Export receptors
CRM1 (chromosome region maintenance 1)
The most extensively characterized export receptor is chromosome region maintenance 1 (CRM1), a member of the karyopherin‐β family of receptor proteins. CRM1 exports proteins that contain leucine‐rich NESs from the nucleus into the cytoplasm 53, 55. CRM1 has 20 HEAT domains that allow RanGTP to bind. Cargo binding takes place outside of this HEAT domain ring in a hydrophobic cleft producing a generic NES docking site 56. CRM1 recognizes NESs that are present in a large range of proteins that are structurally unrelated 72. CRM1 can also be recruited by adaptor molecules in situations where it does not bind directly to a protein that is to be exported from the nucleus (e.g. Exp5 cooperates with CRM1 to export large ribosomal subunits 73). CRM1 also participates in the export of the 40s and 60s pre‐ribosomal subunits as well as essential RNPs 74, 75, 76, 77. With two adaptors [the Cap binding complex (CBC) complex and PHAX] CRM1 is also essential for the maturation of the spliceosomal U snRNPs 78, 79. Furthermore, CRM1 actively maintains the exclusive cytoplasmic localization of RanBP1, RanGAP and other translation factors that contribute to the identity of the nuclear compartment 80, 81, 82, 83. Another key function of CRM1 is its role in SPN1 recycling (an adaptor for U snRNPs) back to the cytoplasm 51. Intriguingly, overexpression of CRM1 has been reported in various tumor types and has been correlated with poor prognosis and resistance to therapy 4, 5. In addition CRM1 is hijacked during infection by many viruses 32, 84, 85, 86.
Other exportins
In addition to CRM1, a number of other alternative export receptors have been characterized. Exportin 2 recycles Imp‐α from the nucleus into the cytoplasm allowing Imp‐α to mediate another round of nuclear import if required 87, 88, 89. Together with the adaptor STRADα exportin 7 regulates the distribution of LKB1 kinase 90 and regulates the leakage of RhoGAP1 and 14‐3‐3‐σ into the nucleus 91. Exportins are also dedicated to RNA transit. For example, exportin‐t is dedicated to export fully mature tRNA with the 5′‐ and 3′‐ends correctly processed 92, 93, 94, 95. A second tRNA (alone or with eEF1A) exporter is exportin 5 which displays a different binding specificity compared to exportin‐t 80, 96. Exportin 5 also exports double strand RNA, pre‐miRNAs 97, 98, 99 and cooperates with CRM1 as described previously. Exportin 4 exports eIF5A and Smad3 but can also act as an importin for Sox‐type transcription factors 100, 101. Importin 13, despite its namesake, also demonstrates a bi‐directional transit capability directing eIF1A nuclear export and the import of the heterodimer component of the exon junction complex Mago‐Y14 102, 103.
Alternative nuclear transport pathways
While most proteins are transported through the NPC by conventional karyopherin‐β mediated mechanisms, alternative nuclear transport pathways have been described for a number of proteins. These karyopherin‐β‐independent pathways include transport by alternative carriers such as the calcium‐binding proteins calmodulin and calreticulin and translocation that seems to be mediated by direct interaction with NPC components, independent of carrier molecules. Importantly, many proteins are transported by more than one mechanism. It has been suggested that these seemingly redundant pathways may ensure the maintenance of cellular functions under conditions in which one pathway is inhibited 104. Calmodulin has been shown to facilitate the nuclear import of the transcription factors SRY and SOX9 by binding to specific sequences in a way similar to NLS binding by karyopherin‐β 105. The nuclear import of these transcription factors is regulated by calcium. Similarly, calreticulin exports the glucocorticoid receptor from the nucleus in a calcium‐dependent manner 106. In addition, calreticulin is involved in the nuclear export of thyroid hormone receptor α1 and viral proteins 107, 108, 109.
Some proteins can enter the nucleus without requiring receptor proteins and there is growing evidence that receptor‐independent nuclear import can be mediated by the direct binding of the transport cargo to FG‐containing Nups. It has been suggested that proteins with armadillo repeats such as β‐catenin, that are structurally related to karyopherin transport receptors, can directly bind to the NPC and are imported into the nucleus independently of conventional NTRs of the karyopherin‐β type. Other proteins such as the tumor suppressor proteins SMAD3 and 4 and the transcription factor PU.1 utilize direct binding to FG‐Nups to cross the NPC. Conversely, some proteins can be transported through their interaction with proteins that contain a functional NLS known as a ‘piggyback’ mechanism. Recently, Speese et al. reported a novel NPC‐independent mechanism for the nuclear export of RNP by nuclear envelope budding akin to nuclear egress of herpes‐type viruses 110. During this mechanism, RNP granules bud into the perinuclear space in a manner dependent on lamin C.
The Ran system
The nucleo‐cytoplasmic transport process mediated by members of the karyopherin protein family requires metabolic energy. The loading and unloading of transport receptors with cargo molecules is controlled by the small Ras‐like GTPase Ran and requires GTP hydrolysis 111. Ran is a 25 kDa protein that exists in two different nucleotide‐bound states: RanGDP and RanGTP 112, 113. Ran hydrolyzes GTP very slowly and interacts with regulatory proteins including RanGAP1 (Ran GTPase activating protein 1) and RanBP1 that significantly increase GTP hydrolysis by Ran 114. Conversely, the Ran regulatory protein RanGTP exchange factor (RanGEF, also termed RCC1 in human cells) accelerates the exchange of nucleotides restoring the pool of the RanGTP. RanGAP1 is exclusively cytoplasmic 115 and RanBP1 is predominantly localized in the cytoplasm 116 whereas RanGEF is nuclear. As a consequence of this strict nuclear localization of RanGEF and cytoplasmic RanGAP1, Ran in complex with GTP is localized mostly in the nucleus and RanGDP (Ran in complex with GDP) in the cytoplasm 62. This RanGTP gradient provides directionality to nuclear‐cytoplasmic transport because importins and exportins differ in the way they utilize the RanGTP gradient 117. Import complexes are dissociated by RanGTP binding in contrast to export complexes which are formed by association with RanGTP. Both importins and exportins bind RanGTP directly 52, 118, 119 and use the metabolic energy supplied by the RanGTPase system for directional transport 120, 121. Importins bind their cargo at low RanGTP level (in the cytoplasm) and traverse the NPC as dimeric complexes transporting cargo (Fig. 2) 118, 119. Exportins act in an opposite manner, recruiting their cargo at high RanGTP levels in the nucleus (Fig. 2) 84, 88. The RanGTP‐exportin‐cargo complex crosses the NPC into the cytoplasm where it disassembles following GTPase activation releasing the transported cargo. The free export in translocates back into the nucleus to mediate another round of export.
Trafficking of macromolecules through the NPC
A number of models have been proposed to describe the molecular mechanism of selective gating through the NPC. The polymer brush model suggests that movements of the unfolded FG‐Nups sweep away macromolecules 11, 122, 123. Conversely, the collapse model, based on atomic force microscopy data 124, 125, suggests that regions of FG repeats may collapse following the binding of transport factors. These transport factors would open up their own passage through the central tube when they pass the meshwork of FG repeats. The ‘hydrophobic gel’ model 126, also called ‘saturated model’ 127, 128, proposed that the phenylalanines in the FG repeat regions are crosslinked with each other and form a dense gel of FG repeat filaments. Transport factors bind to these FG repeats, dissolve the crosslinks and facilitate passage through the nuclear pore. Another model suggested that the FG repeat regions form a layer coating the inner walls of the central tube where non‐binding molecules can only pass through the narrow FG‐Nup‐free middle and where the transport factors enter this layer through binding giving them full access to the tube volume 129. Melcák et al. proposed a model of pore dilatation by intermolecular sliding of Nup58/45 tetramers to adjust the diameter of the transport channel for the passage of cargo 130.
