Abstract
BACKGROUND:
The mammalian immune system is a remarkable sensory system for the detection and neutralization of pathogens. History is replete with the devastating effects of plagues, and the coronavirus disease 2019 (COVID-19) pandemic is a defining global health crisis of our time. Although the development of effective vaccines has saved many lives, the basic workings of the immune system are complex and require the development of animal models, such as inbred mice. Indeed, research in mice has been enormously productive, and the tremendous insights gleaned have resulted in many Nobel prizes and other accolades. However, past results are not necessarily a reliable guide to the future, and a notable limitation of animal models has been their failure to accurately model some human diseases and their inability to predict human immune responses in many cases. With regard to inbred mice, which have been the principal model of choice for immunology, this is likely due to the compromises that were necessary to create a more tractable and reproducible system for experimentation, such as genetic uniformity and lack of pathogen exposure, as well as the fact that mice are evolutionarily quite distinct. These considerations suggest that direct studies of the human immune system are likely to be extremely rewarding, both from a scientific and a medical perspective.
ADVANCES:
In the past decade there has been an explosion of new approaches and technologies to explore the human immune system with unprecedented precision. Insights into the human immune response to vaccination, cancers, and viral infections such as COVID-19 have come from high-throughput “omics” technologies that measure the behavior of genes, mRNA (single-cell transcriptomics), proteins (proteomics), metabolites (metabolomics), cells (mass cytometry), and epigenetic modifications (ATAC-seq), coupled with computational approaches.
OUTLOOK:
Sydney Brenner remarked in 2008, “We don’t have to look for a model organism anymore. Because we are the model organisms.” We propose that studying the immune system in humans, who are genetically diverse and afflicted by a multitude of diseases, offers both a direct link to medicine (i.e., “translation”) and the very real prospect of discovering fundamentally new human biology. New approaches and technology are now making this area much more approachable, but profiling immunity in humans is but the first step. Computational mining of the data and biological validation in animal models or human organoids are essential next steps, in an iterative cycle that seeks to bridge fundamental and applied science, as well as mouse and human immunology, in a seamless continuum of scientific discovery and translational medicine. This will represent a new paradigm for accelerating the development of vaccines and therapeutics.
Graphical Abstract
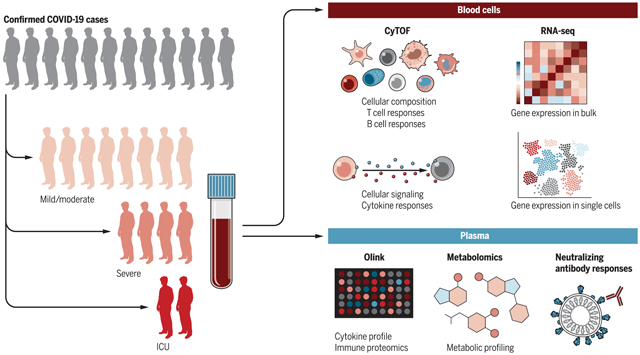
Probing the human immune response to viral infections. Systems biology techniques can be used to probe the human immune response to viral infections and can define molecular signatures that predict disease severity and illuminate the underlying mechanisms of disease.
Although the development of effective vaccines has saved countless lives from infectious diseases, the basic workings of the human immune system are complex and have required the development of animal models, such as inbred mice, to define mechanisms of immunity. More recently, new strategies and technologies have been developed to directly explore the human immune system with unprecedented precision. We discuss how these approaches are advancing our mechanistic understanding of human immunology and are facilitating the development of vaccines and therapeutics for infection, autoimmune diseases, and cancer.
As is the case with other biological disciplines, many of the major paradigms in immunology have emerged from research using animal models. For example, experimental validation of the clonal selection theory, the foundational paradigm on which modern immunology rests (1, 2), came from experiments in rats (3). The role of the thymus in generating T lymphocytes was discovered using mice (4), and studies in chickens led to the discovery of T and B lymphocytes as the organizing principles of the adaptive immune system (5). The role of the innate immune system in sensing pathogens through Toll-like receptors (TLRs) was discovered through studies of mice (6) and fruit flies (7). Dendritic cells (DCs), the central orchestrators of the immune response, were first discovered in mice (8). Indeed, the majority of the Nobel prizes for discoveries in immunology have been awarded for research performed in animals (Table 1).
Table 1. Animal models have enabled many Nobel Prize-winning discoveries in immunology.
Nobel Prize in Medicine or Physiology award dates are in chronological order.
| Discovery | Animal model |
|---|---|
| Development of antitoxins against diphtheria and tetanus: Emil von Behring (1901 Nobel Prize) and Kitasato Shibasaburo | Guinea pigs, goats, horses, mice |
| Discovery of phagocytes and their role in innate immunity: Elie Metchnikoff (1908 Nobel Prize) | Starfish |
| Methods for standardization of antibody activity in immune sera, description of neutralizing and complement-depending effect of antibodies, and enunciation of the “side-chain” theory of the formation of antibodies: Paul Ehrlich (1908 Nobel Prize) | Guinea pigs, horses |
| Discovery of anaphylaxis reactions: Charles Richet (1913 Nobel Prize) | Dogs |
| Discovery of immunological tolerance: Peter Medawar and MacFarlane Burnet (1960 Nobel Prize) | Mice |
| Discovery of major histocompatibility genes: Baruj Benacerraf, Jean Dausset, and George Davis Snell (1980 Nobel Prize) | Mice |
| Method for generating monoclonal antibodies: Cesar Milstein and George Kohler (1984 Nobel Prize) | Mice |
| Discovery of mechanism of generating antibody diversity: Susumu Tonegawa (1987 Nobel Prize) | Mouse embryo |
| Invention of organ transplantation techniques: Joseph Murray and Donnall Thomas (1990 Nobel Prize) | Dogs |
| Discovery of the role of major histocompatibility complex (MHC) molecules in mediating detection and killing virally infected cells: Peter Doherty and Rolf Zinkernagel (1996 Nobel Prize) | Mice |
| Discovery of dendritic cells: Ralph Steinman (2011 Nobel Prize) | Mice |
| Discovery of the role of Toll-like receptors in innate sensing: Bruce Beutler and Jules Hoffman (2011 Nobel Prize) | Mice, fruit flies |
| Discovery of cancer immunotherapy by inhibition of negative immune regulation: James Allison and Tasuku Honjo (2019 Nobel Prize) | Mice |
Yet one must remember that the origin of immunology is rooted deeply in experimentation in humans by Edward Jenner and Louis Pasteur, regarded as the fathers of immunology. Jenner’s first demonstration of the concept of vaccination was performed in 1796, when he inoculated 8-year-old James Phipps with pus from cowpox blisters on the hands of Sarah Nelmes, a milkmaid who had caught cowpox. Phipps developed a fever, but no disease, and was protected from smallpox when subsequently variolated (9). The findings gradually gained wide acceptance, culminating on 8 May 1980 with the declaration by the World Health Assembly that “the world and all its people have won freedom from smallpox.” Pasteur’s human experimentation began almost a century after Jenner, in 1885, when he treated his first human patient, a 9-year-old boy who had been bitten by rabid dogs. The victim received daily injections of progressively more virulent doses of an attenuated form of the virus (10). The boy survived and Pasteur became an international hero. Although Jenner and Pasteur would never receive institutional review board approval to perform these studies today, these experiments in humans can be considered to represent the First Golden Age in human immunology.
In the century and a half that followed Pasteur and Jenner, there has been elegant research on the human immune system, particularly in analyzing the cellular composition and functions of immune cell types isolated from the blood of healthy (11–13) and immune-deficient patients (14–18), yet there have been very few studies that have analyzed immune responses in humans in vivo. Instead, it is research in animal models that has been largely responsible for advancing our mechanistic understanding of the immune system. Indeed, a recurring theme in biology is that biologists have adapted unique features of various animal models to address fundamental mechanistic questions. Thus, neurobiologists have used squid with giant axons for studying synaptic action potential (19), cell biologists have used yeast and sea urchins for studying the cell cycle (20), developmental biologists have used nematodes for studying development (21), and immunologists have relied extensively on genetically inbred strains of mice. In such mice, the immune system can be studied without the confounding effects of genetic variability, and genes can be deleted (22, 23) or overexpressed (24) in a given tissue or cell type. However, these mice are genetically homogeneous and are usually housed in abnormally hygienic and specific pathogen-free (SPF) environments. Hence, they are far from ideal models for the immune systems of humans. Some recent studies have used free-living populations of feral mice or pet store mice (which occupy more natural habitats) to study immune responses (25, 26), but the extent to which the immune states of such mice reflect the immune states of humans is uncertain. Mice and humans diverged evolutionarily 96 million years ago, and although the immune systems of the two species are broadly similar, they differ in many important details (Table 2).
Table 2. The immune systems of mice and humans.
Adapted from (149).
| Mus musculus | Homo sapiens | |
|---|---|---|
| Evolutionary divergence | ~65 to 80 million years | |
| Genome | ~3.1 billion base pairs | |
| Size | Weight: ~20 to 30 g; length: 7.5 to 10 cm | Weight: ~62 kg: height: 163 cm (females), 176 cm (males) |
| Some differences in the innate immune system | TLR7 on pDCs, myeloid DCs (150) | TLR7 only on pDCs and B cells (150) |
| TLR9 on myeloid cells, pDCs and B cells (150) | TLR9 only on pDCs and B cells (150) | |
| TLR10: pseudogene (150) | TLR10: widely expressed (150) | |
| MyD88 knockout mice: impaired immunity to bacteria, viruses, and parasites (150) | MyD88 mutant humans: susceptible to invasive pathogenic bacterial infections, but normal immunity to many bacteria, viruses, fungi, and parasites (151) | |
| Some differences in the adaptive immune system | Ig subclasses: IgA, IgD, IgE, IgG1, IgG2a*, IgG2b, IgG3, IgM (152) *absent in C57BL/6, /10, SJL, and NOD mice, which have IgG2c |
Ig subclasses: IgA1, IgA2, IgD, IgE, IgG1, IgG2, IgG3, IgG4, IgM (152) |
| Ig CDR-H3 region: shorter, less diverse (153) | Ig CDR-H3 region: longer, more diverse (153) | |
| Effect of γc deficiency: loss of T, NK, and B cells | Effect of γc deficiency: loss of T, NK, but normal B cells (154) | |
| Effect of IL-7R deficiency: blocks T and B cell development (154, 155) | Effect of IL-7R deficiency: only blocks T cell development (154, 155) | |
Experiments in mice have led to the development of many successful therapies in humans. For example, basic discoveries in mice enabled the development of anti-tumor necrosis factor (TNF) therapy for rheumatoid arthritis (27) as well as the enormous success of cancer immunotherapy (metastatic melanoma, which was almost incurable previously, can now be in remission in >50% of patients as a result of combined anti-CTLA4 and anti-PD1 therapy) (28). Yet the number of failures to translate successes from animals to humans has been increasing. One of the most notorious examples in recent years is theralizumab, an anti-CD28 antibody (29). Its first trial in humans resulted in hospitalization of all six volunteers, at least four of whom suffered multi-organ dysfunction (29), despite no such adverse effects being observed in preclinical studies in primates. In vaccine development, the world of HIV vaccines suffered a startling setback in 2008 when the first clinical trial of a vaccine designed to elicit cellular immunity against the virus appeared to increase the rate of HIV infection in individuals with prior immunity against the adenovirus 5 vector used in the vaccine (30). Yet this vaccine had shown protective efficacy against a humanized simian immune deficiency virus (SHIV) in primates, as well as some modest protection against the pathogenic SIV (31). These failures symbolize the “valley of death” in drug and vaccine development and underscore the imperative to harness the “human model” at a relatively early stage in the discovery-development pipeline. In this context, recent advances have highlighted new approaches to studying immune responses in humans. In particular, advances in systems biology coupled with human phenotypic data have allowed the delineation of molecular networks that drive human immunity. Here, we review these advances and discuss the limitations and critical challenges that need to be overcome to effectively exploit the human model to advance our understanding of the immune system and design novel vaccines and therapeutics.
