Abstract
Background
A hexanucleotide repeat expansion in the C9ORF72 gene has recently been shown to cause a large proportion of amyotrophic lateral sclerosis (ALS) and fronto-temporal dementia (FTD).
Methods
We screened 4,448 patients diagnosed with ALS and 1,425 patients diagnosed with FTD drawn from diverse populations for the hexanucleotide expansion using a repeat-primed PCR assay. ALS and FTD were diagnosed according to the El Escorial and Lund-Manchester criteria respectively. Familial status was based on self-reported family history of similar neurodegenerative diseases at the time of sample collection. Haplotype data of 262 patients carrying the expansion were compared with the known Finnish founder risk haplotype across the chromosomal locus. Age-related penetrance was calculated by the Kaplan-Meier method using data from 603 individuals carrying the expansion.
Findings
The mutation was observed among 7·0% (n = 236 of 3,377) of Caucasians, 4·1% (n = 2 of 49) of African-Americans, and 8·3% (n = 6 of 72) of Hispanic individuals diagnosed with sporadic ALS, whereas the rate was 6·0% (n = 59 of 981) among Caucasians diagnosed with sporadic FTD. Among Asians, 5·0% (n = 1 of 20) of familial ALS and 66·6% (n = 2 of 3) of familial FTD cases carried the repeat expansion. In contrast, mutations were not observed among patients of Native American (n = 3 sporadic ALS), Indian (n = 31 sporadic ALS, n = 31 sporadic FTD), and Pacific Islander (n = 90 sporadic ALS) ethnicity. All patients with the repeat expansion carried, either partially or fully, the founder haplotype suggesting that the expansion occurred on a single occasion in the past (~1,500 years ago). The pathogenic expansion was non-penetrant below 35 years of age, increasing to 50·0% penetrance by 58 years of age, and was almost fully penetrant by 80 years of age.
Interpretation
We confirm that a common single Mendelian genetic lesion is implicated in a large proportion of sporadic and familial ALS and FTD. Testing for this pathogenic expansion will be important in the management and genetic counseling of patients with these fatal neurodegenerative diseases.
Funding
See Acknowledgements.
Introduction
Amyotrophic lateral sclerosis (ALS, Online Mendelian Inheritance in Man (OMIM) 105400) is a fatal neurodegenerative disease characterized by rapidly progressive paralysis and death from respiratory failure, typically within three years of symptom onset. Approximately five percent of the disease is inherited following a clear Mendelian pattern, whereas the vast majority of cases are classified as sporadic, as they appear to occur randomly in the community.1 Considerable progress has been made in understanding the genetic underpinnings of familial ALS.2 In contrast, the etiology of sporadic or idiopathic ALS is far less well understood. Mutations in the known familial ALS genes, SOD1, FUS, and TDP-43, occur only rarely in sporadic cases (each accounting for less than 1·0%)3–5, and genome-wide association studies have identified a limited number of risk loci that have proven difficult to replicate.6
Frontotemporal dementia (FTD, OMIM 600274) is a degenerative disorder of the frontal and anterior temporal lobes, and is a common form of dementia affecting individuals below the age of 65 years. The syndrome is characterized clinically by initial behavioral disturbances, followed by cognitive decline that leads to dementia and death within a median of seven years from symptom onset. Similar to ALS and other neurodegenerative diseases, a large proportion (~60·0%) of these cases are categorized as sporadic, and the etiology of this idiopathic form of disease is largely unknown.7 There is growing recognition that ALS and FTD represent a continuum of neurological disease that share a common pathology consisting of TDP-43-positive inclusions within the CNS.8
We recently reported that a large hexanucleotide repeat expansion located within the non-coding portion of the C9ORF72 gene is the cause of chromosome 9-linked ALS and FTD.9,10 This genetic lesion accounts for an unprecedented large portion (~40·0%) of both familial ALS and familial FTD cases. The same mutation was present in nearly one quarter of apparently sporadic ALS and FTD cases in the genetically homogeneous Finnish population, and in 4·1%of sporadic ALS cases and 3·0% of sporadic FTD cases from the United States. However, these estimates were based on relatively small cohorts drawn from a limited number of institutions.
These findings prompted us to more accurately estimate the frequency of this C9ORF72 hexanucleotide repeat expansion in a large cohort of European and US patients with sporadic ALS and sporadic FTD. We also examined the occurrence of this mutation in diverse, non-Caucasian populations around the world. To do this, we screened 4,448 patients diagnosed with ALS and 1,425 patients diagnosed with FTD drawn from nineteen distinct regions. Our findings have clinical implications for the diagnosis and management of ALS and FTD patients.
Methods
Samples
Ethnicity and clinical features of the 3,860 sporadic ALS patients, 1,022 sporadic FTD, 588 familial ALS, and 403 familial FTD patients are shown in webappendix pp 5 – 8. Of these, data for 401 Finnish ALS, 75 Finnish FTD, 233 European-ancestry familial ALS, 340 Dutch FTD, and 420 English FTD cases have been already published.10–12
Patients with ALS were diagnosed according to the El Escorial diagnostic criteria13, and patients with FTD were diagnosed according to the Lund-Manchester criteria.14 Patients were classified as familial in nature based on any other member of their family (regardless of relationship level) being diagnosed with ALS and/or FTD, as reported at the time of sample collection. Ethnic and racial classification was based on patient self-report at time of sample collection. The Italian samples represented a population-based cohort that had been collected through the Piemonte ALS Registry, an ongoing population-based epidemiological study of ALS based in northwestern Italy.15 The remaining cohorts were recruited through medical centers and from repositories in various countries.