Nuclear import
The recognition of NLSs by Imp‐α and heterodimerization with Imp‐β initiates the process of nuclear import. The import complex, consisting of the cargo protein and the Imp‐α/β complex, localizes to the nuclear envelope, binds RanGDP and docks at the nuclear pore. The Imp‐α/β complex mediates the binding to and translocation through the NPC. Once translocation through the pore is executed, dissociation of the import complex is stimulated by RanGTP in the nucleus. Importin‐α is recycled back to the cytoplasm through the nuclear exporter CAS, whereas Imp‐β is separately transported back to the cytoplasm together with RanGTP. RanGAP1‐facilitated GTP hydrolysis of Ran on the cytoplasmic side causes the release of Imp‐β for the next cycle.
Nuclear export
The process of nuclear export is carried out according to principles which are analogous to those of nuclear import, using specific nuclear export receptors like CRM1 that recognize NES sequences on cargo proteins 131, 132. The affinity of CRM1 for most NESs is low and formation of the export complex is promoted by RanBP3 which links CRM1 to the chromatin binding protein RCC1 133, 134 and increases the active concentration of RanGTP. This promotes the affinity of the NES cargo for the export receptor 135, 136. This complex moves through the NPC via the interaction with FG repeat proteins 137. Within the cytoplasm, the CRM1 complex binds to the cytoplasmic filament complex (Nup88, Nup214 and Nup358 84, 138, 139, 140, 141) and interacts with RanGAP causing the hydrolysis of GTP which promotes the dissociation of the protein complex. Following this dissociation, the cargo is released in the cytoplasm 36, 142.
Export of RNAs
RNAs transcribed in the nucleus have to be exported, either to fulfill their function in protein synthesis or to mature into functional particles 143. The pre‐mRNA is processed and packaged into messenger ribonucleoprotein (mRNP) complexes to be exported through bulk or specific export. The majority of poly‐A transcripts are exported via the non‐karyopherin heterodimer Nxf1/Nxt1 independently of the RanGTP gradient. Conversely, a specific subset of endogenous transcripts is exported from the nucleus via CRM1. For efficient export, RNAs must undergo processing that includes splicing, 3′‐end formation of the poly‐A tail and the addition of a methyl‐7‐guanosine (m7G) cap structure to their 5′‐ends 144, 145, 146. For the majority of transcripts, the m7G cap recruits the CBC, which then activates export factors that allow the export mRNPs to bind to the NPC and traverse the hydrophobic central channel. Nxf1 (also known as TAP) 147) is the major driver of interaction between the export mRNP and the NPC. The Nxf1/Nxt1 heterodimer is recruited to the mRNP via the transcription‐export (TREX) complex 79, 144, 145, 148, 149, 150. The TREX complex consists of UAP56, REF/a Aly, CIP29 and THO multi‐subunit complex composed of THOC1/Hpr1, hTho2, THOC5, THOC6, THOC7 and Tex1 144, 151, 152, 153. REF/Aly and THO complexes promote the interaction of cargo mRNAs with the Nxf1 receptor 144, 151 that associates mRNPs with the nuclear basket via the ribonucleic acid export protein Rae1 and Nup98 to permit passage through the central channel (Fig. 2) 154, 155. At the cytoplasmic face, the cargo mRNPs are released and the export factors are recycled. RanBP2 associates with Nup88 and Nup214 141 and plays a very important role in cargo release and recycling after bulk mRNA export.
An alternative to the Nxf1‐driven export pathway involves the serine‐ and arginine‐rich SR proteins SRp20 and 9G8 that mediate the export of H2a mRNA and of some spliced transcripts 156, 157. Although the majority of mRNAs use the Nxf1 receptor to cross the NPC, subsets of transcripts are exported via the general CRM1 pathway. Additionally, through protein cofactors, CRM1 is involved in the export of specific types of mRNAs, small nuclear RNAs (U snRNAs) and ribosomal RNAs. CRM1 does not bind RNA directly but via NES‐containing adaptor proteins that bind to RNA or other RNA binding proteins 148. Once this step is completed, CRM1–cargo complexes dissociate, permitting the RNA to enter the cytoplasm and to recycle export factors 148. As in bulk mRNA export, Nup88, Nup214 and RanBP2 play critical roles in the recycling and release steps for CRM1‐dependent export 138, 158.
Regulation of the nucleo‐cytoplasmic trafficking
The nucleo‐cytoplasmic transport processes are regulated by cellular signaling systems via cargo protein modification(s) and the transport machinery, including the transport receptors, the NPC and the Ran system. Indeed targeting nucleo‐cytoplasmic transport has directed the development of new exciting therapeutic avenues to treat cancers that display aberrant protein subcellular localization 159, 160.
Modification of the cargo proteins
Importin‐NLS/exportin‐NES interactions can be modulated by conformational changes in both NLS/NES regions and in the substrate binding site of karyopherins 161. In some cases the three‐dimensional structure of a protein masks its own transport signal as in the case of p105 161, the precursor p50 subunit of NF‐κB. Upon stimulation, p105 is phosphorylated, the C‐terminal part of the protein is degraded and the NLS becomes accessible allowing the p50 protein to be recognized by the Imp‐α/Imp‐β complex 162. Furthermore, phosphorylation within or close to the NLS/NES can promote intramolecular masking. In the case of Hog1p (high osmolarity glycerol pathway‐signaling protein) phosphorylation at Thr174 and Tyr176 renders Hog1p‐NES inaccessible for binding to CRM1 and prevents its export from the nucleus 163. Similarly NF‐AT2 (nuclear factor of activated T cell 2) contains two NLSs that can be masked by phosphorylation at a low calcium concentration. Following a calcium concentration increase, calcineurin is dephosphorylated and NF‐AT2 is imported into the nucleus 164, 165, 166. In the canonical NF‐κB pathway, I‐κB masks the NLS sequence of NF‐κB p65 167. In resting conditions, I‐κB is phosphorylated, ubiquitinated and degraded by proteasome resulting in NLS unmasking and NF‐κB p65 nuclear import 168, 169. One of the multiple regulatory mechanisms of p53 is its homo‐tetramerization that masks one of its NES sequences. Dissociation of this tetramer is required for its nuclear export 170. Binding of proteins to RNA and DNA can also modulate the intermolecular masking of localization signal. For example, the NLS of the Rev protein, implicated in HIV mRNA translocation, is masked when Rev is linked to mRNA and recycling of Rev is possible only after release of the transported mRNA 171. Conversely, nucleo‐cytoplasmic transport can be enhanced by phosphorylation of SV40 virus large T antigen 172 or Pho4 factor 173 increasing the affinity of their NLS/NES signals to the corresponding karyopherin receptors 172.
Viral regulation of nucleo‐cytoplasmic trafficking
Many viruses have evolved elegant strategies to exploit the host nucleo‐cytoplasmic transport pathways to evade the cellular anti‐viral response or to facilitate viral replication. Some viruses such as the vesicular stomatitis virus interact with Nups to inhibit the export of host mRNAs that encode anti‐viral factors and make the translation machinery available for expression of viral mRNAs 174. Similarly, the ICP27 protein of herpes simplex virus interacts with Nup62 and blocks nuclear import of proteins via Imp‐α/β1 and Imp‐β2 pathways 175, 176. Conversely, poliovirus and human rhinovirus block the nuclear import of proteins via the Imp‐α/β1 and Imp‐β2 pathways by viral‐mediated proteolytic cleavage of specific Nups 21, 177, 178, 179, 180, 181, 182, 183. Other viruses inhibit the nuclear import of the STAT proteins that are known to be key regulators of the cell anti‐virus response. Furthermore, the severe acute respiratory syndrome (SARS) virus disrupts the nuclear import of STAT1 by tethering the tyrosine‐phosphorylated STAT1–Imp‐α/Imp‐β complex to endoplasmic reticulum/Golgi membranes 184, 185, 186, 187. Yet another way exploited by viruses to prevent STAT1 nuclear localization is to bind viral proteins to import receptors. The Ebola virus VP24 protein binds Imp‐α to block its interaction with phosphorylated STAT1 and hnRNP C1/C2 188, 189, 190. The L1 protein of the human papilloma virus type 11 binds Imp‐β2/β3 and disrupts cargo import. Encephalomyocarditis virus exerts its inhibitory effect on the nuclear protein import of infected cells via its L protein that hyper‐phosphorylates Nups and binds Ran 191, 192, 193. In addition herpes viruses and HIV promote viral mRNA export by reprogramming the cellular transport pathways. The HIV‐1 Rev protein facilitates nuclear export of unspliced or partially spliced viral mRNAs through the Rev‐responsive element, an RNA signature within these viral mRNAs 85, 194, 195. As a result, Rev‐bound viral RNA binds to CRM1 and RanGTP and is transported through the NPC.
