FIGURE 7.
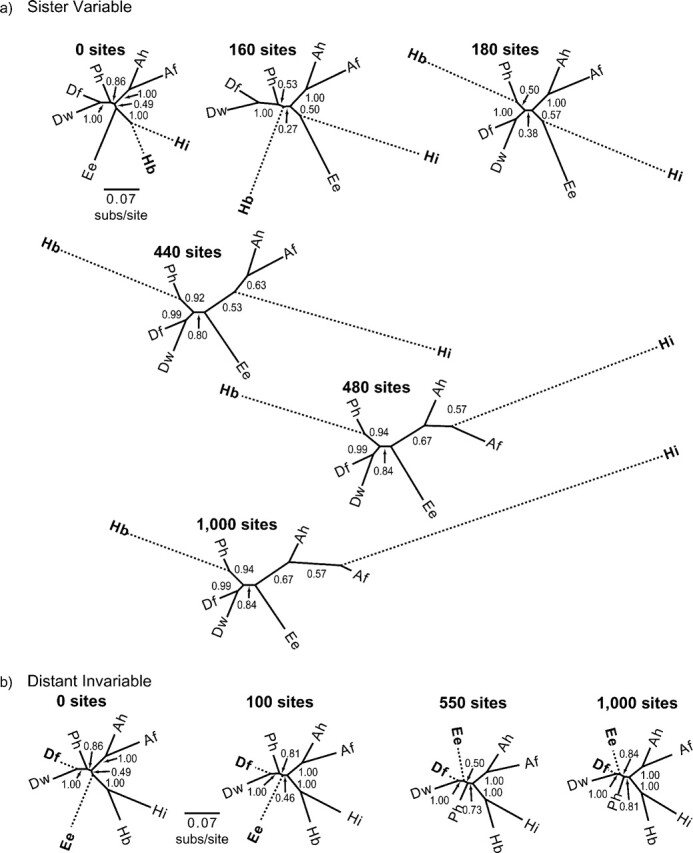
The effect of ambiguous characters on estimates of an empirical phylogeny estimated in a Bayesian framework. In (a), we present results based on an empirical data set with up to 1000 variable sites appended. The character state at each appended site was unambiguous but different for the sister taxa Hydromantes brunus (Hb) and Hydromantes italicus (Hi) and was ambiguous (“?”) for the other 6 taxa: Aneides flavipunctatus (Af), Aneides hardii (Ah), Desmognathus fucus (Df), Desmognathus wrighti (Dw), Ensatina eschscholtzii (Ee), and Phaeognathus hubrichti (Ph). The number of appended sites is given above each phylogeny, and the bipartition posterior probability estimate is given at each internal branch. In (b), we present results based on the same empirical data set but with up to 1000 invariable sites appended. Here, the character state at each appended site was identical for the distant taxa Df and Ee and was ambiguous for the other 6 taxa: Af, Ah, Dw, Hb, Hi, and Ph. Note that when variable sites are added, taxa with unambiguous characters are pushed apart on the phylogeny, whereas when invariable sites are added, taxa with unambiguous characters are pulled together. Topologies estimated in an ML framework matched those estimated in a Bayesian framework.
