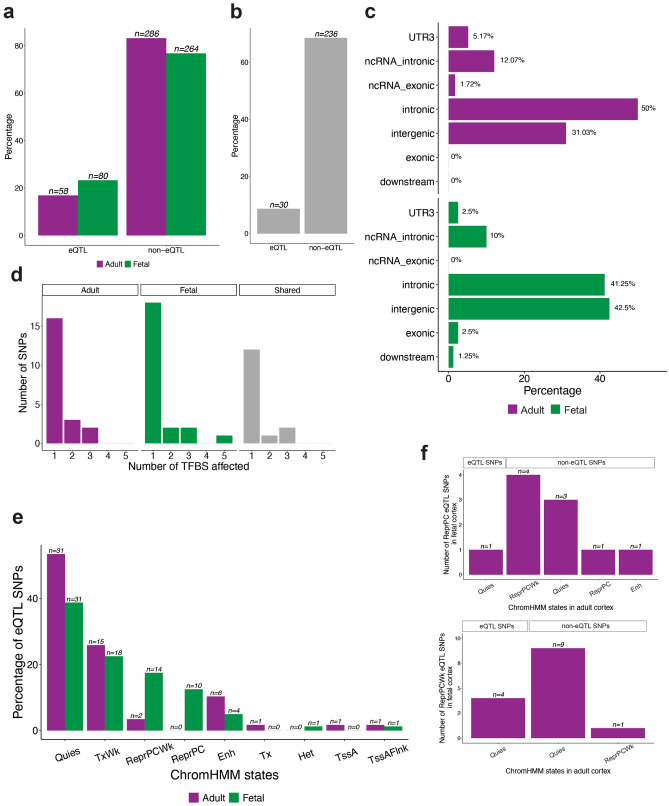
Figure 2.
ASD-associated SNPs are enriched within non-coding putative regulatory regions. (a) Of 344 ASD-associated SNPs represented in both fetal and adult cortex-specific eQTL databases, more SNPs (n = 80) are involved in spatial eQTL-gene interactions in the fetal cortex than in the adult cortex (n = 58). The proportions of eQTL and non-eQTL SNPs are significantly different in fetal and adult cortical tissues (Fisher’s exact test, p = 0.04531). (b) Thirty ASD-associated SNPs are eQTLs in both fetal and adult cortical tissues. (c) All ASD-associated eQTLs in adult cortex (n = 58) and approximately 78 (97.5%) of the ASD-associated eQTLs within the fetal cortex are located within non-coding genomic regions (Supplementary Table 5). (d) 15 and 18 ASD-associated eQTLs affect at least one transcription factor binding sites within the fetal and adult cortical tissues, respectively. (e) Most of the fetal ASD-associated eQTLs are located within quiescent/low transcribed, week repressed PolyComb, repressed PolyComb and weak transcription regions. By contrast, the majority of ASD-associated eQTLs, that were identified in the adult cortex, are located within quiescent/low transcribed and weak transcription regions. Enh enhancers, Het heterochromatin, Quies quiescent/low, ReprPC repressed PolyComb, ReprPCWk week repressed PolyComb, TssA active TSS, TssAFlnk flanking active TSS, Tx strong transcription, TxWk weak transcription. (f) The majority of the fetal ASD-associated eQTLs that are located within weakly repressed PolyComb (ReprPCWk) and repressed PolyComb (ReprPC) regions were not identified as being eQTLs within the adult cortex.