Conclusions
The nuclear transport processes within our cells are governed by several types of protein–protein interactions (e.g. adaptor protein–cargo, adaptor protein–transport receptor, transport receptor–cargo, transport receptor–Ran, transport receptor–Nup), yet only one enzymatic reaction occurs, namely the hydrolysis of GTP by Ran GTPase. While pharmacological targeting of protein–protein interactions has historically been considered challenging, the development of nuclear export inhibitors proves the viability of this approach. Therapeutic agents that attempt to normalize or to target protein localization are aimed at various regulatory components within the transport process, including the upstream regulatory components, the cargo proteins, the transport receptors, the Ran regulators and the NPC itself. In some cases, compounds have been developed to successfully influence subcellular protein distribution in disease states, including CRM1 and Imp‐α/β inhibitors. Continued and intensified research efforts aimed at better understanding the nuclear transport mechanisms, and how they relate to pathogenesis, will probably reveal the identity of novel targets for the treatment of cancer and to subvert viral infections.
Author contributions
BC, RH, NdP and WL wrote the paper. WL coordinated the drafting of the manuscript.
Acknowledgements
The authors would like to acknowledge networking support by the COST Action CM1106 StemChem – ‘Chemical Approaches to Targeting Drug Resistance in Cancer Stem Cells’. Dr Richard Hill is the recipient of a Fundação para a Ciência e a Tecnologia (FCT) 2012 research grant no. SFRH/BPD/84634/2012. We acknowledge Ana Teresa Maia for critical reading of this paper.
References
- 1. Hung MC & Link W (2011) Protein localization in disease and therapy. J Cell Sci 124, 3381–3392. [DOI] [PubMed] [Google Scholar]
- 2. Mor A, White MA & Fontoura BM (2014) Nuclear trafficking in health and disease. Curr Opin Cell Biol 28C, 28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hill R, Cautain B, de Pedro N & Link W (2014) Targeting nucleocytoplasmic transport in cancer therapy. Oncotarget 5, 11–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Turner JG, Dawson J & Sullivan DM (2012) Nuclear export of proteins and drug resistance in cancer. Biochem Pharmacol 83, 1021–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Turner JG & Sullivan DM (2008) CRM1‐mediated nuclear export of proteins and drug resistance in cancer. Curr Med Chem 15, 2648–2655. [DOI] [PubMed] [Google Scholar]
- 6. Caly L, Wagstaff KM & Jans DA (2012) Nuclear trafficking of proteins from RNA viruses: potential target for antivirals? Antiviral Res 95, 202–206. [DOI] [PubMed] [Google Scholar]
- 7. Callan HG, Randall JT & Tomlin SG (1949) An electron microscope study of the nuclear membrane. Nature 163, 280. [DOI] [PubMed] [Google Scholar]
- 8. Field MC & Dacks JB (2009) First and last ancestors: reconstructing evolution of the endomembrane system with ESCRTs, vesicle coat proteins, and nuclear pore complexes. Curr Opin Cell Biol 21, 4–13. [DOI] [PubMed] [Google Scholar]
- 9. Lim RY, Ullman KS & Fahrenkrog B (2008) Biology and biophysics of the nuclear pore complex and its components. Int Rev Cell Mol Biol 267, 299–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT & Matunis MJ (2002) Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol 158, 915–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y & Chait BT (2000) The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol 148, 635–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grossman E, Medalia O & Zwerger M (2012) Functional architecture of the nuclear pore complex. Annu Rev Biophys 41, 557–584. [DOI] [PubMed] [Google Scholar]
- 13. Beck M, Lucic V, Forster F, Baumeister W & Medalia O (2007) Snapshots of nuclear pore complexes in action captured by cryo‐electron tomography. Nature 449, 611–615. [DOI] [PubMed] [Google Scholar]
- 14. Bui KH, von Appen A, DiGuilio AL, Ori A, Sparks L, Mackmull MT, Bock T, Hagen W, Andres‐Pons A, Glavy JS et al (2013) Integrated structural analysis of the human nuclear pore complex scaffold. Cell 155, 1233–1243. [DOI] [PubMed] [Google Scholar]
- 15. Maimon T, Elad N, Dahan I & Medalia O (2012) The human nuclear pore complex as revealed by cryo‐electron tomography. Structure 20, 998–1006. [DOI] [PubMed] [Google Scholar]
- 16. Schwartz TU (2005) Modularity within the architecture of the nuclear pore complex. Curr Opin Struct Biol 15, 221–226. [DOI] [PubMed] [Google Scholar]
- 17. Neumann N, Lundin D & Poole AM (2010) Comparative genomic evidence for a complete nuclear pore complex in the last eukaryotic common ancestor. PLoS One 5, e13241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hoelz A, Debler EW & Blobel G (2011) The structure of the nuclear pore complex. Annu Rev Biochem 80, 613–643. [DOI] [PubMed] [Google Scholar]
- 19. Wente SR (2000) Gatekeepers of the nucleus. Science 288, 1374–1377. [DOI] [PubMed] [Google Scholar]
- 20. Therizols P, Fairhead C, Cabal GG, Genovesio A, Olivo‐Marin JC, Dujon B & Fabre E (2006) Telomere tethering at the nuclear periphery is essential for efficient DNA double strand break repair in subtelomeric region. J Cell Biol 172, 189–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Capelson M, Doucet C & Hetzer MW (2010) Nuclear pore complexes: guardians of the nuclear genome. Cold Spring Harb Symp Quant Biol 75, 585–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Krull S, Dorries J, Boysen B, Reidenbach S, Magnius L, Norder H, Thyberg J & Cordes VC (2010) Protein Tpr is required for establishing nuclear pore‐associated zones of heterochromatin exclusion. EMBO J 29, 1659–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Strambio‐De‐Castillia C, Niepel M & Rout MP (2010) The nuclear pore complex: bridging nuclear transport and gene regulation. Nat Rev Mol Cell Biol 11, 490–501. [DOI] [PubMed] [Google Scholar]
- 24. Van de Vosse DW, Wan Y, Wozniak RW & Aitchison JD (2011) Role of the nuclear envelope in genome organization and gene expression. Wiley Interdiscip Rev Syst Biol Med 3, 147–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brickner DG, Cajigas I, Fondufe‐Mittendorf Y, Ahmed S, Lee PC, Widom J & Brickner JH (2007) H2A.Z‐mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol 5, e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Erkmann JA & Kutay U (2004) Nuclear export of mRNA: from the site of transcription to the cytoplasm. Exp Cell Res 296, 12–20. [DOI] [PubMed] [Google Scholar]
- 27. Devos D, Dokudovskaya S, Williams R, Alber F, Eswar N, Chait BT, Rout MP & Sali A (2006) Simple fold composition and modular architecture of the nuclear pore complex. Proc Natl Acad Sci USA 103, 2172–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mitchell JM, Mansfeld J, Capitanio J, Kutay U & Wozniak RW (2010) Pom121 links two essential subcomplexes of the nuclear pore complex core to the membrane. J Cell Biol 191, 505–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Debler EW, Ma Y, Seo HS, Hsia KC, Noriega TR, Blobel G & Hoelz A (2008) A fence‐like coat for the nuclear pore membrane. Mol Cell 32, 815–826. [DOI] [PubMed] [Google Scholar]