Humans as model organisms in immunology
In human beings, a widely available resource containing a representation of the immune system is blood, which is routinely taken from almost all patients (as well as from healthy individuals) and provides a kaleidoscope of many lineages and differentiation states within the immune system. Because migration of cells is a key feature of the immune system, blood leukocytes (present at 1 to 3 million per milliliter of blood) represent recent emigrants from tissues, including sites of infection or vaccination. This, coupled with the advent of high-throughput “omics” technologies that measure the behavior of genes, mRNA (transcriptomics), proteins (proteomics), metabolites (metabolomics), cells (mass cytometry), and epigenetic modifications (ATAC-seq), has started to provide unprecedented insight into the human immune response in the context of vaccination, infections, cancers, and autoimmunity (Fig. 1). In particular, the development of next-generation sequencing (NGS) has facilitated the comprehensive evaluation of transcripts including noncoding RNAs, microRNAs, and long noncoding RNAs, and the use of ATAC-seq has further allowed definition of chromatin accessibility loci to identify epigenetic regulators of gene expression (32). Furthermore, such NGS permits the characterization of T and B cell repertoires (33, 34), and high-throughput single-cell sequencing is yielding rich new information about the heterogeneity of cell subsets and identification of new populations of cells (35). Single-cell mass cytometry techniques allow analysis of the cellular dynamics of immune responses with an exquisite degree of precision (36) and analysis of the epigenetic landscape of single cells (37) (Fig. 1). Finally, the development of high-throughput platforms has allowed researchers to analyze the functional properties of antigen-specific antibodies isolated from serum of vaccinated or infected humans (38, 39). Specific examples of the application of these technologies are considered below.
Fig. 1. Systems biology approach to probing human immunity.

The human immune system can be perturbed by vaccination, infection, or allergens, or in autoimmunity. Blood samples or fine needle aspirate samples of lymph nodes or other human tissues (e.g., skin biopsies) can be isolated and the immune response analyzed at multiple levels of the hierarchy of biological organization, using a wide array of technologies including transcriptomics, epigenomics, mass cytometry, and metabolomics.
Vaccines as probes for the human immune system
Vaccination represents the most effective way to prevent infectious diseases. But to immunologists, vaccines represent excellent probes of the human immune system. First, vaccines allow a synchronized perturbation of the immune system, such that the immune response can be assessed from the earliest minutes to several decades after vaccination (40). Although the immune system is also perturbed in infections, autoimmune diseases, and cancers, in such cases the precise moment of immune perturbation cannot be known. Second, vaccines represent a diverse array of microbial stimuli (e.g., live viruses, carbohydrate vaccines, vaccines containing adjuvants) that can be used to perturb the immune system (40). Third, vaccines are widely administered to diverse human populations (e.g., neonates versus elderly; undernourished versus obese). For decades, vaccine manufacturers have relied on a single measurement—typically the magnitude of the antigen-specific antibody response (e.g., neutralization antibody titers or binding antibody titers measured by enzyme-linked immunosorbent assay)—to assess the immune response stimulated by vaccination. However, such measurements fail to capture the global architecture of the immune response to vaccination and the multiple immunological mechanisms that can mediate protection against a given pathogen (41).
Recent advances in omics technologies have enabled scientists to probe the immune response to vaccination in a comprehensive way, by analyzing the cellular and molecular networks driving immune response to vaccination. These omics technologies can measure changes in cellular subsets, transcriptome, metabolome, or epigenome, even at the single-cell level. Coupling these with computational approaches developed to analyze and interpret such data has led to the generation of the new field of “systems vaccinology.” The aim of systems vaccinology is to comprehensively analyze the immune response to vaccination with a view to defining new mechanisms and correlates of protective immunity (41, 42).
Initial proof-of-concept studies demonstrating the use of systems biology approaches to identify molecular signatures of vaccination used the yellow fever live-attenuated vaccine YF-17D (43, 44). YF-17D is one of the most successful vaccines ever developed, having been administered to more than 600 million people worldwide with an efficacy of 99% (45); a single immunization results in robust antigen-specific cytotoxic T cell responses and neutralizing antibody responses that can persist for several decades (42). Transcriptomic analysis of peripheral blood mononuclear cells (PBMCs) isolated 3 to 7 days after vaccination of healthy young adults with YF-17D revealed a gene expression profile pattern consisting of genes encoding proteins involved in antiviral sensing and viral immunity, including the type I interferon (IFN) pathway (43, 44). Computational analysis and machine learning approaches revealed signatures of early gene expression, which correlated with and predicted the magnitude of the later antigen-specific CD8+ T cell and neutralizing antibody responses, in an independent study (43).
Subsequently, several groups applied systems-based analysis to study immune responses to vaccination with seasonal influenza (46–50), meningococcal (51), shingles (52), malaria (53, 54), smallpox (55), Ebola (56) vaccines, as well as a candidate vaccine against HIV (57). In addition, several groups analyzed the transcriptional profile at “baseline” (i.e., prior to vaccination) to identify signatures of vaccine responsiveness prior to vaccination (49, 58–61). Furthermore, although until recently most studies have relied on transcriptional profiling to define signatures of vaccine immunity, recent studies have performed integrated analysis of orthogonal datasets to generate an integrated model of vaccine immunity. To understand the mechanisms of immunity induced by the live attenuated shingles vaccine, we constructed a multiscale, multifactorial response network (MMRN) of immunity induced by vaccination in healthy young and older adults. The MMRN revealed striking associations between orthogonal datasets, such as transcriptomic and metabolomics signatures, cell populations, and cytokine levels, and identified immune and metabolic correlates of vaccine immunity (52).
Defining correlates of vaccine efficacy (i.e., the capacity of vaccination to protect from infection) is more challenging. One approach to defining correlates of efficacy is the controlled human infection model (such as those that involve controlled challenge of humans with pathogenic strains of typhoid, influenza, or malaria), in which subjects can be vaccinated and subsequently challenged with the pathogen to define the mechanisms and correlates of vaccine immunity (62). Such studies have already been done in the context of the RTS,S malaria vaccine and have yielded important insights about the mechanisms of protection (53, 54). In particular, the results from these studies reveal that protection from malaria infection can be mediated by multiple immunological mechanisms involving the magnitude of serum circumsporozoite-specific antibody titers as well as the magnitude of antigen-specific CD4+ T cells with the capacity to produce polyfunctional cytokines (53, 54). In the case of phase 2b or phase 3 efficacy trials that involve several thousand subjects, the primary endpoint is efficacy. Typically, the trial design and sample collection schedules do not provide opportunities for retrospective analysis of immune responses. This issue has been brought to the fore recently, as the world races to develop a vaccine against COVID-19. At present, more than 160 vaccine candidates are being developed across the world, and systems vaccinology approaches may be useful to help identify early predictors of efficacy and accelerate the COVID-19 vaccine testing pipeline. Therefore, future efficacy trials should be leveraged to include sample collection schedules that facilitate systems-based immune profiling, with a view to defining mechanisms and correlates of efficacy.
An important question is the extent to which these high-throughput data can yield new insights about the immune system. Several mechanistic insights are emerging from such studies. For example, among the signatures that predicted CD8+ T cell responses to YF-17D was general control nonderepressible 2 kinase (GCN2; also called eukaryotic translation initiation factor 2α kinase 4), a sensor of amino acid starvation and an orchestrator of the integrated stress response (43). Studies with knockout mice showed a key role for YF-17D– induced GCN2 activation in programming DCs to initiate autophagy and enhance antigen presentation to CD4+ and CD8+ T cells (63), as well as in the control of intestinal inflammation (64). These results revealed an unappreciated link between virus-induced integrated stress response in DCs and the adaptive immune response.
Another insight that emerged from the influenza vaccine study was the impact of the gut microbiota on vaccine-induced immunity (65). Transcriptional profiling of immune responses to vaccination with the seasonal influenza vaccine revealed a striking correlation for the expression of the gene encoding TLR5—a receptor for bacterial flagellin—3 to 7 days after vaccination and the ensuing antibody response a month later (47). This prompted mechanistic investigation using mice that were genetically deficient in TLR5. Such studies revealed that TLR5-deficient mice were impaired in the vaccine-specific antibody response induced by vaccination with the seasonal influenza vaccine (66). This effect was shown to depend on sensing of bacterial flagellin by TLR5; consequently, mice administered antibiotics to deplete their microbiota or mice devoid of microbiota from birth (so-called “germ-free mice”) were impaired in their capacity to launch antibody responses to seasonal influenza vaccination (66). This experiment in mice raised the key question regarding whether the microbiota controlled immune responses after vaccination in humans. A recent clinical trial assessed the impact of administering broad-spectrum antibiotics to healthy adults before and after seasonal influenza vaccination; despite a reduction in gut bacterial load by four orders of magnitude and a long-lasting restriction in gut bacterial diversity, antibody responses were unaffected (67). However, in a second trial of subjects with low preexisting antibody titers, there was significant impairment in H1N1-specific neutralization and binding immunoglobulin G1 (IgG1) and IgA responses (67). In addition to these effects on the adaptive immune response, antibiotic administration also enhanced inflammatory signatures (including AP-1/NR4A expression), observed previously in the elderly (49), as well as perturbation of the serum metabolic profile, with a reduction in serum secondary bile acids by three orders of magnitude. The change in serum bile acids was highly correlated with AP-1/NR4A signaling and inflammasome activation. Integrative analysis of multi-omics datasets revealed significant associations between bacterial species and metabolic phenotypes, highlighting a key role for the microbiome in modulating human immunity (67). These studies show that systems approaches can help to identify early “signatures” that could predict the later immunogenicity or efficacy of a vaccine and yield new mechanistic insights.