Of the 2,585 neurologically normal control individuals from Australia (n = 213), Finland (n = 478), Germany (n = 309), Human Gene Diversity Panel (n = 300), Italian (n = 354), Sardinian (n = 87), and the United States (n = 844) that have been screened for the presence of the pathogenic repeat expansion, 1,167 samples were reported in our original Neuron publication.10 Ethics Committees of the respective institutions approved the study, and informed written consent was obtained from all case and control individuals.
Repeat-primed PCR assay
Our previously published repeat-primed PCR assay was used to screen case and control samples for the presence of the chromosome 9p21 GGGGCC hexanucleotide repeat expansion (webappendix p 4).10 This assay is an accurate and rapid system that allows samples to be categorized into those that carry a pathogenic repeat expansion (greater than 30 repeats) and those that carry only wild-type alleles (less than 20 repeats).
Haplotype analysis
We analyzed genome-wide SNP data available to us from 262 patients who carried the repeat expansion. Unphased sample genotype data were compared with the 42-SNP founder risk haplotype that we previously identified in the Finnish population using a custom PERL software script.16
Estimation of mutation age
Mutation ages were estimated for all populations separately using the packageDMLEv2.3 Bayesian linkage disequilibrium gene mapping.17 Mutation ages were iterated for 10,000 burn-in iterations and a further 10,000 iterations of the maximum-likelihood model. To obtain generalizable estimate of age of the repeat per population, median values of binned estimates passing the α threshold of 0·05 per iteration were used.
Statistical analysis
Confidence intervals for proportions were calculated using the Clopper-Pearson exact method. Penetrance of the GGGGCC hexanucleotide repeat expansion in relation to the patients’ age was estimated based on data available for 603 individuals by the Kaplan-Meier method using the survival package within R statistical software (version 2.9.0), but substituting patient age at symptom onset for survival time.18
Role of the funding sources
The sponsors of the study had no role in study design, data collection, analysis, interpretation, writing of the report, or in the decision to submit the paper for publication. All authors had full access to all of the data and held final responsibility for the decision to submit for publication.
Results
Supplementary Figure 1 (webappendix p 11) and Table 1 show the frequency of the C9ORF72 hexanucleotide repeat expansion in patients diagnosed with sporadic ALS and sporadic FTD from different geographical regions of the world. The pathogenic expansion was found in 7·0% of Caucasians diagnosed with sporadic ALS (n = 236 of 3,377 samples from the United States, Europe, Middle East and Australia), 4·1% of African-Americans (n = 2 of 49), and 8·3% of Hispanic ALS samples (n = 6 of 72). The rate of pathogenic expansion was lower among sporadic FTD cases: 6·0% of Caucasian patients carried the mutation (n = 59 of 981 samples). In contrast, the GGGGCC repeat expansion on chromosome 9p21 was not present in sporadic patients of Native American (n = 3 ALS cases), Asian (n = 238 ALS and 10 FTD cases), or Pacific Islander origin (n = 90 ALS cases), though this may reflect the smaller size of the cohorts screened in these populations.
Table 1.
Frequency of the pathogenic GGGGCC hexanucleotide repeat expansion of the C9ORF72 gene in patients diagnosed with sporadic ALS and sporadic FTD classified by region*
| Origin | Sporadic ALS
|
Sporadic FTD
|
||||
|---|---|---|---|---|---|---|
| n | Carriers (%) | 95% CI | n | Carriers (%) | 95% CI | |
| Europe: | ||||||
| Finnish | 289 | 61 (21·1%) | 16·5–26·3 | 48 | 9 (18·8%) | 8·9–32·6 |
| Swedish | - | - | - | 6 | 0 (0·0%) | 0·0–45·9 |
| English | 916 | 62 (6·8%) | 5·2–8·6 | 543 | 31 (5·7%) | 3·9–8·0 |
| German | 421 | 22 (5·2%) | 3·3–7·8 | - | - | - |
| Dutch | - | - | - | 224 | 5 (2·2%) | 0·7–5·1 |
| French | - | - | - | 150 | 14 (9·3%) | 5·2–15·2 |
| Italian | 465 | 19 (4·1%) | 2·5–6·3 | - | - | - |
| Sardinian | 129 | 10 (7·8%) | 3·8–13·8 | 10 | 0 (0·0%) | 0·0–30·8 |
| Moldovan | 3 | 0 (0·0%) | 0·0–70·8 | - | - | - |
| Total (Europe) | 2,223 | 174 (7·8%) | 6·7–9·0 | 981 | 59 (6·0%) | 4·6–7·7 |
| United States: | ||||||
| White | 890 | 48 (5·4%) | 4·0–7·1 | - | - | - |
| Hispanic | 72 | 6 (8·3%) | 3·1–17·3 | - | - | - |
| African American | 49 | 2 (4·1%) | 0·5–14·0 | - | - | - |
| Native American | 3 | 0 (0·0%) | 0·0–70·8 | - | - | - |
| Total (US) | 1,014 | 56 (5·5%) | 4·2–7·1 | - | - | - |
| Global: | ||||||
| Middle Eastern | 1 | 0 (0·0%) | 0·0–97·5 | - | - | - |
| Indian | 31 | 0 (0·0%) | 0·0–11·2 | 31 | 0 (0·0%) | 0·0–11·2 |
| Asian | 238 | 0 (0·0%) | 0·0–1·5 | 10 | 0 (0·0%) | 0·0–30·8 |
| Pacific Islander/Guam | 90 | 0 (0·0%) | 0·0–4·0 | - | - | - |
| Australian | 263 | 14 (5·3%) | 2·9–8·8 | - | - | - |
| Total | 3,860 | 244 (6·3%) | 5·6–7·1 | 1,022 | 59 (5·8%) | 4·4–7·4 |
In addition to the sporadic cases, we screened 588 familial ALS cases and 403 familial FTD cases for the presence of the C9ORF72 repeat expansion (Table 2 and webappendix p 12). Of these, 373 familial ALS cases and 230 familial FTD cases were reported in our original Neuron paper and elsewhere.10–12 All of the cohorts are presented here to provide a comprehensive assessment of the global frequency of the expansion. Overall, 37·6% of familial ALS cases and 25·1% of familial FTD cases carried the genetic lesion, confirming our previous findings that this mutation was responsible for an unparalleled proportion of these diseases. Notably, we identified a single Japanese individual diagnosed with familial ALS, who carried the hexanucleotide repeat expansion. We also found that a familial FTD case from Lund, Sweden, carried the expansion, suggesting that the chromosome 9p21 genetic lesion may be responsible for the geographical cluster of FTD patients observed in that region.19
Table 2.