- 30. Lange A, Mills RE, Lange CJ, Stewart M, Devine SE & Corbett AH (2007) Classical nuclear localization signals: definition, function, and interaction with importin alpha. J Biol Chem 282, 5101–5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dingwall C, Sharnick SV & Laskey RA (1982) A polypeptide domain that specifies migration of nucleoplasmin into the nucleus. Cell 30, 449–458. [DOI] [PubMed] [Google Scholar]
- 32. Fischer U, Huber J, Boelens WC, Mattaj IW & Luhrmann R (1995) The HIV‐1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 82, 475–483. [DOI] [PubMed] [Google Scholar]
- 33. Kalderon D, Roberts BL, Richardson WD & Smith AE (1984) A short amino acid sequence able to specify nuclear location. Cell 39, 499–509. [DOI] [PubMed] [Google Scholar]
- 34. Lee BJ, Cansizoglu AE, Suel KE, Louis TH, Zhang Z & Chook YM (2006) Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell 126, 543–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wen W, Meinkoth JL, Tsien RY & Taylor SS (1995) Identification of a signal for rapid export of proteins from the nucleus. Cell 82, 463–473. [DOI] [PubMed] [Google Scholar]
- 36. Dong X, Biswas A, Suel KE, Jackson LK, Martinez R, Gu H & Chook YM (2009) Structural basis for leucine‐rich nuclear export signal recognition by CRM1. Nature 458, 1136–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Monecke T, Guttler T, Neumann P, Dickmanns A, Gorlich D & Ficner R (2009) Crystal structure of the nuclear export receptor CRM1 in complex with Snurportin1 and RanGTP. Science 324, 1087–1091. [DOI] [PubMed] [Google Scholar]
- 38. Kalderon D, Richardson WD, Markham AF & Smith AE (1984) Sequence requirements for nuclear location of simian virus 40 large‐T antigen. Nature 311, 33–38. [DOI] [PubMed] [Google Scholar]
- 39. Tran EJ, Bolger TA & Wente SR (2007) SnapShot: nuclear transport. Cell 131, 420. [DOI] [PubMed] [Google Scholar]
- 40. Marfori M, Mynott A, Ellis JJ, Mehdi AM, Saunders NF, Curmi PM, Forwood JK, Boden M & Kobe B (2011) Molecular basis for specificity of nuclear import and prediction of nuclear localization. Biochim Biophys Acta 1813, 1562–1577. [DOI] [PubMed] [Google Scholar]
- 41. Lange A, McLane LM, Mills RE, Devine SE & Corbett AH (2010) Expanding the definition of the classical bipartite nuclear localization signal. Traffic 11, 311–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chook YM & Suel KE (2011) Nuclear import by karyopherin‐betas: recognition and inhibition. Biochim Biophys Acta 1813, 1593–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lange A, Mills RE, Devine SE & Corbett AH (2008) A PY‐NLS nuclear targeting signal is required for nuclear localization and function of the Saccharomyces cerevisiae mRNA‐binding protein Hrp1. J Biol Chem 283, 12926–12934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Suel KE & Chook YM (2009) Kap104p imports the PY‐NLS‐containing transcription factor Tfg2p into the nucleus. J Biol Chem 284, 15416–15424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xu D, Farmer A & Chook YM (2010) Recognition of nuclear targeting signals by Karyopherin‐beta proteins. Curr Opin Struct Biol 20, 782–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shin HY & Reich NC (2013) Dynamic trafficking of STAT5 depends on an unconventional nuclear localization signal. J Cell Sci 126, 3333–3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bogerd HP, Fridell RA, Benson RE, Hua J & Cullen BR (1996) Protein sequence requirements for function of the human T‐cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization‐selection assay. Mol Cell Biol 16, 4207–4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Henderson BR & Eleftheriou A (2000) A comparison of the activity, sequence specificity, and CRM1‐dependence of different nuclear export signals. Exp Cell Res 256, 213–224. [DOI] [PubMed] [Google Scholar]
- 49. la Cour T, Kiemer L, Molgaard A, Gupta R, Skriver K & Brunak S (2004) Analysis and prediction of leucine‐rich nuclear export signals. Protein Eng Des Sel 17, 527–536. [DOI] [PubMed] [Google Scholar]
- 50. Kosugi S, Hasebe M, Tomita M & Yanagawa H (2008) Nuclear export signal consensus sequences defined using a localization‐based yeast selection system. Traffic 9, 2053–2062. [DOI] [PubMed] [Google Scholar]
- 51. Paraskeva E, Izaurralde E, Bischoff FR, Huber J, Kutay U, Hartmann E, Luhrmann R & Gorlich D (1999) CRM1‐mediated recycling of snurportin 1 to the cytoplasm. J Cell Biol 145, 255–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fornerod M, Ohno M, Yoshida M & Mattaj IW (1997) CRM1 is an export receptor for leucine‐rich nuclear export signals. Cell 90, 1051–1060. [DOI] [PubMed] [Google Scholar]
- 53. Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M & Nishida E (1997) CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 390, 308–311. [DOI] [PubMed] [Google Scholar]
- 54. Ossareh‐Nazari B, Bachelerie F & Dargemont C (1997) Evidence for a role of CRM1 in signal‐mediated nuclear protein export. Science 278, 141–144. [DOI] [PubMed] [Google Scholar]
- 55. Stade K, Ford CS, Guthrie C & Weis K (1997) Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 90, 1041–1050. [DOI] [PubMed] [Google Scholar]
- 56. Guttler T, Madl T, Neumann P, Deichsel D, Corsini L, Monecke T, Ficner R, Sattler M & Gorlich D (2010) NES consensus redefined by structures of PKI‐type and Rev‐type nuclear export signals bound to CRM1. Nat Struct Mol Biol 17, 1367–1376. [DOI] [PubMed] [Google Scholar]
- 57. Kosugi S, Hasebe M, Entani T, Takayama S, Tomita M & Yanagawa H (2008) Design of peptide inhibitors for the importin alpha/beta nuclear import pathway by activity‐based profiling. Chem Biol 15, 940–949. [DOI] [PubMed] [Google Scholar]
- 58. Fu SC, Huang HC, Horton P & Juan HF (2013) ValidNESs: a database of validated leucine‐rich nuclear export signals. Nucleic Acids Res 41, D338–D343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Fu SC, Imai K & Horton P (2011) Prediction of leucine‐rich nuclear export signal containing proteins with NESsential. Nucleic Acids Res 39, e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Prieto G, Fullaondo A & Rodriguez JA (2014) Prediction of nuclear export signals using weighted regular expressions (Wregex). Bioinformatics 30, 1220–1227. [DOI] [PubMed] [Google Scholar]
- 61. Kosugi S, Yanagawa H, Terauchi R & Tabata S (2014) NESmapper: accurate prediction of leucine‐rich nuclear export signals using activity‐based profiles. PLoS Comput Biol 10, e1003841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cook A, Bono F, Jinek M & Conti E (2007) Structural biology of nucleocytoplasmic transport. Annu Rev Biochem 76, 647–671. [DOI] [PubMed] [Google Scholar]
- 63. Mosammaparast N & Pemberton LF (2004) Karyopherins: from nuclear‐transport mediators to nuclear‐function regulators. Trends Cell Biol 14, 547–556. [DOI] [PubMed] [Google Scholar]
- 64. Gorlich D, Dabrowski M, Bischoff FR, Kutay U, Bork P, Hartmann E, Prehn S & Izaurralde E (1997) A novel class of RanGTP binding proteins. J Cell Biol 138, 65–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Conti E & Izaurralde E (2001) Nucleocytoplasmic transport enters the atomic age. Curr Opin Cell Biol 13, 310–319. [DOI] [PubMed] [Google Scholar]
- 66. Conti E, Muller CW & Stewart M (2006) Karyopherin flexibility in nucleocytoplasmic transport. Curr Opin Struct Biol 16, 237–244. [DOI] [PubMed] [Google Scholar]