Probing the human immune system in infections
Despite the invention of vaccines and antibiotics, infectious diseases continue to exert a major toll on human health, as evidenced by the recent COVID-19 pandemic (68). In 2016, lower respiratory infections, diarrheal diseases, tuberculosis (TB), and HIV/AIDS accounted for nearly 7 million deaths per year (69), and these diseases alone were the leading cause of disability-adjusted life years (DALY), accounting for 12.1% of all DALYs (69). Animal models have been developed for several infectious diseases and have yielded many valuable insights about mechanisms of immunity, yet differences in pathogenesis between these models and the human model have posed challenges.
In the past decade or so, there has been a growing effort to study immunity to infections in humans. An obvious limitation here, unlike the case with vaccination, is that the precise moment of natural infection cannot be known, thus posing an obstacle to analyzing immune responses during the earliest stages of infection. Nonetheless, there are some notable examples of addressing this issue, such as the FRESH study (Females Rising through Education, Support, and Health), which is a longitudinal study to identify and analyze participants immediately after they are infected with HIV (70). The FRESH program is located in the Umlazi township of South Africa, where the HIV prevalence rate in young women is as high as 66% at age 23. Investigators are collecting blood samples from 300 noninfected women aged 18 to 23, with a view to studying the earliest events that occur after natural infection with HIV (70). This FRESH study is providing key mechanistic insights into the biological factors that predispose to HIV infection. For example, the composition and diversity of the cervicovaginal microbiome were found to have a noticeable effect on the local host inflammatory response and the risk of HIV acquisition (71). In addition, there was profound CD8+ T cell activation and proliferation (>70% of CD8+ T cells in some individuals, the majority being HIV-specific by tetramer staining) in untreated acute infection. These cells rapidly became proapoptotic and were defective at producing antiviral cytokines such as IFN-γ, which suggested that HIV infection caused abnormal activation of CD8+ T cells (72), different from that observed with live viral vaccines such as YF-17D or smallpox (73).
Another area that has emerged in recent years is the profiling of transcriptional gene expression signatures in the blood to enhance our understanding of the host immune response to infection (74–84). Mycobacterium tuberculosis has been a scourge of humanity since ancient times, having survived more than 70,000 years, and currently one-third of the world’s population are carriers of the latent pathogen and are at risk for developing active disease (85). One of its mysteries is that an estimated 10% of infected adults succumb to full-blown TB, but most are able to control the disease throughout their lives (85). Blood transcriptional analysis of tuberculosis has revealed IFN-inducible genes whose expression diminished after successful treatment (76). Coupled with sensitive radiography techniques, blood transcriptomics have revealed heterogeneity in patients with active tuberculosis and asymptomatic people with latent tuberculosis, suggesting a continuum of infection and immune states (76). A recent study using high-dimensional mass cytometry demonstrated that latent TB is associated with enhanced cytotoxic responses and natural killer (NK) cells (86). Finally, a meta-analysis of 24 datasets containing 3083 transcriptome profiles from whole blood or PBMC samples of healthy controls or patients with active or latent TB from 14 countries yielded a three-gene set in whole blood that was robustly diagnostic for active tuberculosis, suggesting the clinical utility of such a signature (87, 88).
An especially fruitful area of research has been the analysis of immune responses in HIV-infected individuals, which has provided new insights about antigen design strategies for vaccination. A small proportion of HIV-infected individuals develop broadly cross-reactive neutralizing antibody responses that can neutralize a broad array of HIV viruses. Monoclonal antibodies have been isolated from such subjects and have been used to define critical antigenic epitopes that represent the targets of the broadly cross-reactive neutralizing antibodies (89–91).
Probing the human immune system in autoimmunity
Autoimmune diseases represent a diverse family of some 80 distinct diseases, such as rheumatoid arthritis, type I diabetes, and systemic lupus erythematosus (SLE), which afflict 5 to 9% of the global population (92). In healthy individuals, the immune system is considered to be “tolerant” of proteins present within the host’s own body, so-called “self” proteins. However, in humans who develop autoimmune diseases, there is a complex and dynamic interplay between host genes and the environment that results in the loss of immune tolerance to “self antigens,” leading to the development of autoreactive T and B cells that attack the body’s own tissues (93). Such autoreactivity can be organ-specific (e.g., rheumatoid arthritis) or systemic (e.g., SLE).
Preclinical animal models of human autoimmune diseases have provided important clues to these diseases in the form of possible mechanisms and etiology. Studies using mouse models of spontaneous autoimmune diseases [e.g., the experimental autoimmune encephalomyelitis (EAE) model for multiple sclerosis (MS); NOD mice for diabetes; the MRL/lpr model for lupus] have provided powerful tools to assess mechanisms and test novel therapeutic concepts (94, 95). Furthermore, mice genetically deficient in particular genes have established critical regulators of autoimmune processes, and the use of engineered models of autoimmune disease, such as mice transgenic for T and B cell receptors specific for autoantigens, has yielded a wealth of mechanistic insight (94, 95). However, the failure to translate promising therapies from these animal models to humans has led to a reassessment of the extent to which such models reflect the complexity and heterogeneity of the clinical presentation of the human disease. For example, of the ~200 treatments that prevent or delay the development of type 1 diabetes in the NOD mouse model, only a few have been shown to have clinical promise, and in only a fraction of patients (94–96). Some of the failures in translation were likely due to differences in the experimental design in the preclinical versus clinical studies, but species differences in the immune systems and human variations have likely been a major impediment.
The discovery of TNF-α as a central regulator in rheumatoid arthritis and the development of therapeutic anti-TNF-α antibodies have established a new paradigm for the treatment of autoimmune diseases (97). Ironically, it was an experiment with human tissues—the evaluation of cytokine expression in human rheumatoid synovium—that represented the key experiment in defining the cytokines that were overexpressed in the disease site (98). Many cytokines were overexpressed, so which of those represented the best therapeutic target was unclear. This was analyzed by blocking various cytokines in rheumatoid synovial cultures, which revealed that blockade of TNF-α reduced interleukin-1 (IL-1) synthesis as well as all other pro-inflammatory cytokines found in joints. This was the first demonstration that TNF-α could be an important therapeutic target (98). Concurrent experiments in mice supported the concept of a TNF-α-dependent cytokine cascade (99). Subsequent experiments in animal models demonstrated that administration of anti-TNF-α monoclonal antibody ameliorated both inflammation and joint damage in the collagen type II model of arthritis (100). This provided the rationale for anti-TNF therapy in rheumatoid arthritis, which has heralded a revolution in the introduction of protein therapeutics, termed “biologics.”
Genome-wide association studies (GWASs) have been performed over the past decade, with the goal of analyzing single-nucleotide polymorphisms (SNPs) in genes associated with a disease in large patient populations, and have revealed the polygenetic basis of many autoimmune diseases (101). Originally it was hoped that genetic studies of autoimmune diseases would discover one or a few defective genes that could explain the pathology and be useful diagnostically. This launched a number of GWASs, but unfortunately these efforts have shown that many loci contribute very small effects. However, some alleles. such as the human leukocyte antigen (HLA) alleles, do show statistically significant associations. Because different HLA alleles bind to distinct antigenic peptides, and because these HLA-peptide complexes are specific targets for T cell recognition, this points to a potential role for T cells in initiating and maintaining the disease (102).
Transcriptomics of patient PBMCs and whole blood have revealed alterations in the type I IFN network in SLE, Sjögren’s syndrome, inflammatory myopathies, and a subset of systemic sclerosis patients. SLE is an autoimmune disease characterized by loss of tolerance to nucleic acids, including double-stranded DNA and ribonucleoproteins, with diverse clinical manifestation. Recently, Pascual and colleagues analyzed transcriptional changes in the blood of 158 pediatric lupus patients (103). Using computational approaches, they identified transcriptional signatures of the type I IFN and plasmablast responses as the most robust biomarker of disease activity. They detected increased enrichment of neutrophil transcripts during progression to active nephritis. Strikingly, they were able to stratify the patients into seven groups according to distinct molecular correlates of disease activity. This study reveals the molecular heterogeneity underlying disease manifestation in SLE and suggests that future trials should implement “precision medicine” therapies that target the specific molecular anomalies in genetically and clinically complex autoimmune diseases.
Such approaches have also revealed a key role for IL-1β in systemic-onset juvenile idiopathic arthritis and have led to the development of anti-IL-1-based therapeutics (104). In the case of MS, IFN-β therapy has been used as a drug therapy, but despite its clinical use, the treatment modality involved a protracted regimen of uncertain clinical benefit (105). Transcriptomics has revealed transcriptional responses to IFN-β1α and has enabled the stratification of patients according to therapeutic response (106). Furthermore, systems approaches have been used to probe the immune response in patients with celiac disease, an intestinal autoimmune disease. Administration of gluten to patients with celiac disease not only induced the expansion and mobilization into the circulatory system of gliadin-specific CD4 T cells 6 days after challenge, but also induced CD8 T cells and γδ T cells with oligoclonal T cell receptors with similar kinetics (107). This same pattern of co-mobilized CD4+, CD8+, and γδ T cells was seen in the blood and central nervous system tissue of mice induced to develop EAE, which suggests that this may be a general feature of autoimmune diseases. Surprisingly, upon analysis of the specificities and functional properties of these different T cells, the CD4+ and γδ T cells were found to be pathogenic, whereas the CD8+ cells were found to be suppressive and able to kill the CD4+ cells in vitro (108). This indicates a complex dynamic in autoimmunity that will be fascinating to investigate further.
Clearly, analysis of the human immune system in the context of autoimmune diseases is yielding novel insights about the mechanisms and potential targets for therapy. Future studies could be aimed at using systems-based approaches to define the immune mechanisms and gene-environment interactions that lead to autoimmunity. However, unlike with vaccination, it has been challenging to assess the earliest stages of the response, and thus the etiology of the disease has been difficult to ascertain. One solution to this problem may be to establish human cohort studies such as the TEDDY study (The Environmental Determinants of Diabetes in the Young), a multi-center prospective cohort study that was designed to follow children with and without a family history of type 1 diabetes to understand the environmental factors that contribute to the disease (109, 110). In this study, 424,790 newborn children younger than 4 months were screened for high-risk HLA alleles, of whom 21,589 had the qualifying haplotypes and 8676 were enrolled. Of this group, 672 developed persistent antibodies to insulin, glutamic acid decarboxylase, or islet antigen 2, and these subjects are being followed for 15 years. It is predicted that 390 children will develop diabetes. Blood samples are being collected and banked from the day of birth and every 3 months, and stool samples collected (for microbiome analysis) quarterly, throughout the 15-year period (110). Clinical metadata (infections, medication, immunizations), exposure to dietary and other environmental factors, negative life events, family history, tap water, and measurements of psychological stress will also be collected. The major goals of the TEDDY study are to discover genes associated with the development of autoantibodies and type 1 diabetes, to identify environmental exposures that modify the risk of autoimmunity, and to use omics technologies to define signatures of the transcriptome, metabolome, epigenome, or microbiome that correlate with and predict autoantibody formation and disease progression (110). Such system analysis will not only yield novel biomarkers but also reveal mechanistic insights about the sequalae of immunological events, starting at the very earliest stages, that culminate in disease. Studies such as TEDDY also provide unique opportunities for discovering novel autoantigens—for example, by analyzing the B cell repertoire and isolating monoclonal antibodies from plasmablasts, or analyzing the T cell receptor repertoire using the recently discovered GLIPH algorithm (grouping of lymphocyte interactions by paratope hotspots) (34).