Frequency of the pathogenic GGGGCC hexanucleotide repeat expansion of the C9ORF72 gene in patients diagnosed with familial ALS and familial FTD classified by region*
| Origin | Familial ALS
|
Familial FTD
|
||||
|---|---|---|---|---|---|---|
| n | Carriers (%) | 95% CI | n | Carriers (%) | 95% CI | |
| Europe: | ||||||
| Finnish | 112 | 52 (46·4%) | 37·0–56·1 | 27 | 13 (48·1%) | 28·7–68·0 |
| Swedish | - | - | - | 1 | 1 (100·0%) | 2·5–100·0 |
| English | 98 | 45 (45·9%) | 35·8–56·3 | 170 | 28 (16·5%) | 11·2–22·9 |
| Irish | 1 | 1 (100·0%) | 2·5–100·0 | - | - | |
| German | 69 | 15 (21·7%) | 12·7–33·3 | 29 | 4 (13·8%) | 3·9–31·7 |
| Dutch | - | - | - | 116 | 30 (25·9%) | 18·2–34·8 |
| French | - | - | - | 50 | 22 (44·0%) | 30·0–58·7 |
| Italian | 90 | 34 (37·8%) | 27·8–48·6 | - | - | - |
| Sardinian | 19 | 11 (57·9%) | 33·5–79·7 | 7 | 1 (14·3%) | 0·4–57·9 |
| Total (Europe) | 389 | 158 (40·6%) | 35·7–45·7 | 400 | 99 (24·8%) | 20·6–29·3 |
| United States: | ||||||
| White | 163 | 59 (36·2%) | 28·8–44·1 | - | - | - |
| Total (US) | 163 | 59 (36·2%) | 28·8–44·1 | - | - | - |
| Global: | ||||||
| Middle Eastern | 2 | 0 (0·0%) | 0·0–84·2 | - | - | - |
| Israeli | 14 | 3 (21·4%) | 4·7–50·8 | - | - | - |
| Asian | 20 | 1 (5·0%) | 0·1–24·9 | 3 | 2 (66·6%) | 9·4–99·2 |
| Total | 588 | 221 (37·6%) | 33·7–41·6 | 403 | 101 (25·1%) | 20·9–29·6 |
Of the 2,585 neurologically normal control samples screened for the C9ORF72 repeat expansion, five (0·2%) were found to be carriers: two were previously reported elderly Finnish samples10, and the remaining three were German and American individuals under the age of 40 (webappendix p 9).
Within Europe, the highest mutation frequency was observed in the Finnish population (21·1% of sporadic ALS patients and 18·8% of sporadic FTD patients).10 Approximately 6·0% of sporadic ALS patients from Germany and England carried the expansion, whereas Mediterranean Italian ALS patients had a lower rate (4·1%). Interestingly, 7·8% of sporadic ALS patients from the isolated island population of Sardinia were found to have the mutation, and the Dutch population had the lowest rate observed among European countries (2·2% of sporadic FTD cases). Both the Australian and US white populations had an intermediate rate with ~5·0% of their sporadic ALS patients carrying the pathogenic repeat expansion, perhaps reflecting the population and immigration histories of these continents.