- 67. Goldfarb DS, Corbett AH, Mason DA, Harreman MT & Adam SA (2004) Importin alpha: a multipurpose nuclear‐transport receptor. Trends Cell Biol 14, 505–514. [DOI] [PubMed] [Google Scholar]
- 68. Isgro TA & Schulten K (2005) Binding dynamics of isolated nucleoporin repeat regions to importin‐beta. Structure 13, 1869–1879. [DOI] [PubMed] [Google Scholar]
- 69. Harel A & Forbes DJ (2004) Importin beta: conducting a much larger cellular symphony. Mol Cell 16, 319–330. [DOI] [PubMed] [Google Scholar]
- 70. Macara IG (2001) Transport into and out of the nucleus. Microbiol Mol Biol Rev 65, 570–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bayliss R, Littlewood T & Stewart M (2000) Structural basis for the interaction between FxFG nucleoporin repeats and importin‐beta in nuclear trafficking. Cell 102, 99–108. [DOI] [PubMed] [Google Scholar]
- 72. Xu D, Grishin NV & Chook YM (2012) NESdb: a database of NES‐containing CRM1 cargoes. Mol Biol Cell 23, 3673–3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wild T, Horvath P, Wyler E, Widmann B, Badertscher L, Zemp I, Kozak K, Csucs G, Lund E & Kutay U (2010) A protein inventory of human ribosome biogenesis reveals an essential function of exportin 5 in 60S subunit export. PLoS Biol 8, e1000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ciufo LF & Brown JD (2000) Nuclear export of yeast signal recognition particle lacking srp54p by the Xpo1p/Crm1p NES‐dependent pathway. Curr Biol 10, R882. [DOI] [PubMed] [Google Scholar]
- 75. Gadal O, Strauss D, Kessl J, Trumpower B, Tollervey D & Hurt E (2001) Nuclear export of 60s ribosomal subunits depends on Xpo1p and requires a nuclear export sequence‐containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol Cell Biol 21, 3405–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ho JH, Kallstrom G & Johnson AW (2000) Nmd3p is a Crm1p‐dependent adapter protein for nuclear export of the large ribosomal subunit. J Cell Biol 151, 1057–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Thomas F & Kutay U (2003) Biogenesis and nuclear export of ribosomal subunits in higher eukaryotes depend on the CRM1 export pathway. J Cell Sci 116, 2409–2419. [DOI] [PubMed] [Google Scholar]
- 78. Izaurralde E, Lewis J, Gamberi C, Jarmolowski A, McGuigan C & Mattaj IW (1995) A cap‐binding protein complex mediating U snRNA export. Nature 376, 709–712. [DOI] [PubMed] [Google Scholar]
- 79. Ohno M, Segref A, Bachi A, Wilm M & Mattaj IW (2000) PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell 101, 187–198. [DOI] [PubMed] [Google Scholar]
- 80. Bohnsack MT, Regener K, Schwappach B, Saffrich R, Paraskeva E, Hartmann E & Gorlich D (2002) Exp5 exports eEF1A via tRNA from nuclei and synergizes with other transport pathways to confine translation to the cytoplasm. EMBO J 21, 6205–6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Feng W, Benko AL, Lee JH, Stanford DR & Hopper AK (1999) Antagonistic effects of NES and NLS motifs determine S. cerevisiae Rna1p subcellular distribution. J Cell Sci 112(Pt 3), 339–347. [DOI] [PubMed] [Google Scholar]
- 82. Maurer P, Redd M, Solsbacher J, Bischoff FR, Greiner M, Podtelejnikov AV, Mann M, Stade K, Weis K & Schlenstedt G (2001) The nuclear export receptor Xpo1p forms distinct complexes with NES transport substrates and the yeast Ran binding protein 1 (Yrb1p). Mol Biol Cell 12, 539–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Richards SA, Lounsbury KM, Carey KL & Macara IG (1996) A nuclear export signal is essential for the cytosolic localization of the Ran binding protein, RanBP1. J Cell Biol 134, 1157–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti KG, Fransen J & Grosveld G (1997) The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO J 16, 807–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Malim MH, Hauber J, Le SY, Maizel JV & Cullen BR (1989) The HIV‐1 rev trans‐activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature 338, 254–257. [DOI] [PubMed] [Google Scholar]
- 86. Malim MH, McCarn DF, Tiley LS & Cullen BR (1991) Mutational definition of the human immunodeficiency virus type 1 Rev activation domain. J Virol 65, 4248–4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kunzler M & Hurt EC (1998) Cse1p functions as the nuclear export receptor for importin alpha in yeast. FEBS Lett 433, 185–190. [DOI] [PubMed] [Google Scholar]
- 88. Kutay U, Bischoff FR, Kostka S, Kraft R & Gorlich D (1997) Export of importin alpha from the nucleus is mediated by a specific nuclear transport factor. Cell 90, 1061–1071. [DOI] [PubMed] [Google Scholar]
- 89. Solsbacher J, Maurer P, Bischoff FR & Schlenstedt G (1998) Cse1p is involved in export of yeast importin alpha from the nucleus. Mol Cell Biol 18, 6805–6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Dorfman J & Macara IG (2008) STRADalpha regulates LKB1 localization by blocking access to importin‐alpha, and by association with Crm1 and exportin‐7. Mol Biol Cell 19, 1614–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Mingot JM, Bohnsack MT, Jakle U & Gorlich D (2004) Exportin 7 defines a novel general nuclear export pathway. EMBO J 23, 3227–3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Arts GJ, Fornerod M & Mattaj IW (1998) Identification of a nuclear export receptor for tRNA. Curr Biol 8, 305–314. [DOI] [PubMed] [Google Scholar]
- 93. Arts GJ, Kuersten S, Romby P, Ehresmann B & Mattaj IW (1998) The role of exportin‐t in selective nuclear export of mature tRNAs. EMBO J 17, 7430–7441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Hellmuth K, Lau DM, Bischoff FR, Kunzler M, Hurt E & Simos G (1998) Yeast Los1p has properties of an exportin‐like nucleocytoplasmic transport factor for tRNA. Mol Cell Biol 18, 6374–6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Kutay U, Lipowsky G, Izaurralde E, Bischoff FR, Schwarzmaier P, Hartmann E & Gorlich D (1998) Identification of a tRNA‐specific nuclear export receptor. Mol Cell 1, 359–369. [DOI] [PubMed] [Google Scholar]
- 96. Calado A, Treichel N, Muller EC, Otto A & Kutay U (2002) Exportin‐5‐mediated nuclear export of eukaryotic elongation factor 1A and tRNA. EMBO J 21, 6216–6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Bohnsack MT, Czaplinski K & Gorlich D (2004) Exportin 5 is a RanGTP‐dependent dsRNA‐binding protein that mediates nuclear export of pre‐miRNAs. RNA 10, 185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Lund E, Guttinger S, Calado A, Dahlberg JE & Kutay U (2004) Nuclear export of microRNA precursors. Science 303, 95–98. [DOI] [PubMed] [Google Scholar]
- 99. Yi R, Qin Y, Macara IG & Cullen BR (2003) Exportin‐5 mediates the nuclear export of pre‐microRNAs and short hairpin RNAs. Genes Dev 17, 3011–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Gontan C, Guttler T, Engelen E, Demmers J, Fornerod M, Grosveld FG, Tibboel D, Gorlich D, Poot RA & Rottier RJ (2009) Exportin 4 mediates a novel nuclear import pathway for Sox family transcription factors. J Cell Biol 185, 27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Kurisaki A, Kurisaki K, Kowanetz M, Sugino H, Yoneda Y, Heldin CH & Moustakas A (2006) The mechanism of nuclear export of Smad3 involves exportin 4 and Ran. Mol Cell Biol 26, 1318–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Bono F, Cook AG, Grunwald M, Ebert J & Conti E (2010) Nuclear import mechanism of the EJC component Mago‐Y14 revealed by structural studies of importin 13. Mol Cell 37, 211–222. [DOI] [PubMed] [Google Scholar]