Probing the human immune system in cancer
The development of checkpoint blockade inhibitors has revolutionized cancer immunotherapy (28) and has led to durable survival outcomes in some patients with metastatic disease (111). Combination therapy of previously untreated advanced melanoma patients with nivolumab and ipilimumab (monoclonal antibodies that respectively target the PD-1 and CTLA-4 receptors on T cells) resulted in significantly greater objective-response rates and progression-free survival relative to ipilimumab monotherapy alone (112, 113). The development of these immune therapy regimens occurred as a result of fundamental discoveries using the mouse model (114, 115). However, a major issue with checkpoint inhibitors is that only a fraction of cancer patients experience durable clinical benefit (28). Thus, the identification of molecular signatures that can be used to predict nonresponsiveness to checkpoint inhibitors would be valuable in identifying patients who would be viable candidates for treatment. In this context, tissue-based immune monitoring is providing important insights into the cellular and molecular mechanisms of immune checkpoint therapy. For example, in a trial of ipilimumab, analysis of the RNA transcriptome in pre- and post-treatment tumor tissues revealed major transcriptional changes related to T cell signaling and activation after CTLA-4 blockade. In particular, ICOS+ T cells were increased in tumor tissues from patients with ipilimumab, and the increase in ICOS+ T cells was accompanied by similar increases in the blood (116). To test the hypothesis that ICOS+ CD4 T cells may play a role in the therapeutic effect of CTLA-4 blockade, similar studies were conducted in mice; although anti-CTLA-4 treatment was shown to induce tumor rejection in 80 to 90% of mice, the efficacy was less than 50% in ICOS-deficient mice (116).
In addition to tissue-based immune monitoring, emerging studies demonstrate that analysis of peripheral blood can yield new insights into mechanisms and biomarkers of tumor immune therapy. A recent study used immune profiling of blood from patients with stage IV melanoma before and after treatment with anti-PD-1 antibody (pembro-lizumab); the clinical failure in many patients was not due to a lack of invigoration of exhausted T cells, but rather due to the high tumor burden prior to anti-PD-1 therapy (117). Thus, the magnitude of reinvigoration of circulating exhausted T cells determined in relation to pretreatment tumor burden correlated with clinical response.
These studies highlight the power of human immune profiling technologies to identify signatures and mechanisms of antitumor immunity. In this context, emerging evidence suggests that patients receiving immunotherapy can be stratified into responders versus nonresponders based on the composition of their gut microbiomes (118–120). Future work should thus be aimed at using multi-omics technologies to delineate the transcriptional, metabolomic, epigenetic, microbiome, and cellular networks that regulate antitumor immunity, with a view to obtaining molecular signatures of responsiveness to cancer immunotherapy, similar to what has been accomplished with analysis of vaccination-induced responses (52, 67).
Challenges in the pursuit of human immunology in the 21st century
Human immunology is being rapidly revitalized by spectacular advances in technology and their application to the study of immune responses in humans. Although the preceding discussion has focused on vaccines, infections, autoimmunity, and cancer, systems-based approaches are also beginning to be used to probe the immune response in asthma and allergic diseases in, for example, children before, during, and after placebo-controlled oral challenges to peanuts (121, 122) or in the context of transplantation (123, 124). Yet the field is just beginning, and major challenges need to be addressed if it is to be a success. These include scientific, practical, and cultural challenges, as discussed below.
Scientific challenges: The obstinately diverse species
One major scientific challenge is dealing with diversity in the human model. Peter Medawar once called us the obstinately diverse species (125), a reference to our diverse genetic makeup. However, profound demographic, socioeconomic, and environmental changes have resulted in even further diversification with respect to health and age distributions. For example, The State of Food Security and Nutrition in the World 2019 states that maternal and child undernutrition contributes to 45% of deaths in children under 5, yet overweight and obesity are increasing in almost all countries and contribute to 4 million deaths globally (126). Indeed, paradoxically many populations are afflicted by the dual burdens of undernutrition and overweight. In 2018, Africa and Asia bore the greatest share of all forms of malnutrition, accounting for 90% of all stunted and wasted children but also nearly 75% of all overweight children worldwide (126). Disparity in life expectancy is also wide; as of 2018, Monaco had an average life expectancy of 89.52 years, whereas Chad was lower at 49.81 years (127), only moderately better than the 35 years in 17th-century Britain. These wide disparities no doubt impinge on the physiological states of humans and on variations in their immunological states in particular. This in turn is likely to affect responses to vaccination or susceptibility to inflammatory diseases.
The immunological state of an individual can vary longitudinally with time as a result of environmental changes, aging, or developmental changes in early life. In this context, Olin et al. used mass cytometry to analyze the changes in blood leukocyte composition in 100 newborn children during the first 3 months of life. The blood leukocyte composition of preterm and term children differed at birth but converged on a shared trajectory (128). Alternatively, there can be variations in the immunological states in individuals within a given population (because of genetic, microbiome, and environmental differences) (129, 130), or there can be variations in the immunological states between different populations (Fig. 2). This diversity is, however, an excellent source of natural variation or perturbation that enables effective interrogation of the human immune system in a population to draw predictive insights, and is thus a major advantage of the human model (131). To address this issue, Carr et al. used systems approaches to establish the cellular profiles of 670 healthy individuals and demonstrated high interindividual variation but low longitudinal variation at the level of cellular subset composition. Influenza vaccination induced expansion of antigen-specific B cell clones, but the cellular subset structure was elastic and returned to the individual’s unique baseline state. The largest influence on immune variation between individuals was cohabitation, with 50% less variation between individuals who share an environment than between people in the wider population (130). Such variations can lead to drastic differences in the responsiveness of the immune systems, such as wide differences in the immunogenicity and efficacy of vaccines. Therefore, a major challenge for human immunology is to embrace this diversity by probing the immune system in diverse human populations, with a view to obtaining mechanistic insights about the factors that lead to this variation. To understand this diversity, it must be accurately captured and recorded. This means aiming to record and standardize as much clinical metadata as possible during human studies. This can be facilitated through close collaboration between immunologists/basic scientists and clinicians. As precision medicine and wearable health technology advance, more and more of these data will become available (132, 133).
Fig. 2. Variations in the immunological states of humans.
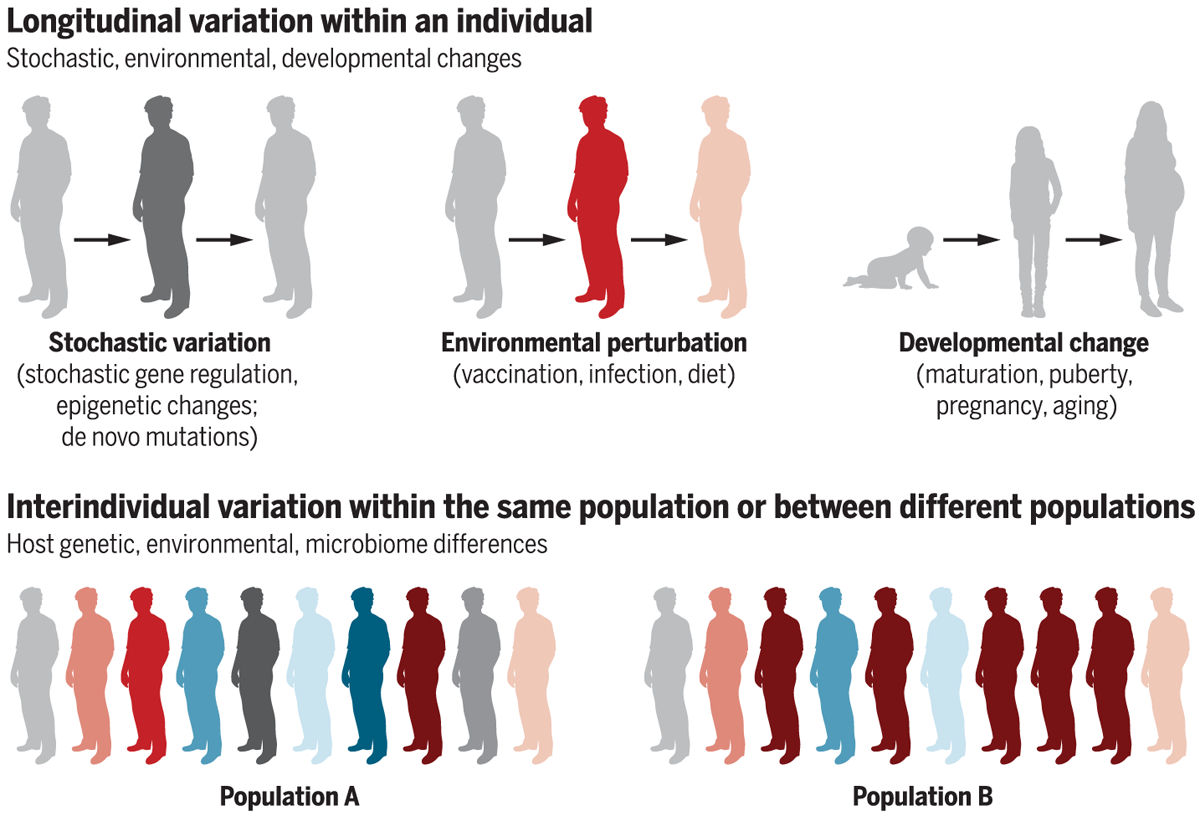
The immunological state of an individual can vary with time as a result of stochastic changes, environmental perturbations (e.g., vaccination, infection, exposure to allergens), or developmental changes. Alternatively, the immunological state of individuals in a given population, or individuals in different populations, may vary because of differences in host genetics, the environment, or individuals’ microbiomes. Such human variation can lead to different outcomes in responses to vaccination, therapies, or susceptibilities to infections and inflammatory disorders.