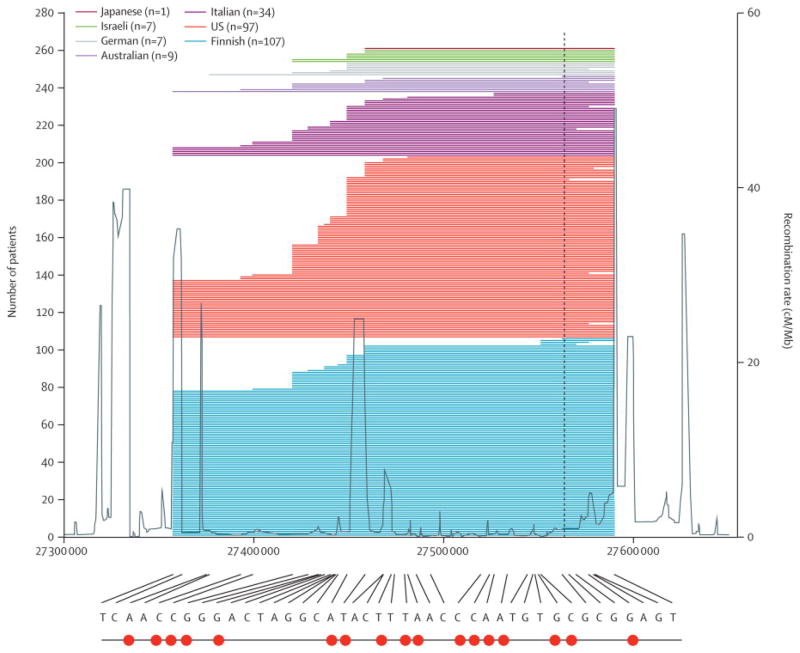
We previously reported the identification of a 42-SNP founder haplotype across the same 232kb block of chromosome 9p21 region where the pathogenic hexanucleotide expansion was ultimately found.16,20 For the current paper, we analyzed genome-wide SNP chip data from 262 patients that carried the C9ORF72 hexanucleotide repeat expansion, including 107 Finnish ALS cases, 97 US cases, 34 Italian cases, 9 Australian cases, 7 German cases, 7 Israeli cases and 1 Japanese case. This analysis showed that each case carrying the pathogenic GGGGCC repeat expansion also shared the Finnish founder risk haplotype, at least in part (Figure 1A). Furthermore, sporadic and familial cases carried the same founder risk haplotype. These findings suggest that the pathogenic hexanucleotide repeat expansion in the C9ORF72 gene may have occurred on a single occasion in human history and subsequently disseminated throughout these populations. Analysis of haplotype sharing between these cases estimated the age of the C9ORF72 repeat expansion to be ~1,500 years old (median = 100·5 generations, interquartile range = 57·6 – 127·6 generations, assuming one generation represents 15 years, Figure 1B).
Figure 1.
(A) Finnish risk haplotypes across the region in 262 ALS patients carrying the pathogenic C9ORF72 hexanucleotide repeat expansion. The previously identified Finnish risk haplotype across the chromosome 9p21 region is shown at the bottom (27,357,278 – 27,589,746 bp; NCBI build 36; 42 SNPs).16 Underneath the haplotype is a binary representation of the same data, with red circles at SNP positions where the haplotype has the less common allele at that site. In the graph, individual patients are shown as horizontal lines representing the extent to which they share the risk haplotype. Blue horizontal lines represent Finnish patients (n = 107), orange lines represent US ALS cases (n = 97), red lines represent Italian cases (n = 34), violet lines represent Australian cases (n = 9), grey lines represent German cases (n = 7), yellow lines represent Israeli cases (n = 7) and the green line represents the single Japanese patient. The vertical black dashed line indicates the location of the C9ORF72 hexanucleotide repeat expansion. Recombination rates (cM/Mb) from phase 2 CEPH samples of HapMap are represented by the grey line. (B) Clade diagram showing age of the pathogenic GGCCCC hexanucleotide repeat expansion of the C9ORF72 gene, and the approximate age of divergence of the Finnish, US and Italian populations carrying this mutation.
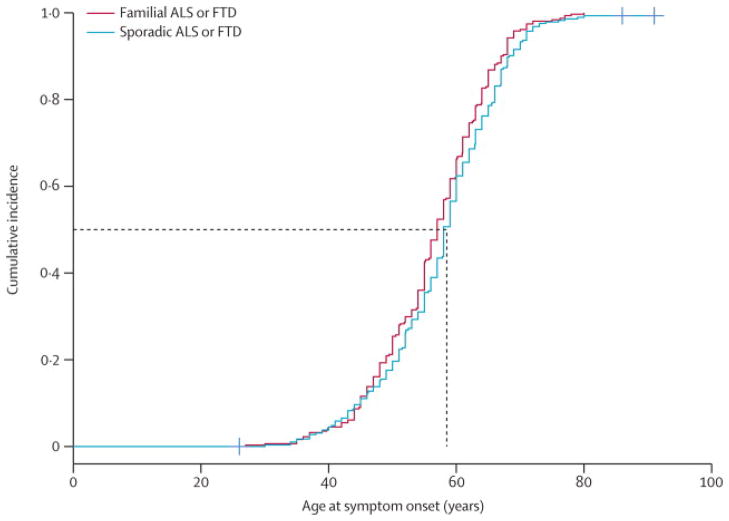
Age-related penetrance of the C9ORF72 hexanucleotide repeat expansion was calculated based on the ages of 603 mutant-gene carriers (Figure 2). The pathogenic expansion was non-penetrant in carriers below 35 years of age, increasing to 50·0% penetrance by 58 years of age, and was almost fully penetrant by 80 years of age. No difference was observed in disease penetrance according to familial status; by ALS or FTD diagnosis; by gender; or, according to symptom onset in ALS or FTD patients (webappendix p 13).
Figure 2.