- 103. Mingot JM, Kostka S, Kraft R, Hartmann E & Gorlich D (2001) Importin 13: a novel mediator of nuclear import and export. EMBO J 20, 3685–3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Wagstaff KM & Jans DA (2009) Importins and beyond: non‐conventional nuclear transport mechanisms. Traffic 10, 1188–1198. [DOI] [PubMed] [Google Scholar]
- 105. Argentaro A, Sim H, Kelly S, Preiss S, Clayton A, Jans DA & Harley VR (2003) A SOX9 defect of calmodulin‐dependent nuclear import in campomelic dysplasia/autosomal sex reversal. J Biol Chem 278, 33839–33847. [DOI] [PubMed] [Google Scholar]
- 106. Holaska JM, Black BE, Love DC, Hanover JA, Leszyk J & Paschal BM (2001) Calreticulin Is a receptor for nuclear export. J Cell Biol 152, 127–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Alefantis T, Flaig KE, Wigdahl B & Jain P (2007) Interaction of HTLV‐1 Tax protein with calreticulin: implications for Tax nuclear export and secretion. Biomed Pharmacother 61, 194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Bunn CF, Neidig JA, Freidinger KE, Stankiewicz TA, Weaver BS, McGrew J & Allison LA (2001) Nucleocytoplasmic shuttling of the thyroid hormone receptor alpha. Mol Endocrinol 15, 512–533. [DOI] [PubMed] [Google Scholar]
- 109. Grespin ME, Bonamy GM, Roggero VR, Cameron NG, Adam LE, Atchison AP, Fratto VM & Allison LA (2008) Thyroid hormone receptor alpha1 follows a cooperative CRM1/calreticulin‐mediated nuclear export pathway. J Biol Chem 283, 25576–25588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Speese SD, Ashley J, Jokhi V, Nunnari J, Barria R, Li Y, Ataman B, Koon A, Chang YT, Li Q et al (2012) Nuclear envelope budding enables large ribonucleoprotein particle export during synaptic Wnt signaling. Cell 149, 832–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Sorokin AV, Kim ER & Ovchinnikov LP (2007) Nucleocytoplasmic transport of proteins. Biochemistry (Mosc) 72, 1439–1457. [DOI] [PubMed] [Google Scholar]
- 112. Bourne HR, Sanders DA & McCormick F (1991) The GTPase superfamily: conserved structure and molecular mechanism. Nature 349, 117–127. [DOI] [PubMed] [Google Scholar]
- 113. Moore MS (1998) Ran and nuclear transport. J Biol Chem 273, 22857–22860. [DOI] [PubMed] [Google Scholar]
- 114. Bischoff FR, Krebber H, Smirnova E, Dong W & Ponstingl H (1995) Co‐activation of RanGTPase and inhibition of GTP dissociation by Ran‐GTP binding protein RanBP1. EMBO J 14, 705–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Hopper AK, Traglia HM & Dunst RW (1990) The yeast RNA1 gene product necessary for RNA processing is located in the cytosol and apparently excluded from the nucleus. J Cell Biol 111, 309–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Kunzler M, Gerstberger T, Stutz F, Bischoff FR & Hurt E (2000) Yeast Ran‐binding protein 1 (Yrb1) shuttles between the nucleus and cytoplasm and is exported from the nucleus via a CRM1 (XPO1)‐dependent pathway. Mol Cell Biol 20, 4295–4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Kalab P, Pralle A, Isacoff EY, Heald R & Weis K (2006) Analysis of a RanGTP‐regulated gradient in mitotic somatic cells. Nature 440, 697–701. [DOI] [PubMed] [Google Scholar]
- 118. Gorlich D, Pante N, Kutay U, Aebi U & Bischoff FR (1996) Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J 15, 5584–5594. [PMC free article] [PubMed] [Google Scholar]
- 119. Rexach M & Blobel G (1995) Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell 83, 683–692. [DOI] [PubMed] [Google Scholar]
- 120. Melchior F, Paschal B, Evans J & Gerace L (1993) Inhibition of nuclear protein import by nonhydrolyzable analogues of GTP and identification of the small GTPase Ran/TC4 as an essential transport factor. J Cell Biol 123, 1649–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Moore MS & Blobel G (1993) The GTP‐binding protein Ran/TC4 is required for protein import into the nucleus. Nature 365, 661–663. [DOI] [PubMed] [Google Scholar]
- 122. Lim RY, Huang NP, Koser J, Deng J, Lau KH, Schwarz‐Herion K, Fahrenkrog B & Aebi U (2006) Flexible phenylalanine‐glycine nucleoporins as entropic barriers to nucleocytoplasmic transport. Proc Natl Acad Sci USA 103, 9512–9517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Rout MP, Aitchison JD, Magnasco MO & Chait BT (2003) Virtual gating and nuclear transport: the hole picture. Trends Cell Biol 13, 622–628. [DOI] [PubMed] [Google Scholar]
- 124. Lim RY, Fahrenkrog B, Koser J, Schwarz‐Herion K, Deng J & Aebi U (2007) Nanomechanical basis of selective gating by the nuclear pore complex. Science 318, 640–643. [DOI] [PubMed] [Google Scholar]
- 125. Lim RY, Koser J, Huang NP, Schwarz‐Herion K & Aebi U (2007) Nanomechanical interactions of phenylalanine‐glycine nucleoporins studied by single molecule force‐volume spectroscopy. J Struct Biol 159, 277–289. [DOI] [PubMed] [Google Scholar]
- 126. Ribbeck K & Gorlich D (2002) The permeability barrier of nuclear pore complexes appears to operate via hydrophobic exclusion. EMBO J 21, 2664–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Frey S & Gorlich D (2007) A saturated FG‐repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell 130, 512–523. [DOI] [PubMed] [Google Scholar]
- 128. Frey S, Richter RP & Gorlich D (2006) FG‐rich repeats of nuclear pore proteins form a three‐dimensional meshwork with hydrogel‐like properties. Science 314, 815–817. [DOI] [PubMed] [Google Scholar]
- 129. Peters R (2005) Translocation through the nuclear pore complex: selectivity and speed by reduction‐of‐dimensionality. Traffic 6, 421–427. [DOI] [PubMed] [Google Scholar]
- 130. Melcák I, Hoelz A & Blobel G (2007) Structure of Nup58/45 suggests flexible nuclear pore diameter by intermolecular sliding. Science 315, 1729–1732. [DOI] [PubMed] [Google Scholar]
- 131. Bednenko J, Cingolani G & Gerace L (2003) Nucleocytoplasmic transport: navigating the channel. Traffic 4, 127–135. [DOI] [PubMed] [Google Scholar]
- 132. Arnaoutov A, Azuma Y, Ribbeck K, Joseph J, Boyarchuk Y, Karpova T, McNally J & Dasso M (2005) Crm1 is a mitotic effector of Ran‐GTP in somatic cells. Nat Cell Biol 7, 626–632. [DOI] [PubMed] [Google Scholar]
- 133. Kehlenbach RH, Assheuer R, Kehlenbach A, Becker J & Gerace L (2001) Stimulation of nuclear export and inhibition of nuclear import by a Ran mutant deficient in binding to Ran‐binding protein 1. J Biol Chem 276, 14524–14531. [DOI] [PubMed] [Google Scholar]
- 134. Nemergut ME, Lindsay ME, Brownawell AM & Macara IG (2002) Ran‐binding protein 3 links Crm1 to the Ran guanine nucleotide exchange factor. J Biol Chem 277, 17385–17388. [DOI] [PubMed] [Google Scholar]