The confounding effects of genes can be addressed to some degree by performing studies in monozygotic twins. Immune studies along these lines have shown some strong genetic influences in young children receiving vaccines, but these genetic dependencies dwindle with age. One study reported that 70% of variation had little or no genetic dependency, reinforcing the idea that this is an adaptive system, adapting to microbial exposure similar to the way the nervous system responds to sensory input (61). An additional way to analyze the effects of genes is to study immune responses in subjects with monogenic lesions that consist of highly penetrant genetic mutations that exhibit large phenotypic effects. Such human knockouts, such as individuals with a common stop codon polymorphism in the ligand-binding domain of TLR5 (TLR5392STOP), are unable to mediate flagellin signaling and are susceptible to infection with a flagellated organism, Legionella pneumophila (134). Given the aforementioned studies demonstrating a role for suboptimal antibody response of TLR5 knockout mice to seasonal influenza vaccination (66), it would be of interest to use high-throughput systems approaches to profile immune responses in such human mutants. Indeed, associations of SNPs in genes involved in innate and adaptive immunity and with measles and rubella vaccine responses have been described (135).
In the case of microbial imprinting, one particularly striking example of microbial influence is the case of cytomegalovirus (CMV), a type of herpes virus that has representatives throughout the animal world. Monozygotic twins who were discordant for CMV had many alterations in their immune system; almost 60% of 200+ variables indicated that this virus has an unusual ability to change many aspects of the immune system (61). Other studies have shown that CMV+ young adults have better antibody responses to a flu vaccine than age-matched CMV comparators, and that even more profound effects can be seen in mice infected with mCMV prior to influenza infection, with substantially reduced flu titers, indicating a profound protective effect (136). This result suggests that CMV is largely beneficial to the immune defense, which may explain its ubiquity around the world and in many organisms.
Scientific challenges: From data to knowledge to understanding
Sydney Brenner has said that systems biology is “low input, high throughput, no output” biology (137). A major challenge in the field of systems immunology in humans, and indeed for the field of systems biology in general, is learning how to extract knowledge and understanding from the vast sea of data that will be generated using profiling studies. Identification of omics-based signatures that predict susceptibility to autoimmune disease or vaccine response does not provide any mechanistic insight per se. The concept of knowledge-based (“gnostic”) predictors uses groups of biologically related genes that are coordinately regulated in response to infection, autoimmune disease, or vaccination (138). Although such tools are of great value, it is experimental validation of the signatures that will ultimately yield new biological insight. Data generated in the human model will enable the formulation of novel hypotheses about mechanisms of immune regulation. Mechanistic validation of this hypothesis will require the use of animal models such as gene knockout mice. An example of this approach involves the TLR5-microbiome-influenza vaccination study discussed above (47, 66, 67). The first clue came from systems analysis of immune responses to influenza vaccination in humans, which identified TLR5 as an early correlate of the later antibody titers to vaccination (47), raising the hypothesis that TLR5 could be functionally relevant in modulating antibody responses to influenza vaccination. This was tested in mice, where flagellin in the gut microbiota was shown to signal through TLR5 and provide adjuvant signals to influenza vaccination (66). The relevance of the gut microbiota as an adjuvant to antibody responses to influenza vaccination was subsequently shown to be important in humans in a clinical trial (67). Therefore, this represents an iterative cycle of inquiry that started with correlative observations in a human study, which led to a mechanistic study in mice, which was then validated in a perturbation study in humans.
Such examples illustrate that animal models are key for validating hypotheses generated in human studies. However, this approach of functionally validating genes, one gene knockout mouse at a time, does not have the throughput to rapidly validate the large number of genes that are typically contained in signatures. Therefore, additional approaches for validating hypotheses generated from human studies such as human organoid cultures offer promise in this regard. This, coupled with recent development of CRISPR-based high-throughput screens in primary cells, offers a potential way to rapidly screen and validate immune signatures (139). In addition, recent advances in genetics and microbiome research are facilitating the design of better mouse models that may more closely mimic humans. For example, mice might better mimic humans if their genomes have been engineered to include modified single bases in coding and noncoding regions or even entire networks (140), or if they are infection-experienced (25, 26), transplanted with human microbiota (141, 142), or genetically outbred and collaboratively crossed (143). Such models would represent an invaluable tool for the mechanistic validation of results observed in humans.
Practical and cultural challenges
A particular challenge to human studies is access to human samples. Although most human studies to date have focused on analyzing blood samples, emerging studies are also analyzing immune responses in other tissues, including the draining lymph nodes (using fine needle aspirates) (144), bone marrow (to assess long-lived bone marrow plasma cells) (145), and even liver (using fine needle aspirate) (146). Thus, establishment of systems for sample banking and management could facilitate human immunology studies tremendously. Another practical challenge is standardization of methods and techniques to profile immune responses in well-characterized human cohorts. An excellent example of this is the NIH-funded Human Immunology Project Consortium (HIPC), a collaborative network of researchers established to characterize the human immune system before and after vaccination or infection (www.immuneprofiling.org/hipc/page/show). Through HIPC, well-characterized human cohorts are studied using omics technologies and multiple computational methods. Cross-center and cross-assay analyses enable rigorous standardization of assays and analytical methods.
In addition, there are numerous cultural challenges. The academic ideal has been that of the lone hero/heroine going into a cave and slaying some ferocious dragon of a problem. But science and medicine are actually quite collaborative and team-dependent, and there has been a cultural inertia in academia to reward (or even acknowledge) this. Yet the medical and science worlds are just too complex and not getting any simpler, and to this must be added the computational, statistical, and technical expertise of the team. Ironically, in biotech and pharmaceutical companies, it is a given that tackling important disease areas requires a team approach. Here, the academic approach seems hopelessly outdated and is especially so when faced with the range of skill sets needed to advance human immunology; no one group can do this alone. This also means ensuring that group efforts are rewarded with a fair distribution of the credit in terms of promotions and funding. However, the emphasis on team effort and technology should not detract from the enormous value of individual creativity. To a large extent, scientific discovery has always been, and will continue to be, driven by the creativity and conceptual ideas of individual scientists. Technology, while having a transformative impact on biology, will never be a substitute for biological insight. Therefore, academic institutions and grant-awarding bodies should help to build a culture that stimulates and rewards the creative impulses and aspirations of individual scientists while nurturing a spirit of team science and robust collaboration.
An important cultural challenge is the notion that systems-based approaches to profile human immune response are not hypothesis-driven. This criticism reflects a long-standing philosophical debate on the nature of the scientific method itself. It is unquestionable that hypothesis-driven deductive research has represented the bedrock of the modern scientific method, and Popper has even argued that the so called “hypothetico-deductive” method alone is sufficient for the progress of science (147). Even so, it is clear that observational science or exploratory research that is not necessarily aimed at testing a specific hypothesis has led to major scientific advances. An example of this concerns Darwin’s observations in his field notebooks, without any apparent focus, during his voyage on the HMS Beagle to the Galapagos, which subsequently led to the theory of evolution by natural selection. Another example involves Galileo’s discovery of the four large moons of Jupiter and the rings of Saturn when he peered through the telescope. A more recent example is the collection of data by the Hubble Telescope about the origins of the universe. In the biological sciences, the omics revolution has stimulated a whole new era of science in which the starting point is data collection. Systems-based approaches represent hypothesis-enabling research rather than hypothesis-driven research.
The way of the future: Human immunology in the 21st century
Sydney Brenner remarked in 2008: “We don’t have to look for a model organism anymore. Because we are the model organisms” (148). We propose that the immune system, above all other physiological systems, is uniquely amenable to discovering new human biology. Why? Because cells of the immune system are easily accessible in blood samples. Each drop of blood provides a snapshot of many lineages, as well as dozens of differentiation and activation states. Moreover, other immunological tissues such as lymph nodes, skin, rectal mucosae, and even liver can be safely sampled. This is clearly not the case with research on the human brain, heart, or nervous system. Also, cells of the immune system are uniquely sensitive to deliberate and controlled perturbation; vaccination and infection cause profound changes in their transcriptional, epigenetic, and metabolic profiles.
As discussed above, we are firmly convinced that human immunology will form an ever-larger part of the field going forward. This does not mean that the work with mice will disappear. It is too valuable a system built up over 70 years for that; it presents too many excellent opportunities for well-controlled, mechanistic experiments and will remain an absolutely essential part of biology. But exploration of the immune system in humans offers both a much more direct link to medicine (e.g., translation) and the very real prospect of discovering new immunological phenomena, with thousands of diseases and the genetic diversity that come with a more true-to-life immune system. Thus, despite the challenges of human immunology, new approaches and technology are now making this area much more accessible, and the yields— both scientific and translational—are well worth the effort. However, profiling immunity in humans is but the first step. Computational mining of the data and biological validation in animal models are essential next steps, in an iterative cycle that seeks to bridge fundamental and applied science, mouse and human immunology, in a seamless continuum of scientific discovery and translational medicine (Fig.3). Then, humans will occupy their rightful place in the pantheon of model organisms, and the legacy of Jenner and Pasteur may be regained.
Fig. 3. From bedside to bench to bedside: The iterative cycle of human immunology.

Historically, drugs and vaccines have been developed by the “bench-to-bedside” model, which is a linear process that starts with basic research in animal models and culminates in clinical trials. New advances in systems biology have revitalized human immunology and our capacity to profile human immunity with great precision. Thus, the discovery process can now begin with the human model, where systems-based approaches can be used to analyze the immune response (e.g., response to a vaccine or an infection). The ensuing high-throughput data can then be mined computationally to generate novel hypotheses about the underlying biological mechanisms, which can in turn be tested in animal models. The insights gained can then guide the design of new therapeutics and vaccines. This new framework represents an iterative cycle that seeks to bridge basic and applied science, as well as mouse and human immunology, in a continuum of scientific discovery and translational medicine.
ACKNOWLEDGMENTS
We thank G. Nossal and B. Rouse for comments on the manuscript. B.P. thanks members of our lab for excellent feedback.
Funding: Supported by grants from NIH and the Bill & Melinda Gates Foundation to B.P. and M.M.D. M.M.D. acknowledges the generous support of HHMI.
Footnotes
Competing interests: The authors have no competing interests.