Age-related penetrance of the GGGGCC hexanucleotide repeat expansion in the C9ORF72 gene based on an analysis of 603 mutant-gene carriers (n = 212 familial ALS patients, n = 234 sporadic ALS, n = 99 familial FTD, n = 53 sporadic FTD, and n = 5 neurologically normal controls). The age-related penetrance (i.e., the proportion of mutant-gene carriers with manifestations of the disease by a given age) rose steadily, from 10% in the <45 years-old group, to ~100% by the age of 80 years. The dotted lines represent the age at which 50% of the cohort developed symptoms.
Clinical details of patients carrying the hexanucleotide repeat expansion are given in Table 3. ALS patients with the pathogenic repeat expansion were more likely to be female (χ2= 11·2, 1 degree of freedom (df), p-value = 0·0008), have a family history of disease (χ2= 589·0, 1 df, p-value < 0·0001), and to have bulbar-onset disease (χ2= 10·7, 1 df, p-value = 0·0011) compared to cases that did not carry the expansion. FTD patients were also more likely to have a family history of disease (χ2= 106·0, 1 df, p-value < 0·0001), and to present with behavioural variant FTD (χ2= 25·1, 2 df, p-value < 0·0001).
Table 3.
Demographic and clinical features of patients classified by diagnosis and by carrier status for the GGGGCC hexanucleotide repeat expansion in the C9ORF72 gene
| ALS
|
FTD
|
|||
|---|---|---|---|---|
| With expansion§ | Without expansion¶ | With expansion° | Without Expansion* | |
| N | 465 | 3,983 | 160 | 1,265 |
| Age at onset (range) | 56·8 (27·0–80·0) | 58·7 (4·0–93·0) | 57·5 (30·0–76·3) | 60·0 (23·0–87·0) |
| Male (%) | 232 (50·1%) | 2,251 (58·4%) | 87 (54·4%) | 683 (55·4%) |
| Positive family history (%) | 221 (47·5%) | 367 (9·2%) | 101 (63·1%) | 302 (23·9%) |
| Presentation: | ||||
| Bulbar (%) | 139 (33·1%) | 933 (26·0%) | - | - |
| Limb (%) | 281 (66·9%) | 2,655 (74·0%) | - | - |
| Behavioural (%) | - | - | 106 (85·5%) | 685 (65·6%) |
| PNFA (%) | - | - | 11 (8·9%) | 165 (15·8%) |
| SD (%) | - | - | 7 (5·6%) | 195 (18·6%) |
Data were not available for age at onset (n = 19 patients), and site of onset (n = 45).
Data were not available for age at onset (n = 305 patients), gender (n = 130), and site of onset (n = 395).
Data were not available for age at onset (n = 8), and site of onset (n = 36).
Data were not available for age at onset (n = 71), gender (n = 32), and site of onset (n = 220).
Discussion
Our data show that the C9ORF72 hexanucleotide repeat expansion is the most frequent cause of sporadic ALS and sporadic FTD identified to date, accounting for ~5·0–7·0% of all Caucasian cases in our large series of patients. These frequency rates were moderately higher than published estimates that were based on smaller cohorts obtained from a single institution.9 Prior to the identification of the genetic lesion underlying chromosome 9-linked ALS/FTD, the most common genetic cause of sporadic ALS was mutations in the SOD1 gene (accounting for 0·7% of cases in a population-based cohort)3, whereas mutations in the PGRN gene were the most common cause of sporadic FTD (3·0–4·0% in clinic referral series).21 The high frequency of the pathogenic expansion in our patient cohort is consistent with previous GWAS studies that identified the association signal on chromosome 9p21 as the only replicable locus in the sporadic form of ALS and FTD.16,22–24 Our findings confirm the central importance of genetics in the pathogenesis of the idiopathic form of these fatal neurodegenerative diseases.
Our haplotype data suggest that the pathogenic GGGGCC hexanucleotide repeat expansion in the C9ORF72 gene arose from a single mutational event16,20 that occurred ~1,500 years ago. The geographical distribution of the mutation suggests that the mutation appeared in Northern Europe and spread from there. Alternatively, the high frequencies in Finland and other isolated populations could be explained by the history of these communities. Finland and Sardinia are isolated areas, and have genetically homogeneous populations that originated from a small number of founders.25 Genetic drift has had a large influence on allele frequencies in these populations and could explain the high occurrence of the mutation in these geographical isolates.
Recognizing that all cases carrying the C9ORF72 repeat expansion share a common ancestor has important implications for the interpretation of global frequency data of this mutation. Though the hexanucleotide repeat expansion is commonest among European Caucasians, it is also present in African American, Hispanic, and Middle Eastern populations. This likely reflects the scale and nature of past human migration and intermarriage involving these ethnicities. Similarly, the relative absence of the pathogenic hexanucleotide repeat in the Indian subcontinent, Asia, and the Pacific Islands may be explained by the greater physical distances of these regions from Europe, and the consequent lack of admixture between these populations. Interestingly, the single Japanese patient that we identified as a carrier of the C9ORF72 expansion carried the Finnish risk haplotype, reinforcing the notion that the expansion occurred on a single occasion in the past.