- 135. Englmeier L, Fornerod M, Bischoff FR, Petosa C, Mattaj IW & Kutay U (2001) RanBP3 influences interactions between CRM1 and its nuclear protein export substrates. EMBO Rep 2, 926–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Lindsay ME, Holaska JM, Welch K, Paschal BM & Macara IG (2001) Ran‐binding protein 3 is a cofactor for Crm1‐mediated nuclear protein export. J Cell Biol 153, 1391–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Yang W, Gelles J & Musser SM (2004) Imaging of single‐molecule translocation through nuclear pore complexes. Proc Natl Acad Sci USA 101, 12887–12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Hutten S & Kehlenbach RH (2006) Nup214 is required for CRM1‐dependent nuclear protein export in vivo . Mol Cell Biol 26, 6772–6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Kehlenbach RH, Dickmanns A, Kehlenbach A, Guan T & Gerace L (1999) A role for RanBP1 in the release of CRM1 from the nuclear pore complex in a terminal step of nuclear export. J Cell Biol 145, 645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Askjaer P, Bachi A, Wilm M, Bischoff FR, Weeks DL, Ogniewski V, Ohno M, Niehrs C, Kjems J, Mattaj IW et al (1999) RanGTP‐regulated interactions of CRM1 with nucleoporins and a shuttling DEAD‐box helicase. Mol Cell Biol 19, 6276–6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Bernad R, van der Velde H, Fornerod M & Pickersgill H (2004) Nup358/RanBP2 attaches to the nuclear pore complex via association with Nup88 and Nup214/CAN and plays a supporting role in CRM1‐mediated nuclear protein export. Mol Cell Biol 24, 2373–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Dong X, Biswas A & Chook YM (2009) Structural basis for assembly and disassembly of the CRM1 nuclear export complex. Nat Struct Mol Biol 16, 558–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Rodriguez MS, Dargemont C & Stutz F (2004) Nuclear export of RNA. Biol Cell 96, 639–655. [DOI] [PubMed] [Google Scholar]
- 144. Carmody SR & Wente SR (2009) mRNA nuclear export at a glance. J Cell Sci 122, 1933–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Kohler A & Hurt E (2007) Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol 8, 761–773. [DOI] [PubMed] [Google Scholar]
- 146. Siddiqui N & Borden KL (2012) mRNA export and cancer. Wiley Interdiscip Rev RNA 3, 13–25. [DOI] [PubMed] [Google Scholar]
- 147. Fried H & Kutay U (2003) Nucleocytoplasmic transport: taking an inventory. Cell Mol Life Sci 60, 1659–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Hutten S & Kehlenbach RH (2007) CRM1‐mediated nuclear export: to the pore and beyond. Trends Cell Biol 17, 193–201. [DOI] [PubMed] [Google Scholar]
- 149. Murdoch K, Loop S, Rudt F & Pieler T (2002) Nuclear export of 5S rRNA‐containing ribonucleoprotein complexes requires CRM1 and the RanGTPase cycle. Eur J Cell Biol 81, 549–556. [DOI] [PubMed] [Google Scholar]
- 150. Rouquette J, Choesmel V & Gleizes PE (2005) Nuclear export and cytoplasmic processing of precursors to the 40S ribosomal subunits in mammalian cells. EMBO J 24, 2862–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Katahira J (2012) mRNA export and the TREX complex. Biochim Biophys Acta 1819, 507–513. [DOI] [PubMed] [Google Scholar]
- 152. Luna R, Rondon AG & Aguilera A (2012) New clues to understand the role of THO and other functionally related factors in mRNP biogenesis. Biochim Biophys Acta 1819, 514–520. [DOI] [PubMed] [Google Scholar]
- 153. Rodriguez‐Navarro S & Hurt E (2011) Linking gene regulation to mRNA production and export. Curr Opin Cell Biol 23, 302–309. [DOI] [PubMed] [Google Scholar]
- 154. Fontoura BM, Dales S, Blobel G & Zhong H (2001) The nucleoporin Nup98 associates with the intranuclear filamentous protein network of TPR. Proc Natl Acad Sci USA 98, 3208–3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Ren Y, Seo HS, Blobel G & Hoelz A (2010) Structural and functional analysis of the interaction between the nucleoporin Nup98 and the mRNA export factor Rae1. Proc Natl Acad Sci USA 107, 10406–10411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Huang Y & Steitz JA (2005) SRprises along a messenger's journey. Mol Cell 17, 613–615. [DOI] [PubMed] [Google Scholar]
- 157. Long JC & Caceres JF (2009) The SR protein family of splicing factors: master regulators of gene expression. Biochem J 417, 15–27. [DOI] [PubMed] [Google Scholar]
- 158. Walther TC, Pickersgill HS, Cordes VC, Goldberg MW, Allen TD, Mattaj IW & Fornerod M (2002) The cytoplasmic filaments of the nuclear pore complex are dispensable for selective nuclear protein import. J Cell Biol 158, 63–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Cautain B, de Pedro N, Murillo Garzon V, Munoz de Escalona M, Gonzalez Menendez V, Tormo JR, Martin J, El Aouad N, Reyes F, Asensio F et al (2014) High‐content screening of natural products reveals novel nuclear export inhibitors. J Biomol Screen 19, 57–65. [DOI] [PubMed] [Google Scholar]
- 160. Hill R, Cautain B, de Pedro N & Link W (2014) Targeting nucleocytoplasmic transport in cancer therapy. Oncotarget 5, 11–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Craig E, Zhang ZK, Davies KP & Kalpana GV (2002) A masked NES in INI1/hSNF5 mediates hCRM1‐dependent nuclear export: implications for tumorigenesis. EMBO J 21, 31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Riviere Y, Blank V, Kourilsky P & Israel A (1991) Processing of the precursor of NF‐kappa B by the HIV‐1 protease during acute infection. Nature 350, 625–626. [DOI] [PubMed] [Google Scholar]
- 163. Ferrigno P, Posas F, Koepp D, Saito H & Silver PA (1998) Regulated nucleo/cytoplasmic exchange of HOG1 MAPK requires the importin beta homologs NMD5 and XPO1. EMBO J 17, 5606–5614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Beals CR, Clipstone NA, Ho SN & Crabtree GR (1997) Nuclear localization of NF‐ATc by a calcineurin‐dependent, cyclosporin‐sensitive intramolecular interaction. Genes Dev 11, 824–834. [DOI] [PubMed] [Google Scholar]
- 165. Erickson ES, Mooren OL, Moore D, Krogmeier JR & Dunn RC (2006) The role of nuclear envelope calcium in modifying nuclear pore complex structure. Can J Physiol Pharmacol 84, 309–318. [DOI] [PubMed] [Google Scholar]
- 166. Sarma A & Yang W (2011) Calcium regulation of nucleocytoplasmic transport. Protein Cell 2, 291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Beg AA, Ruben SM, Scheinman RI, Haskill S, Rosen CA & Baldwin AS Jr (1992) I kappa B interacts with the nuclear localization sequences of the subunits of NF‐kappa B: a mechanism for cytoplasmic retention. Genes Dev 6, 1899–1913. [DOI] [PubMed] [Google Scholar]
- 168. Traenckner EB, Wilk S & Baeuerle PA (1994) A proteasome inhibitor prevents activation of NF‐kappa B and stabilizes a newly phosphorylated form of I kappa B‐alpha that is still bound to NF‐kappa B. EMBO J 13, 5433–5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Zanella F, Dos Santos NR & Link W (2013) Moving to the Core: Spatiotemporal Analysis of Forkhead box O (FOXO) and Nuclear factor‐kappaB (NF‐kappaB) nuclear translocation. Traffic 14, 247–258. [DOI] [PubMed] [Google Scholar]
- 170. Stommel JM, Marchenko ND, Jimenez GS, Moll UM, Hope TJ & Wahl GM (1999) A leucine‐rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J 18, 1660–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Fineberg K, Fineberg T, Graessmann A, Luedtke NW, Tor Y, Lixin R, Jans DA & Loyter A (2003) Inhibition of nuclear import mediated by the Rev‐arginine rich motif by RNA molecules. Biochemistry 42, 2625–2633. [DOI] [PubMed] [Google Scholar]
- 172. Hubner S, Xiao CY & Jans DA (1997) The protein kinase CK2 site (Ser111/112) enhances recognition of the simian virus 40 large T‐antigen nuclear localization sequence by importin. J Biol Chem 272, 17191–17195. [DOI] [PubMed] [Google Scholar]
- 173. Komeili A & O'Shea EK (1999) Roles of phosphorylation sites in regulating activity of the transcription factor Pho4. Science 284, 977–980. [DOI] [PubMed] [Google Scholar]