REFERENCES AND NOTES
- 1.Burnet FM, A modification of Jerne’s theory of antibody production using the concept of clonal selection. Aust. J. Sci 20, 67–69 (1957). [DOI] [PubMed] [Google Scholar]
- 2.Talmage DW, Allergy and immunology. Annu. Rev. Med 8, 239–256 (1957). doi: 10.1146/annurev.me.08.020157.001323 [DOI] [PubMed] [Google Scholar]
- 3.Nossal GJ, Lederberg J, Antibody production by single cells. Nature 181, 1419–1420 (1958). doi: 10.1038/1811419a0 [DOI] [PubMed] [Google Scholar]
- 4.Miller JF, Immunological function of the thymus. Lancet 278, 748–749 (1961). doi: 10.1016/S0140-6736(61)90693-6 [DOI] [PubMed] [Google Scholar]
- 5.Cooper MD, Peterson RD, Good RA, Delineation of the Thymic and Bursal Lymphoid Systems in the Chicken. Nature 205, 143–146 (1965). doi: 10.1038/205143a0 [DOI] [PubMed] [Google Scholar]
- 6.Poltorak A et al. , Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: Mutations in Tlr4 gene. Science 282, 2085–2088 (1998). doi: 10.1126/science.282.5396.2085 [DOI] [PubMed] [Google Scholar]
- 7.Lemaitre B, Nicolas E, Michaut L, Reichhart JM,Hoffmann JA, Pillars article: The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996. 86: 973–983. J. Immunol 188, 5210–5220 (2012). [PubMed] [Google Scholar]
- 8.Steinman RM, Cohn ZA, Identification of a novel cell type in peripheral lymphoid organs of mice. I. Morphology, quantitation, tissue distribution. J. Exp. Med 137, 1142–1162 (1973). doi: 10.1084/jem.137.5.1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riedel S, Edward Jenner and the history of smallpox and vaccination. Baylor Univ. Med. Cent. Proc 18, 21–25 (2005). doi: 10.1080/08998280.2005.11928028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rappuoli R, Inner Workings: 1885, the first rabies vaccination in humans. Proc. Natl. Acad. Sci. U.S.A 111, 12273 (2014). doi: 10.1073/pnas.1414226111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lanzavecchia A, Antigen-specific interaction between T and B cells. Nature 314, 537–539 (1985). doi: 10.1038/314537a0 [DOI] [PubMed] [Google Scholar]
- 12.Sallusto F, Lanzavecchia A, Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor α.J. Exp. Med 179, 1109–1118 (1994). doi: 10.1084/jem.179.4.1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caux C, Dezutter-Dambuyant C, Schmitt D, Banchereau J, GM-CSF and TNF-α cooperate in the generation of dendritic Langerhans cells. Nature 360, 258–261 (1992). doi: 10.1038/360258a0 [DOI] [PubMed] [Google Scholar]
- 14.Cooper MD, Chase HP, Lowman JT, Krivit W, Good RA, Wiskott-Aldrich syndrome. An immunologic deficiency disease involving the afferent limb of immunity. Am. J. Med 44, 499–513 (1968). doi: 10.1016/0002-9343(68)90051-X [DOI] [PubMed] [Google Scholar]
- 15.Altare F et al. , Impairment of mycobacterial immunity in human interleukin-12 receptor deficiency. Science 280, 1432–1435 (1998). doi: 10.1126/science.280.5368.1432 [DOI] [PubMed] [Google Scholar]
- 16.Jouanguy E et al. , A human IFNGR1 small deletion hotspot associated with dominant susceptibility to mycobacterial infection. Nat. Genet 21, 370–378 (1999). doi: 10.1038/7701 [DOI] [PubMed] [Google Scholar]
- 17.Lane HC et al. , Correlation between immunologic function and clinical subpopulations of patients with the acquired immune deficiency syndrome. Am. J. Med 78, 417–422 (1985). doi: 10.1016/0002-9343(85)90332-8 [DOI] [PubMed] [Google Scholar]
- 18.Lane HC, Fauci AS, Immunologic abnormalities in the acquired immunodeficiency syndrome. Annu. Rev. Immunol 3, 477–500 (1985). doi: 10.1146/annurev.iy.03.040185.002401 [DOI] [PubMed] [Google Scholar]
- 19.Hodgkin AL, Huxley AF, A quantitative description of membrane current and its application to conduction and excitation in nerve. J. Physiol 117, 500–544 (1952). doi: 10.1113/jphysiol.1952.sp004764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Milestones in cell division. Nat. Cell Biol 3, E265 (2001). doi: 10.1038/ncb1201-e265 [DOI] [PubMed] [Google Scholar]
- 21.Brenner S, In the beginning was the worm…. Genetics 182,413–415 (2009). doi: 10.1534/genetics.109.104976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watts G, Nobel prize is awarded for work leading to “knockout mouse”. BMJ 335, 740 (2007). doi: 10.1136/bmj.39364.367361.DB [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rajewsky K, From a dream to reality. Eur. J. Immunol 37 (suppl. 1), S134–S137 (2007). doi: 10.1002/eji.200737819 [DOI] [PubMed] [Google Scholar]
- 24.Brinster RL et al. , Somatic expression of herpes thymidine kinase in mice following injection of a fusion gene into eggs. Cell 27, 223–231 (1981). doi: 10.1016/0092-8674(81)90376-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beura LK et al. , Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature 532, 512–516 (2016). doi: 10.1038/nature17655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reese TA et al. , Sequential Infection with Common Pathogens Promotes Human-like Immune Gene Expression and Altered Vaccine Response. Cell Host Microbe 19, 713–719 (2016). doi: 10.1016/j.chom.2016.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feldmann M, Maini SR, Role of cytokines in rheumatoid arthritis: An education in pathophysiology and therapeutics. Immunol. Rev 223, 7–19 (2008). doi: 10.1111/j.1600-065X.2008.00626.x [DOI] [PubMed] [Google Scholar]
- 28.Sharma P, Allison JP, The future of immune checkpoint therapy. Science 348, 56–61 (2015). doi: 10.1126/science.aaa8172 [DOI] [PubMed] [Google Scholar]
- 29.Suntharalingam G et al. , Cytokine storm in a phase 1 trial of the anti-CD28 monoclonal antibody TGN1412. N. Engl. J. Med 355, 1018–1028 (2006). doi: 10.1056/NEJMoa063842 [DOI] [PubMed] [Google Scholar]
- 30.Buchbinder SP et al. , Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): A double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet 372, 1881–1893 (2008). doi: 10.1016/S0140-6736(08)61591-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sekaly RP, The failed HIV Merck vaccine study: A step back or a launching point for future vaccine development? J. Exp. Med 205, 7–12 (2008). doi: 10.1084/jem.20072681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davis MM, Tato CM, Furman D, Systems immunology: Just getting started. Nat. Immunol 18, 725–732 (2017). doi: 10.1038/ni.3768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang N et al. , Lineage structure of the human antibody repertoire in response to influenza vaccination. Sci. Transl. Med 5, 171ra19 (2013). doi: 10.1126/scitranslmed.3004794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Glanville J et al. , Identifying specificity groups in the T cell receptor repertoire. Nature 547, 94–98 (2017). doi: 10.1038/nature22976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stubbington MJT, Rozenblatt-Rosen O, Regev A, Teichmann SA, Single-cell transcriptomics to explore the immune system in health and disease. Science 358, 58–63 (2017). doi: 10.1126/science.aan6828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bendall SC et al. , Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 332, 687–696 (2011). doi: 10.1126/science.1198704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheung P et al. , Single-Cell Chromatin Modification Profiling Reveals Increased Epigenetic Variations with Aging. Cell 173, 1385–1397.e14 (2018). doi: 10.1016/j.cell.2018.03.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu LL et al. , A Functional Role for Antibodies in Tuberculosis. Cell 167, 433–443.e14 (2016). doi: 10.1016/j.cell.2016.08.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jennewein MF et al. , Fc Glycan-Mediated Regulation of Placental Antibody Transfer. Cell 178, 202–215.e14 (2019). doi: 10.1016/j.cell.2019.05.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pulendran B, Systems vaccinology: Probing humanity’s diverse immune systems with vaccines. Proc. Natl. Acad. Sci. U.S.A 111, 12300–12306 (2014). doi: 10.1073/pnas.1400476111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pulendran B, Li S, Nakaya HI, Systems vaccinology. Immunity 33, 516–529 (2010). doi: 10.1016/j.immuni.2010.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pulendran B, Learning immunology from the yellow fever vaccine: Innate immunity to systems vaccinology. Nat. Rev. Immunol 9, 741–747 (2009). doi: 10.1038/nri2629 [DOI] [PubMed] [Google Scholar]
- 43.Querec TD et al. , Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat. Immunol 10, 116–125 (2009). doi: 10.1038/ni.1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gaucher D et al. , Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J. Exp. Med 205, 3119–3131 (2008). doi: 10.1084/jem.20082292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Collins ND, Barrett AD, Live Attenuated Yellow Fever 17D Vaccine: A Legacy Vaccine Still Controlling Outbreaks In Modern Day. Curr. Infect. Dis. Rep 19, 14 (2017). doi: 10.1007/s11908-017-0566-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bucasas KL et al. , Early patterns of gene expression correlate with the humoral immune response to influenza vaccination in humans. J. Infect. Dis 203, 921–929 (2011). doi: 10.1093/infdis/jiq156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakaya HI et al. , Systems biology of vaccination for seasonal influenza in humans. Nat. Immunol 12, 786–795 (2011). doi: 10.1038/ni.2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Furman D et al. , Apoptosis and other immune biomarkers predict influenza vaccine responsiveness. Mol. Syst. Biol 9, 659 (2013). doi: 10.1038/msb.2013.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakaya HI et al. , Systems Analysis of Immunity to Influenza Vaccination across Multiple Years and in Diverse Populations Reveals Shared Molecular Signatures. Immunity 43, 1186–1198 (2015). doi: 10.1016/j.immuni.2015.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakaya HI et al. , Systems biology of immunity to MF59-adjuvanted versus nonadjuvanted trivalent seasonal influenza vaccines in early childhood. Proc. Natl. Acad. Sci. U.S.A 113, 1853–1858 (2016). doi: 10.1073/pnas.1519690113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li S et al. , Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nat. Immunol 15, 195–204 (2014). doi: 10.1038/ni.2789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li S et al. , Metabolic Phenotypes of Response to Vaccination in Humans. Cell 169, 862–877.e17 (2017). doi: 10.1016/j.cell.2017.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kazmin D et al. , Systems analysis of protective immune responses to RTS, S malaria vaccination in humans. Proc. Natl. Acad. Sci. U.S.A 114, 2425–2430 (2017). doi: 10.1073/pnas.1621489114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van den Berg RA et al. , Predicting RTS, S Vaccine-Mediated Protection from Transcriptomes in a Malaria-Challenge Clinical Trial. Front. Immunol 8, 557 (2017). doi: 10.3389/fimmu.2017.00557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reif DM et al. , Integrated analysis of genetic and proteomic data identifies biomarkers associated with adverse events following smallpox vaccination. Genes Immun 10, 112–119 (2009). doi: 10.1038/gene.2008.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rechtien A et al. , Systems Vaccinology Identifies an Early Innate Immune Signature as a Correlate of Antibody Responses to the Ebola Vaccine rVSV-ZEBOV. Cell Rep 20, 2251–2261 (2017). doi: 10.1016/j.celrep.2017.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fourati S et al. , Integrated systems approach defines the antiviral pathways conferring protection by the RV144 HIV vaccine. Nat. Commun 10, 863 (2019). doi: 10.1038/s41467-019-08854-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsang JS et al. , Global analyses of human immune variation reveal baseline predictors of postvaccination responses. Cell 157, 499–513 (2014). doi: 10.1016/j.cell.2014.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fourati S et al. , Pre-vaccination inflammation and B-cell signalling predict age-related hyporesponse to hepatitis B vaccination. Nat. Commun 7, 10369 (2016). doi: 10.1038/ncomms10369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.HIPC-CHI Signatures Project Team, HIPC-I Consortium, Multicohort analysis reveals baseline transcriptional predictors of influenza vaccination responses. Sci. Immunol 2, eaal4656 (2017). doi: 10.1126/sciimmunol.aal4656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brodin P et al. , Variation in the human immune system is largely driven by non-heritable influences. Cell 160, 37–47 (2015). doi: 10.1016/j.cell.2014.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Metzger WG, Ehni HJ, Kremsner PG, Mordmüller BG, Experimental infections in humans-historical and ethical reflections. Trop. Med. Int. Health 24, 1384–1390 (2019). doi: 10.1111/tmi.13320 [DOI] [PubMed] [Google Scholar]
- 63.Ravindran R et al. , Vaccine activation of the nutrient sensor GCN2 in dendritic cells enhances antigen presentation. Science 343, 313–317 (2014). doi: 10.1126/science.1246829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ravindran R et al. , The amino acid sensor GCN2 controls gut inflammation by inhibiting inflammasome activation. Nature 531, 523–527 (2016). doi: 10.1038/nature17186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pulendran B, Immunology taught by vaccines. Science 366, 1074–1075 (2019). doi: 10.1126/science.aau6975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Oh JZ et al. , TLR5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity 41, 478–492 (2014). doi: 10.1016/j.immuni.2014.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hagan T et al. , Antibiotics-Driven Gut Microbiome Perturbation Alters Immunity to Vaccines in Humans. Cell 178, 1313–1328.e13 (2019). doi: 10.1016/j.cell.2019.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gates B, Responding to Covid-19—A Once-in-a-Century Pandemic? N. Engl. J. Med 382, 1677–1679 (2020). doi: 10.1056/NEJMp2003762 [DOI] [PubMed] [Google Scholar]
- 69.World Health Organization, Global Health Estimates 2016: Deaths by Cause, Age, Sex, by Country and by Region, 2000–2016 (2018); www.who.int/healthinfo/global_burden_disease/estimates/en/.