The sharing of a common risk haplotype across the C9ORF72 gene in both sporadic and familial ALS cases indicates that these apparently sporadic cases are in fact cryptically related familial cases. This scenario can occur for many reasons: first, lack of knowledge of the pedigree on the part of the patient or neurologist; second, previous generations may have died at a young age prior to the onset of neurological symptoms. The median age at onset among patients with the expansion was 57 years of age, and life expectancy in the United States only exceeded this point in the early 1940’s26; and finally, decreased penetrance of the mutation, where not all individuals carrying the expansion manifest a clinical phenotype, may be a contributing factor in apparently sporadic disease. Indeed, we have observed symptom onset in the ninth decade of life among cases carrying the expansion, and we have encountered a small number of elderly, neurologically normal individuals carrying the expansion. Thus, the penetrance of this mutation appears to be complete only at a late stage of life, an observation that is particularly relevant to genetic counseling of healthy individuals carrying the expansion. The molecular biological substrate underlying this variability in age at onset is currently unclear: it may be driven by differences in expansion lengths between patients, by age-related methylation across the locus, or by genetic factors elsewhere in the genome.
We compared our results with those of previous studies that reported the frequency of the C9ORF72 hexanucleotide repeat expansion in the pathogenesis of ALS and FTD (panel). Data were available from seven studies (webappendix p 10). Our study represents one of the largest cohort of ALS and FTD cases screened to date, and also provides an initial report of the frequency of the pathogenic repeat expansion in non-Caucasian patients, a detailed examination of the haplotype across the locus, and aninitial estimate of age-related disease penetrance in a large group of individuals carrying the expansion.
Our data have important implications for the clinical care of patients diagnosed with ALS and FTD. The current clinical standard is to offer genetic testing to patients reporting a family history of ALS and/or FTD27, and to reassure patients classified as having sporadic disease that their relatives are not at increased risk of neurodegeneration. A paper recently published in this journal suggested, based on analysis of 191 Irish ALS patients, that genetic testing for the C9ORF72 repeat expansion is unnecessary in affected individuals without a family history of disease or significant cognitive impairment.28 In direct contradiction to this, it is the opinion of this article’s authors that genetic testing is a valuable tool in the accurate diagnosis of both conditions and in the decision making process for patients and their families. The discrepancy between the Byrne et al and our study may stem from differences in how sporadic and familial disease were defined in the two studies. Accumulating data, such as for the 5,873 ALS and FTD cases from diverse populations reported here, is an important step towards answering this key question in patient management. Given the large number of patients that carry the repeat expansion, the field should, at the very least, consider a focused debate on this issue.
Our paper has some limitations. First, the number of patients from certain geographical regions was small, and the mutational frequencies may change for those ethnicities as additional cases are screened. Nevertheless, our data on more than 5,000 cases provides a reasonable estimation of C9ORF72 global frequency. Second, although we have examined the chromosome 9p21 haplotype in a large and diverse cohort of individuals carrying the pathogenic expansion, additional testing of carriers may reveal other haplotypes, thereby indicating that the expansion arose on more than one occasion. Even in this instance, our data indicates that the vast majority of expansion carriers share a common ancestor.16,20 Third, age-related penetrance estimates were generated based on data from retrospective cohorts, which potentially leads to over-estimation of penetrance. Additional prospective studies are necessary to confirm these estimates. Finally, case classification as familial or sporadic was based on clinical questioning at sample collection. The level of scrutiny may have varied across centers and countries, but it was not feasible to re-collect this information for these existing cohorts.
In conclusion, the hexanucleotide C9ORF72 repeat expansion causes 5·0–7·0% of apparently sporadic ALS and FTD cases in Caucasians, making it by far the most common cause of the sporadic form of these fatal neurodegenerative diseases identified to date. In addition, our data indicate that the pathogenic expansion on chromosome 9p21 may have occurred on a single occasion, possibly in Scandinavia, and that the modern-day global distribution of the mutation perhaps reflects the genetic legacy of their descendants. Furthermore, the finding of the C9ORF72 pathogenic hexanucleotide repeat expansion in nearly one in fifteen apparently sporadic patients raises the question as to whether genetic testing should be offered to patients regardless of family history of disease.
Supplementary Material
Research in context.
Systematic review
We searched Medline (2011–12) to identify relevant publications, and selected studies that reported the GGGGCC hexanucleotide repeat expansion of the C9ORF72 gene as being relevant to the pathogenesis of amyotrophic lateral sclerosis (ALS) or frontotemporal dementia (FTD). On the basis of these criteria, seven studies were selected for further assessment. We compared the number of patients screened for the pathogenic repeat expansion, as well as the phenotype and racial origin of the screened patients reported by these studies (webappendix p 10).
Interpretation
We report the frequency of the C9ORF72 repeat expansion in a large cohort of sporadic ALS and sporadic FTD published to date. We also screened a large number of non-Caucasian patients for the expansion, and present frequency data for the mutation in these populations. Weconfirmed that the C9ORF72 repeat expansion explains a substantial proportion of sporadic ALS (~7·0%) and sporadic FTD (~6·0%)cases in Caucasian populations. Our study also found that sporadic and familial cases carrying the expansion share a founder risk haplotype, suggesting that these cases have a common ancestor and that the original mutational event that led to the repeat expansion occurred only once in the past (estimated to be ~1,500 years ago). We provide initial estimates of age-related penetrance based on 603 individuals carrying the expansion, finding that 50% of carriers manifest disease by 58 years of age, and that penetrance is near complete by 85 years of age.
