FIGURE 2.
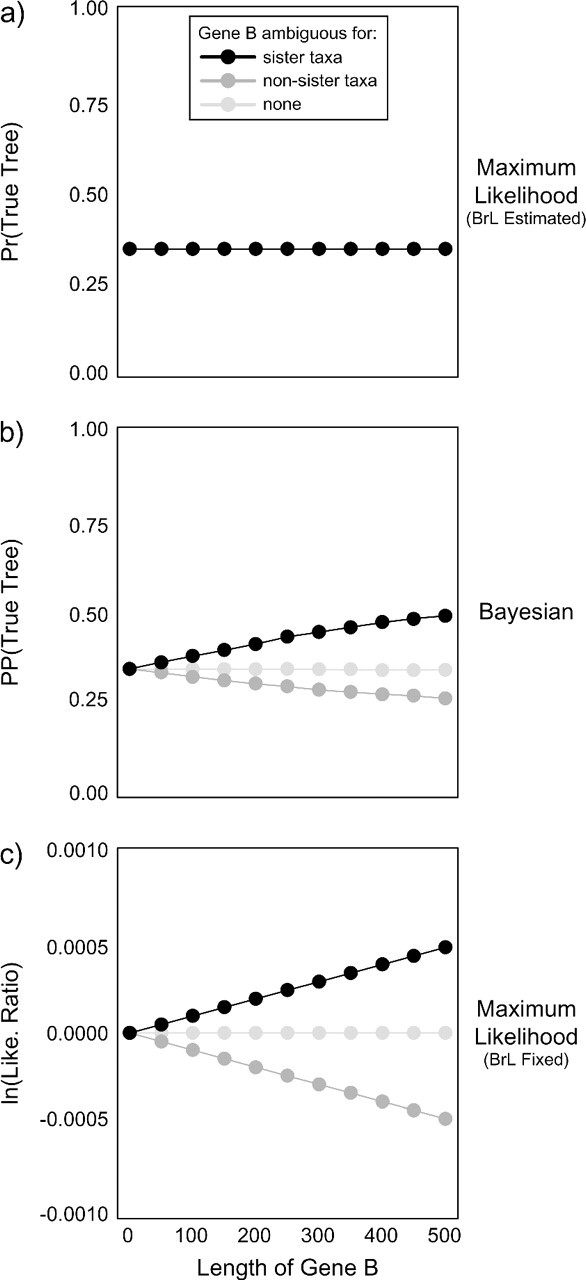
The effect of ambiguous characters on topological support when both genes are effectively invariable (rate = 0.000015 substitutions per site per My). On each graph, we plot the support for the true tree as a function of the length of Gene B. Each point represents the mean across 100 replicate data sets. In an ML framework (a), ambiguous characters do not affect topological support (Pr: calculated as the proportion of 100 replicates in which the true tree was chosen, with a value of 1/3 given to replicates with equal support across topologies) when branch lengths can be collapsed to polytomies. In a Bayesian framework (b), topological support (PP) changes as a function of the length of Gene B and whether Gene B is ambiguous for sister (black) or nonsister (dark gray) taxa. When Gene B is unambiguous (light gray), topological support is unaffected by the length of Gene B. When branch lengths are forced to take a small but nonzero value (10−6 substitutions/site) in an ML framework (c), ambiguous characters bias topological support (measured as the ratio of likelihood scores for the true to one of the false trees) in the manner seen in the Bayesian framework. Note that in a Bayesian framework, the flat prior on bifurcating topologies requires branch lengths to take a nonzero value. PP = posterior probability; Pr = probability.