- 70.Ndung’u T, Dong KL, Kwon DS, Walker BD, A FRESH approach: Combining basic science and social good. Sci. Immunol 3, eaau2798 (2018). doi: 10.1126/sciimmunol.aau2798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Anahtar MN et al. , Cervicovaginal bacteria are a major modulator of host inflammatory responses in the female genital tract. Immunity 42, 965–976 (2015). doi: 10.1016/j.immuni.2015.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ndhlovu ZM et al. , Magnitude and Kinetics of CD8+ T Cell Activation during Hyperacute HIV Infection Impact Viral Set Point. Immunity 43, 591–604 (2015). doi: 10.1016/j.immuni.2015.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Miller JD et al. , Human effector and memory CD8+ T cell responses to smallpox and yellow fever vaccines. Immunity 28, 710–722 (2008). doi: 10.1016/j.immuni.2008.02.020 [DOI] [PubMed] [Google Scholar]
- 74.Ramilo O et al. , Gene expression patterns in blood leukocytes discriminate patients with acute infections. Blood 109, 2066–2077 (2007). doi: 10.1182/blood-2006-02-002477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Andres-Terre M et al. , Integrated, Multi-cohort Analysis Identifies Conserved Transcriptional Signatures across Multiple Respiratory Viruses. Immunity 43, 1199–1211 (2015). doi: 10.1016/j.immuni.2015.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Berry MP et al. , An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature 466, 973–977 (2010). doi: 10.1038/nature09247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Singhania A, Wilkinson RJ, Rodrigue M, Haldar P, O’Garra A, The value of transcriptomics in advancing knowledge of the immune response and diagnosis in tuberculosis. Nat. Immunol 19, 1159–1168 (2018). doi: 10.1038/s41590-018-0225-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Maertzdorf J et al. , Functional correlations of pathogenesis-driven gene expression signatures in tuberculosis. PLOS ONE 6, e26938 (2011). doi: 10.1371/journal.pone.0026938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Maertzdorf J et al. , Common patterns and disease-related signatures in tuberculosis and sarcoidosis. Proc. Natl. Acad. Sci. U.S.A 109, 7853–7858 (2012). doi: 10.1073/pnas.1121072109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kwissa M et al. , Dengue virus infection induces expansion of a CD14+CD16+ monocyte population that stimulates plasmablast differentiation. Cell Host Microbe 16, 115–127 (2014). doi: 10.1016/j.chom.2014.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tran TM, Crompton PD, Decoding the complexities of human malaria through systems immunology. Immunol. Rev 293, 144–162 (2020). doi: 10.1111/imr.12817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lindner SE et al. , Transcriptomics and proteomics reveal two waves of translational repression during the maturation of malaria parasite sporozoites. Nat. Commun 10, 4964 (2019). doi: 10.1038/s41467-019-12936-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tran TM et al. , A Molecular Signature in Blood Reveals a Role for p53 in Regulating Malaria-Induced Inflammation. Immunity 51, 750–765.e10 (2019). doi: 10.1016/j.immuni.2019.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Robinson M et al. , A 20-Gene Set Predictive of Progression to Severe Dengue. Cell Rep. 26, 1104–1111.e4 (2019). doi: 10.1016/j.celrep.2019.01.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Philips JA, Ernst JD, Tuberculosis pathogenesis and immunity. Annu. Rev. Pathol 7, 353–384 (2012). doi: 10.1146/annurev-pathol-011811-132458 [DOI] [PubMed] [Google Scholar]
- 86.Roy Chowdhury R et al. , A multi-cohort study of the immune factors associated with M. tuberculosis infection outcomes. Nature 560, 644–648 (2018). doi: 10.1038/s41586-018-0439-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sweeney TE, Braviak L, Tato CM, Khatri P, Genome-wide expression for diagnosis of pulmonary tuberculosis: A multicohort analysis. Lancet Respir. Med 4, 213–224 (2016). doi: 10.1016/S2213-2600(16)00048-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Warsinske H, Vashisht R, Khatri P, Host-response-based gene signatures for tuberculosis diagnosis: A systematic comparison of 16 signatures. PLOS Med. 16, e1002786 (2019). doi: 10.1371/journal.pmed.1002786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Burton DR, What Are the Most Powerful Immunogen Design Vaccine Strategies? Reverse Vaccinology 2.0 Shows Great Promise. Cold Spring Harb. Perspect. Biol 9, a030262 (2017). doi: 10.1101/cshperspect.a030262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mascola JR, Haynes BF, HIV-1 neutralizing antibodies: Understanding nature’s pathways. Immunol. Rev 254, 225–244 (2013). doi: 10.1111/imr.12075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Freund NT et al. , Coexistence of potent HIV-1 broadly neutralizing antibodies and antibody-sensitive viruses in a viremic controller. Sci. Transl. Med 9, eaal2144 (2017). doi: 10.1126/scitranslmed.aal2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ramos PS, Shedlock AM, Langefeld CD, Genetics of autoimmune diseases: Insights from population genetics. J. Hum. Genet 60, 657–664 (2015). doi: 10.1038/jhg.2015.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Theofilopoulos AN, Kono DH, Baccala R, The multiple pathways to autoimmunity. Nat. Immunol 18, 716–724 (2017). doi: 10.1038/ni.3731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Roep BO, Buckner J, Sawcer S, Toes R, Zipp F, The problems and promises of research into human immunology and autoimmune disease. Nat. Med 18, 48–53 (2012). doi: 10.1038/nm.2626 [DOI] [PubMed] [Google Scholar]
- 95.Roep BO, Atkinson M, von Herrath M, Satisfaction (not) guaranteed: Re-evaluating the use of animal models of type 1 diabetes. Nat. Rev. Immunol 4, 989–997 (2004). doi: 10.1038/nri1502 [DOI] [PubMed] [Google Scholar]
- 96.Herold KC et al. , An Anti-CD3 Antibody, Teplizumab, in Relatives at Risk for Type 1 Diabetes. N. Engl. J. Med 381, 603–613 (2019). doi: 10.1056/NEJMoa1902226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Feldmann M, Maini RN, TNF defined as a therapeutic target for rheumatoid arthritis and other autoimmune diseases. Nat. Med 9, 1245–1250 (2003). doi: 10.1038/nm939 [DOI] [PubMed] [Google Scholar]
- 98.Brennan FM, Chantry D, Jackson A, Maini R, Feldmann M, Inhibitory effect of TNFα antibodies on synovial cell interleukin-1 production in rheumatoid arthritis. Lancet 334, 244–247 (1989). doi: 10.1016/S0140-6736(89)90430-3 [DOI] [PubMed] [Google Scholar]
- 99.Fong Y et al. , Antibodies to cachectin/tumor necrosis factor reduce interleukin 1β and interleukin 6 appearance during lethal bacteremia. J. Exp. Med 170, 1627–1633 (1989).doi: 10.1084/jem.170.5.1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Williams RO, Feldmann M, Maini RN, Anti-tumor necrosis factor ameliorates joint disease in murine collagen-induced arthritis. Proc. Natl. Acad. Sci. U.S.A 89, 9784–9788 (1992). doi: 10.1073/pnas.89.20.9784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Inshaw JRJ, Cutler AJ, Burren OS, Stefana MI, Todd JA, Approaches and advances in the genetic causes of autoimmune disease and their implications. Nat. Immunol 19, 674–684 (2018). doi: 10.1038/s41590-018-0129-8 [DOI] [PubMed] [Google Scholar]
- 102.Banchereau R, Cepika AM, Pascual V, Systems approaches to human autoimmune diseases. Curr. Opin. Immunol 25, 598–605 (2013). doi: 10.1016/j.coi.2013.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Banchereau R et al. , Personalized Immunomonitoring Uncovers Molecular Networks that Stratify Lupus Patients. Cell 165, 1548–1550 (2016). doi: 10.1016/j.cell.2016.05.057 [DOI] [PubMed] [Google Scholar]
- 104.Allantaz F et al. , Blood leukocyte microarrays to diagnose systemic onset juvenile idiopathic arthritis and follow the response to IL-1 blockade. J. Exp. Med 204, 2131–2144 (2007). doi: 10.1084/jem.20070070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jakimovski D, Kolb C, Ramanathan M, Zivadinov R, Weinstock-Guttman B, Interferon β for Multiple Sclerosis. Cold Spring Harb. Perspect. Med 8, a032003 (2018). doi: 10.1101/cshperspect.a032003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Stürzebecher S et al. , Expression profiling identifies responder and non-responder phenotypes to interferon-β in multiple sclerosis. Brain 126, 1419–1429 (2003). doi: 10.1093/brain/awg147 [DOI] [PubMed] [Google Scholar]
- 107.Han A et al. , Dietary gluten triggers concomitant activation of CD4+ and CD8+ αβ T cells and γδ T cells in celiac disease. Proc. Natl. Acad. Sci. U.S.A 110, 13073–13078 (2013). doi: 10.1073/pnas.1311861110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Saligrama N et al. , Opposing T cell responses in experimental autoimmune encephalomyelitis. Nature 572, 481–487 (2019). doi: 10.1038/s41586-019-1467-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rewers M et al. , The Environmental Determinants of Diabetes in the Young (TEDDY) Study: 2018 Update. Curr. Diab. Rep 18, 136 (2018). doi: 10.1007/s11892-018-1113-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Elding Larsson H, A Swedish approach to the prevention of type 1 diabetes. Pediatr. Diabetes 17 (suppl. 22), 73–77 (2016). doi: 10.1111/pedi.12325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Schadendorf D et al. , Pooled Analysis of Long-Term Survival Data From Phase II and Phase III Trials of Ipilimumab in Unresectable or Metastatic Melanoma. J. Clin. Oncol 33, 1889–1894 (2015). doi: 10.1200/JCO.2014.56.2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Postow MA et al. , Nivolumab and ipilimumab versus ipilimumab in untreated melanoma. N. Engl. J. Med 372, 2006–2017 (2015). doi: 10.1056/NEJMoa1414428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hodi FS et al. , Nivolumab plus ipilimumab or nivolumab alone versus ipilimumab alone in advanced melanoma (CheckMate 067): 4-year outcomes of a multicentre, randomised, phase 3 trial. Lancet Oncol 19, 1480–1492 (2018). doi: 10.1016/S1470-2045(18)30700-9 [DOI] [PubMed] [Google Scholar]