Acknowledgments
This work was supported in part by the Intramural Research Programs of the NIH, National Institute on Aging (Z01-AG000949-02), and NINDS. The work was also supported by the Packard Center for ALS Research at Hopkins (B.J.T.), the ALS Association (B.J.T., A.C.), Microsoft Research (B.J.T., P.J.T.), AriSLA (B.J.T., A.C., M.Sabatelli), Hersenstichting Nederland Fellowship project B08.03 and the Neuroscience Campus Amsterdam (J.S.-S.), Nuts Ohra Fonds (J.v.S.), Stichting Dioraphte (J.v.S. - Grant 09020300), the UK MND Association (H.M. - MNDA Grant 6057, J.H., R.W.O., K.E.M.), The Medical Research Council UK (J.H., S.P.B.), the Wellcome Trust (J.H.), The Oxford NIHR Biomedical Research Centre (O.A.), the Helsinki University Central Hospital, the Finnish Academy (P.J.T.), the Finnish Medical Society Duodecim, Kuopio University, the Italian Health Ministry (Ricerca Sanitaria Finalizzata 2007, to A.C.), Fondazione Vialli e Mauro ONLUS (A.C.), Federazione Italiana Giuoco Calcio (A.C., M.S., B.J.T.) and Compagnia di San Paolo (A.C., G.R.), the French Agency for Research (ANR-08-MNPS-009-01) (A.B. and I.L.B.), France Alzheimer - Union Nationale des Associations Alzheimer (I.L.B.) and Institut de France Subvention de la Fondation Thierry et Annick DESMAREST (I.L.B), and the European Community’s Health Seventh Framework Programme (FP7/2007-2013) under grant agreements 259867 (A.C.) and 259867 (M.S., C.D.), Deutsche Forschungsgemeinschaft (M.S. - Grant SFB 581, TP4). DNA samples for this study were obtained in part from the NINDS repository at the Coriell Cell Repositories (http://www.coriell.org/), and from the Australian MND DNA Bank, which is funded by NHMRC grant 402703. We thank the DNA extraction and storage facility of the National Institute for Health and Welfare/FIMM, Helsinki, Finland and the Institute for Ageing and Health, Campus for Ageing and Vitality, Newcastle University, Newcastle upon Tyne, UK for their help in extraction of DNA from ALS patients. We also thank the patients and research subjects who contributed samples for this study.
Footnotes
Contributors
EM, AER, KM, NN, AW, SR, JSS, YA, JOJ, DGH, SA, and JK did laboratory-based experiments and data analysis, and revised the report. ED, MSendtner, RP, RWO, KCS, HH, JDR, KEM, HP, KT, OA, MSabatelli, GM, MC, FG, ACalvo, EE, GB, GLF, AMR, HL, LM, VED, and CD collected data from and characterised patients, and revised the manuscript. MAN analysed the data and revised the report. SMead, JQT, VMVD, GDS, C-SL, T-HY, HI, YT, ST, ILB, AB, and PS supervised laboratory-based experiments, and revised the manuscript. AChio, GR, JvS, NW, JH, PJT, PH, HRM and SP-B designed the study, supervised laboratory-based experiments, and revised the manuscript. BJT designed the study, supervised laboratory-based experiments, did the data analysis, and drafted the manuscript.
Conflicts of interest
Jeffrey Rothstein is Director of the Packard Center for ALS Research at Hopkins, while Pentti Tienari, Peter Heutink, Huw Morris, Stuart Pickering-Brown and Bryan Traynor have a patent pending on the discovery of the hexanucleotide repeat expansion of the C9ORF72 gene. None of the other authors has any conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Logroscino G, Traynor BJ, Hardiman O, et al. Incidence of amyotrophic lateral sclerosis in Europe. J Neurol Neurosurg Psychiatry. 2010;81:385–90. doi: 10.1136/jnnp.2009.183525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Valdmanis PN, Daoud H, Dion PA, Rouleau GA. Recent advances in the genetics of amyotrophic lateral sclerosis. Curr Neurol Neurosci Rep. 2009;9:198–205. doi: 10.1007/s11910-009-0030-9. [DOI] [PubMed] [Google Scholar]
- 3.Chiò A, Traynor BJ, Lombardo F, et al. Prevalence of SOD1 mutations in the Italian ALS population. Neurology. 2008;70:533–37. doi: 10.1212/01.wnl.0000299187.90432.3f. [DOI] [PubMed] [Google Scholar]
- 4.Guerreiro RJ, Schymick JC, Crews C, Singleton A, Hardy J, Traynor BJ. TDP-43 is not a common cause of sporadic amyotrophic lateral sclerosis. PLoS One. 2008;3:e2450. doi: 10.1371/journal.pone.0002450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lai SL, Abramzon Y, Schymick JC, et al. FUS mutations in sporadic amyotrophic lateral sclerosis. Neurobiol Aging. 2011;32(550):e1–4. doi: 10.1016/j.neurobiolaging.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dion PA, Daoud H, Rouleau GA. Genetics of motor neuron disorders: new insights into pathogenic mechanisms. Nat Rev Genet. 2009;10:769–82. doi: 10.1038/nrg2680. [DOI] [PubMed] [Google Scholar]
- 7.Ratnavalli E, Brayne C, Dawson K, Hodges JR. The prevalence of frontotemporal dementia. Neurology. 2002;58:1615–21. doi: 10.1212/wnl.58.11.1615. [DOI] [PubMed] [Google Scholar]