- 174. von Kobbe C, van Deursen JM, Rodrigues JP, Sitterlin D, Bachi A, Wu X, Wilm M, Carmo‐Fonseca M & Izaurralde E (2000) Vesicular stomatitis virus matrix protein inhibits host cell gene expression by targeting the nucleoporin Nup98. Mol Cell 6, 1243–1252. [DOI] [PubMed] [Google Scholar]
- 175. Chen IH, Sciabica KS & Sandri‐Goldin RM (2002) ICP27 interacts with the RNA export factor Aly/REF to direct herpes simplex virus type 1 intronless mRNAs to the TAP export pathway. J Virol 76, 12877–12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Malik P, Tabarraei A, Kehlenbach RH, Korfali N, Iwasawa R, Graham SV & Schirmer EC (2012) Herpes simplex virus ICP27 protein directly interacts with the nuclear pore complex through Nup62, inhibiting host nucleocytoplasmic transport pathways. J Biol Chem 287, 12277–12292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Belov GA, Lidsky PV, Mikitas OV, Egger D, Lukyanov KA, Bienz K & Agol VI (2004) Bidirectional increase in permeability of nuclear envelope upon poliovirus infection and accompanying alterations of nuclear pores. J Virol 78, 10166–10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Castello A, Izquierdo JM, Welnowska E & Carrasco L (2009) RNA nuclear export is blocked by poliovirus 2A protease and is concomitant with nucleoporin cleavage. J Cell Sci 122, 3799–3809. [DOI] [PubMed] [Google Scholar]
- 179. Ghildyal R, Jordan B, Li D, Dagher H, Bardin PG, Gern JE & Jans DA (2009) Rhinovirus 3C protease can localize in the nucleus and alter active and passive nucleocytoplasmic transport. J Virol 83, 7349–7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Gustin KE & Sarnow P (2001) Effects of poliovirus infection on nucleo‐cytoplasmic trafficking and nuclear pore complex composition. EMBO J 20, 240–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Gustin KE & Sarnow P (2002) Inhibition of nuclear import and alteration of nuclear pore complex composition by rhinovirus. J Virol 76, 8787–8796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Park KS, Han BG, Lee KH, Kim DS, Kim JM, Jeon H, Kim HS, Suh SW, Lee EH, Kim SY et al (2009) Depletion of nucleophosmin via transglutaminase 2 cross‐linking increases drug resistance in cancer cells. Cancer Lett 274, 201–207. [DOI] [PubMed] [Google Scholar]
- 183. Watters K & Palmenberg AC (2011) Differential processing of nuclear pore complex proteins by rhinovirus 2A proteases from different species and serotypes. J Virol 85, 10874–10883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Frieman M, Yount B, Heise M, Kopecky‐Bromberg SA, Palese P & Baric RS (2007) Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J Virol 81, 9812–9824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Hussain S, Perlman S & Gallagher TM (2008) Severe acute respiratory syndrome coronavirus protein 6 accelerates murine hepatitis virus infections by more than one mechanism. J Virol 82, 7212–7222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Kopecky‐Bromberg SA, Martinez‐Sobrido L, Frieman M, Baric RA & Palese P (2007) Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J Virol 81, 548–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Shuai K & Liu B (2003) Regulation of JAK‐STAT signalling in the immune system. Nat Rev Immunol 3, 900–911. [DOI] [PubMed] [Google Scholar]
- 188. Ertel KJ, Brunner JE & Semler BL (2010) Mechanistic consequences of hnRNP C binding to both RNA termini of poliovirus negative‐strand RNA intermediates. J Virol 84, 4229–4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Mateo M, Reid SP, Leung LW, Basler CF & Volchkov VE (2010) Ebolavirus VP24 binding to karyopherins is required for inhibition of interferon signaling. J Virol 84, 1169–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Shabman RS, Gulcicek EE, Stone KL & Basler CF (2011) The Ebola virus VP24 protein prevents hnRNP C1/C2 binding to karyopherin alpha1 and partially alters its nuclear import. J Infect Dis 204 (Suppl 3), S904–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Bardina MV, Lidsky PV, Sheval EV, Fominykh KV, van Kuppeveld FJ, Polyakov VY & Agol VI (2009) Mengovirus‐induced rearrangement of the nuclear pore complex: hijacking cellular phosphorylation machinery. J Virol 83, 3150–3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Lidsky PV, Hato S, Bardina MV, Aminev AG, Palmenberg AC, Sheval EV, Polyakov VY, van Kuppeveld FJ & Agol VI (2006) Nucleocytoplasmic traffic disorder induced by cardioviruses. J Virol 80, 2705–2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Porter FW & Palmenberg AC (2009) Leader‐induced phosphorylation of nucleoporins correlates with nuclear trafficking inhibition by cardioviruses. J Virol 83, 1941–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Malim MH, Bohnlein S, Hauber J & Cullen BR (1989) Functional dissection of the HIV‐1 Rev trans‐activator–derivation of a trans‐dominant repressor of Rev function. Cell 58, 205–214. [DOI] [PubMed] [Google Scholar]
- 195. Malim MH, Tiley LS, McCarn DF, Rusche JR, Hauber J & Cullen BR (1990) HIV‐1 structural gene expression requires binding of the Rev trans‐activator to its RNA target sequence. Cell 60, 675–683. [DOI] [PubMed] [Google Scholar]
- 196. Hall MN, Craik C & Hiraoka Y (1990) Homeodomain of yeast repressor alpha 2 contains a nuclear localization signal. Proc Natl Acad Sci USA 87, 6954–6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Makkerh JP, Dingwall C & Laskey RA (1996) Comparative mutagenesis of nuclear localization signals reveals the importance of neutral and acidic amino acids. Curr Biol 6, 1025–1027. [DOI] [PubMed] [Google Scholar]
- 198. Weighardt F, Biamonti G & Riva S (1995) Nucleo‐cytoplasmic distribution of human hnRNP proteins: a search for the targeting domains in hnRNP A1. J Cell Sci 108 (Pt 2), 545–555. [DOI] [PubMed] [Google Scholar]
- 199. Truant R & Cullen BR (1999) The arginine‐rich domains present in human immunodeficiency virus type 1 Tat and Rev function as direct importin beta‐dependent nuclear localization signals. Mol Cell Biol 19, 1210–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Lee DC & Aitchison JD (1999) Kap104p‐mediated nuclear import. Nuclear localization signals in mRNA‐binding proteins and the role of Ran and Rna. J Biol Chem 274, 29031–29037. [DOI] [PubMed] [Google Scholar]
- 201. Lam MH, Hu W, Xiao CY, Gillespie MT & Jans DA (2001) Molecular dissection of the importin beta1‐recognized nuclear targeting signal of parathyroid hormone‐related protein. Biochem Biophys Res Commun 282, 629–634. [DOI] [PubMed] [Google Scholar]
- 202. Huber J, Cronshagen U, Kadokura M, Marshallsay C, Wada T, Sekine M & Luhrmann R (1998) Snurportin1, an m3G‐cap‐specific nuclear import receptor with a novel domain structure. EMBO J 17, 4114–4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Wen W, Harootunian AT, Adams SR, Feramisco J, Tsien RY, Meinkoth JL & Taylor SS (1994) Heat‐stable inhibitors of cAMP‐dependent protein kinase carry a nuclear export signal. J Biol Chem 269, 32214–32220. [PubMed] [Google Scholar]
- 204. Fukuda M, Gotoh I, Gotoh Y & Nishida E (1996) Cytoplasmic localization of mitogen‐activated protein kinase kinase directed by its NH2‐terminal, leucine‐rich short amino acid sequence, which acts as a nuclear export signal. J Biol Chem 271, 20024–20028. [DOI] [PubMed] [Google Scholar]