- 114.Leach DR, Krummel MF, Allison JP, Enhancement of antitumor immunity by CTLA-4 blockade. Science 271, 1734–1736 (1996). doi: 10.1126/science.271.5256.1734 [DOI] [PubMed] [Google Scholar]
- 115.Nishimura H, Minato N, Nakano T, Honjo T, Immunological studies on PD-1 deficient mice: Implication of PD-1 as a negative regulator for B cell responses. Int. Immunol 10, 1563–1572 (1998). doi: 10.1093/intimm/10.10.1563 [DOI] [PubMed] [Google Scholar]
- 116.Liakou CI et al. , CTLA-4 blockade increases IFNγ-producing CD4+ICOShi cells to shift the ratio of effector to regulatory T cells in cancer patients. Proc. Natl. Acad. Sci. U.S.A 105, 14987–14992 (2008). doi: 10.1073/pnas.0806075105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Huang AC et al. , T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature 545, 60–65 (2017). doi: 10.1038/nature22079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Routy B et al. , Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359, 91–97 (2018). doi: 10.1126/science.aan3706 [DOI] [PubMed] [Google Scholar]
- 119.Matson V et al. , The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 359, 104–108 (2018). doi: 10.1126/science.aao3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gopalakrishnan V et al. , Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359, 97–103 (2018). doi: 10.1126/science.aan4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tyler SR, Bunyavanich S, Leveraging -omics for asthma endotyping. J. Allergy Clin. Immunol 144, 13–23 (2019). doi: 10.1016/j.jaci.2019.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Watson CT et al. , Integrative transcriptomic analysis reveals key drivers of acute peanut allergic reactions. Nat. Commun 8, 1943 (2017). doi: 10.1038/s41467-017-02188-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Stapleton CP, Conlon PJ, Phelan PJ, Using omics to explore complications of kidney transplantation. Transpl. Int 31, 251–262 (2018). doi: 10.1111/tri.13067 [DOI] [PubMed] [Google Scholar]
- 124.Keating BJ, Pereira AC, Snyder M, Piening BD, Applying genomics in heart transplantation. Transpl. Int 31, 278–290 (2018). doi: 10.1111/tri.13119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Medawar P, Reith Lectures 1959: The Future of Man, Lecture 4: The Genetic System of Man. BBC broadcast transcript, 13 December 1959; http://downloads.bbc.co.uk/rmhttp/radio4/transcripts/1959_reith4.pdf.
- 126.Food and Agriculture Organization of the United Nations, The State of Food Security and Nutrition in the World 2019: Safeguarding Against Economic Slowdowns and Downturns (2019); https://docs.wfp.org/api/documents/WFP-0000106760/download/?_ga=2.22220859.1213858381.1573050906-1201814519.1573050906.
- 127.Central Intelligence Agency, The World Factbook; https://www.cia.gov/library/publications/resources/the-world-factbook/index.html.
- 128.Olin A et al. , Stereotypic Immune System Development in Newborn Children. Cell 174, 1277–1292.e14 (2018).doi: 10.1016/j.cell.2018.06.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Patin E et al. , Natural variation in the parameters of innate immune cells is preferentially driven by genetic factors. Nat. Immunol 19, 302–314 (2018). doi: 10.1038/s41590-018-0049-7 [DOI] [PubMed] [Google Scholar]
- 130.Carr EJ et al. , The cellular composition of the human immune system is shaped by age and cohabitation. Nat. Immunol 17, 461–468 (2016). doi: 10.1038/ni.3371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tsang JS, Utilizing population variation, vaccination, and systems biology to study human immunology. Trends Immunol. 36, 479–493 (2015). doi: 10.1016/j.it.2015.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Schüssler-Fiorenza Rose SM et al. , A longitudinal bigdata approach for precision health. Nat. Med 25, 792–804 (2019). doi: 10.1038/s41591-019-0414-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jiang C et al. , Dynamic Human Environmental Exposome Revealed by Longitudinal Personal Monitoring. Cell 175, 277–291.e31 (2018). doi: 10.1016/j.cell.2018.08.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hawn TR et al. , A common dominant TLR5 stop codon polymorphism abolishes flagellin signaling and is associated with susceptibility to legionnaires’ disease. J. Exp. Med 198, 1563–1572 (2003). doi: 10.1084/jem.20031220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ovsyannikova IG, Salk HM, Larrabee BR, Pankratz VS, Poland GA, Single-nucleotide polymorphism associations in common with immune responses to measles and rubella vaccines. Immunogenetics 66, 663–669 (2014). doi: 10.1007/s00251-014-0796-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Furman D et al. , Cytomegalovirus infection enhancesthe immune response to influenza. Sci. Transl. Med 7, 281ra43 (2015). doi: 10.1126/scitranslmed.aaa2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Brenner S, Sequences and consequences. Philos. Trans. R. Soc. London Ser. B 365, 207–212 (2010). doi: 10.1098/rstb.2009.0221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Haining WN, Pulendran B, Identifying gnostic predictors of the vaccine response. Curr. Opin. Immunol 24, 332–336 (2012). doi: 10.1016/j.coi.2012.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Knott GJ, Doudna JA, CRISPR-Cas guides the future of genetic engineering. Science 361, 866–869 (2018). doi: 10.1126/science.aat5011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhu F, Nair RR, Fisher EMC, Cunningham TJ, Humanising the mouse genome piece by piece. Nat. Commun 10, 1845 (2019). doi: 10.1038/s41467-019-09716-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Rosshart SP et al. , Wild Mouse Gut Microbiota Promotes Host Fitness and Improves Disease Resistance. Cell 171, 1015–1028.e13 (2017). doi: 10.1016/j.cell.2017.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Rosshart SP et al. , Laboratory mice born to wild mice have natural microbiota and model human immune responses. Science 365, eaaw4361 (2019). doi: 10.1126/science.aaw4361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Noll KE, Ferris MT, Heise MT, The Collaborative Cross: A Systems Genetics Resource for Studying Host-Pathogen Interactions. Cell Host Microbe 25, 484–498 (2019). doi: 10.1016/j.chom.2019.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hill DL et al. , The adjuvant GLA-SE promotes human Tfh cell expansion and emergence of public TCRβ clonotypes. J. Exp. Med 216, 1857–1873 (2019). doi: 10.1084/jem.20190301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Halliley JL et al. , Long-Lived Plasma Cells Are Contained within the CD19−CD38hiCD138+ Subset in Human Bone Marrow. Immunity 43, 132–145 (2015). doi: 10.1016/j.immuni.2015.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gill US et al. , Fine needle aspirates comprehensively sample intrahepatic immunity. Gut 68, 1493–1503 (2019). doi: 10.1136/gutjnl-2018-317071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Popper K, The Logic of Scientific Discovery (Routledge, 1959). [Google Scholar]
- 148.“Sydney Brenner urges cancer researchers to consider ‘bedside to bench’ approach”; www.genomeweb.com/archive/sydney-brenner-urges-cancer-researchers-consider-bedside-bench-approach#.XcnGQi2ZNlM.
- 149.Mestas J, Hughes CCW, Of Mice and Not Men: Differences between Mouse and Human Immunology. J. Immunol 172, 2731–2738 (2004). doi: 10.1182/blood.V96.8.2803 [DOI] [PubMed] [Google Scholar]
- 150.Iwasaki A, Medzhitov R, Toll-like receptor control of the adaptive immune responses. Nat. Immunol 5, 987–995 (2004). doi: 10.1038/ni1112 [DOI] [PubMed] [Google Scholar]
- 151.Casanova JL, Abel L, Quintana-Murci L, Human TLRs and IL-1Rs in host defense: Natural insights from evolutionary, epidemiological, and clinical genetics. Annu. Rev. Immunol 29, 447–491 (2011). doi: 10.1146/annurev-immunol-030409-101335 [DOI] [PubMed] [Google Scholar]
- 152.Dekkers G et al. , Affinity of human IgG subclasses to mouse Fc gamma receptors. mAbs 9, 767–773 (2017).doi: 10.1080/19420862.2017.1323159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zemlin M et al. , Expressed murine and human CDR-H3 intervals of equal length exhibit distinct repertoires that differ in their amino acid composition and predicted range of structures. J. Mol. Biol 334, 733–749 (2003). doi: 10.1016/j.jmb.2003.10.007 [DOI] [PubMed] [Google Scholar]
- 154.Fischer A et al. , Naturally occurring primary deficiencies of the immune system. Annu. Rev. Immunol 15, 93–124 (1997). doi: 10.1146/annurev.immunol.15.1.93 [DOI] [PubMed] [Google Scholar]
- 155.Roifman CM, Zhang J, Chitayat D, Sharfe N, A partial deficiency of interleukin-7Rα is sufficient to abrogate T-cell development and cause severe combined immunodeficiency. Blood 96, 2803–2807 (2000). doi: 10.1182/blood.V96.8.2803 [DOI] [PubMed] [Google Scholar]