- 8.Neumann M, Sampathu DM, Kwong LK, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–3. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 9.Dejesus-Hernandez M, Mackenzie IR, Boeve BF, et al. Expanded GGGGCC Hexanucleotide Repeat in Noncoding Region of C9ORF72 Causes Chromosome 9p-Linked FTD and ALS. Neuron. 2011;72:245–56. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Renton AE, Majounie E, Waite A, et al. A Hexanucleotide Repeat Expansion in C9ORF72 Is the Cause of Chromosome 9p21-Linked ALS-FTD. Neuron. 2011;72:257–68. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simón-Sánchez J, Dopper EGP, Cohn-Hokke PE, et al. The clinical and pathological phenotype of C9orf72 hexanucleotide repeat expansions. Brain. 2012 doi: 10.1093/brain/awr353. published online February 2, 2012. [DOI] [PubMed] [Google Scholar]
- 12.Snowden JS, Rollinson S, Thompson JC, et al. Distinct clinical and pathological characteristics of frontotemporal dementia associated with C9ORF72 mutations. Brain. 2012 doi: 10.1093/brain/awr355. published online February 2, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brooks BR. El Escorial World Federation of Neurology criteria for the diagnosis of amyotrophic lateral sclerosis. Subcommittee on Motor Neuron Diseases/Amyotrophic Lateral Sclerosis of the World Federation of Neurology Research Group on Neuromuscular Diseases and the El Escorial “Clinical limits of amyotrophic lateral sclerosis” workshop contributors. J Neurol Sci. 1994;124 (Suppl):96–107. doi: 10.1016/0022-510x(94)90191-0. [DOI] [PubMed] [Google Scholar]
- 14.Clinical and neuropathological criteria for frontotemporal dementia. The Lund and Manchester Groups. J Neurol Neurosurg Psychiatry. 1994;57:416–8. doi: 10.1136/jnnp.57.4.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Traynor BJ, Nalls M, Lai SL, et al. Kinesin-associated protein 3 (KIFAP3) has no effect on survival in a population-based cohort of ALS patients. Proc Natl Acad Sci USA. 2010;107:12335–8. doi: 10.1073/pnas.0914079107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laaksovirta H, Peuralinna T, Schymick JC, et al. Chromosome 9p21 in amyotrophic lateral sclerosis in Finland: a genome-wide association study. Lancet Neurol. 2010;9:978–85. doi: 10.1016/S1474-4422(10)70184-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reeve JP, Rannala B. DMLE+: Bayesian linkage disequilibrium gene mapping. Bioinformatics. 2002;18:894–5. doi: 10.1093/bioinformatics/18.6.894. [DOI] [PubMed] [Google Scholar]
- 18.Bender BU, Eng C, Olschewski M, et al. VHL c. 505 T>C mutation confers a high age-related penetrance bu no increased overall mortality. J Med Genet. 2001;38:508–14. doi: 10.1136/jmg.38.8.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Passant U, Gustafson L, Brun A. Spectrum of frontal lobe dementia in a Swedish family. Dementia. 1993;4:160–2. doi: 10.1159/000107316. [DOI] [PubMed] [Google Scholar]
- 20.Mok K, Traynor B, Schymick J, et al. The chromosome 9 ALS and FTD locus is probably derived from a single founder. Neurobiol Aging. 2011 doi: 10.1016/j.neurobiolaging.2011.08.005. published online August 5 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le Ber I, van der Zee J, Hannequin D, et al. Progranulin null mutations in both sporadic and familial frontotemporal dementia. Hum Mutat. 2007;28:846–55. doi: 10.1002/humu.20520. [DOI] [PubMed] [Google Scholar]
- 22.Van es MA, Veldink JH, Saris CG, et al. Genome-wide association study identifies 19p13.3 (UNC13A) and 9p21. 2 as susceptibility loci for sporadic amyotrophic lateral sclerosis. Nature Genet. 2009;41:1083–7. doi: 10.1038/ng.442. [DOI] [PubMed] [Google Scholar]
- 23.Shatunov A, Mok K, Newhouse S, et al. Chromosome 9p21 in sporadic amyotrophic lateral sclerosis in the UK and seven other countries: a genome-wide association study. Lancet Neurol. 2010;9:986–94. doi: 10.1016/S1474-4422(10)70197-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Deerlin VM, Sleiman PM, Martinez-Lage M, et al. Common variants at 7p21 are associated with frontotemporal lobar degeneration with TDP-43 inclusions. Nat Genet. 2010;42:234–9. doi: 10.1038/ng.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kristiansson K, Naukkarinen J, Peltonen L. Isolated populations and complex disease gene identification. Genome Biol. 2008;9:109. doi: 10.1186/gb-2008-9-8-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bureau USC. Statistical Abstract of the United States: 2012. 131. Washington, DC: 2011. [Google Scholar]
- 27.The EFNS Task Force on Diagnosis and Management of Amyotrophic Lateral Sclerosis. EFNS guidelines on the Clinical Management of Amyotrophic Lateral Sclerosis (MALS) – revised report of an EFNS task force. Eur J Neurol. 2011 doi: 10.1111/j.1468–1331.2011.03501.x. [DOI] [PubMed] [Google Scholar]
- 28.Byrne S, Elamin M, Bede P, et al. Cognitive and clinical characteristics of patients with amyotrophic lateral sclerosis carrying a C9orf72 repeat expansion: a population-based cohort study. Lancet Neurol. 2012 doi: 10.1016/S1474-4422(12)70014–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.